Abstract
Genetic changes and environmental differences result in cellular heterogeneity among cancer cells within the same tumor, thereby complicating treatment outcomes. Recent advances in single-cell technologies have opened new avenues to characterize the intra-tumor cellular heterogeneity, identify rare cell types, measure mutation rates, and, ultimately, guide diagnosis and treatment. In this paper, we review the recent single-cell technological and computational advances at the genomic, transcriptomic, and proteomic levels, and discuss their applications in cancer research.
Keywords: Single-cell analysis, Cellular heterogeneity, Cancer, Genomics, Transcriptomics, Proteomics
Cancer is a disease of multitudes
Cells are the basic units of life in which the blueprint of the genome is transcribed and translated into biological functions. Cellular heterogeneity, which results from mutation, differences in gene regulation, stochastic variation, or environmental perturbations, is reflected at the genomic, transcriptomic, and proteomic levels. Such heterogeneity is increasingly appreciated as a factor of cancer treatment failure and disease recurrence, since a treatment that targets one tumor cell population may not be effective against another [1]. Not only is cancer itself a complex disease made up of a collection of individually distinct pathologies, but also within each tumor, there is significant heterogeneity among different cells. Current theories propose that cancer development involves both a process of clonal evolution from mutated cells of origin, and a differentiation hierarchy from cancer stem cells [2]. It is increasingly clear that traditional bulk experiments, which only measure the average profile of the population, have limitations in characterizing complex diseases like cancer.
Single cells have been studied since the invention of the microscope, but it is not until recently that genome-scale approaches have been applied to single-cell biology [3–7] (Box 1). For example, microfluidic-based single-cell sorting methods [8,9], high-throughput multiplexed quantitative PCR (qPCR) [10–14] or sequencing approaches [15–23], mass cytometry-based proteomic strategies [24–26], and data analysis methods [27–30] provided an unprecedented opportunity to identify rare cell types, such as cancer stem cells, and to investigate the dynamic processes of cell-fate transitions.
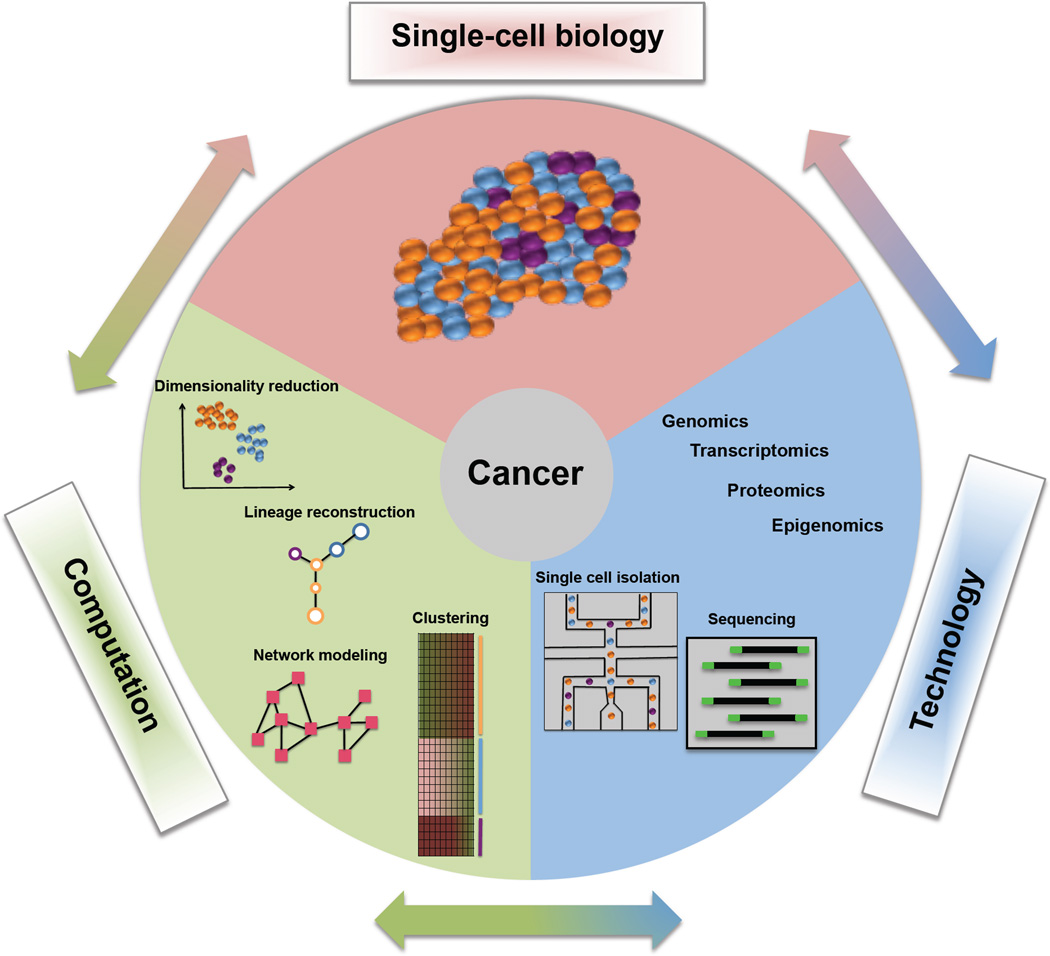
One of the important application areas of single-cell analysis is in cancer genomics (Figure 1). Recently, a number of studies have applied single-cell analysis to characterize the cellular heterogeneity in different cancers [13,23,31–33]. The comprehensive knowledge about cellular heterogeneity will not only provide fundamental insights into development and other biological processes, but also have important applications in therapy, as drug resistance is often caused by heterogeneous response at the cellular level.
Figure 1. An overview of single-cell cancer genomics.
Single-cell technologies are used to generate genomic, transcriptomic, and proteomic data from cancer cells. These data are analyzed by computational methods to identify clusters, lineages, and networks, which in turn generate new biological hypotheses. Biological discoveries in turn guide development of new technologies and computational approaches. The figure also shows a toy example with a heterogeneous cancer sample containing three cell types (orange, blue, and purple). An integrated single-cell analysis is used to identify the cell-types, lineages, and network profiles.
In this paper, we review the recent technological and computational advances in single-cell analysis, and discuss their applications in cancer genomics. We conclude by offering a personal view of the potential challenges and future prospects for this field.
Technological developments in single-cell analysis
Methods for single-cell measurement, such as flow cytometry [34], RNA fluorescence in situ hybridization (FISH) [35,36], and dynamic profiling of fluorescent fusion proteins [37] were developed years ago and are routinely used in modern labs. However, these traditional methods provide limited information from single-cell samples as only several genes or proteins can be profiled at the same time. In the past few years, a new wave of technologies has emerged in the areas of single-cell isolation, nucleic acid amplification and genomic/transcriptomic/proteomic profiling (Table 1). These new methods significantly increased the throughput and scale of single-cell analysis.
Table 1.
Advanced single-cell technologies for genomic, transcriptomic, and proteomic analysis.
| Method | Amplification | Application | Coverage | References |
|---|---|---|---|---|
| Genomic analysis | ||||
| GenomePlex PCR | Multiplexed PCR | Copy number | Low coverage | [32] |
| MDA | MDA | Genome/exome | High coverage | [38,39] |
| MALBAC | MALBAC | Copy number/genome | High coverage and uniform amplification |
[41,42] |
| Transcriptomic analysis | ||||
| Single-cell qPCR | Multiplexed PCR | Transcriptome | Targeted regions | [11,13] |
| Tang et al. method | PolyA tailing | Transcriptome | 3' bias | [17,18] |
| Smart-seq | Template-switching | Transcriptome | Full-length | [16] |
| CEL-seq | IVT | Transcriptome | 3' bias | [45] |
| CytoSeq | Multiplexed PCR | High-throughput transcriptome | Targeted regions | [47] |
| inDrop | IVT | High-throughput transcriptome | 3’ bias | [48] |
| Drop-seq | Template-switching | High-throughput transcriptome | 3’ bias | [49] |
| Proteomic analysis | ||||
| Mass Cytometry | N/A | Proteomic analysis | Targeted proteins | [24] |
| MIBI | N/A | Proteomic analysis with spatial information |
Targeted proteins | [58] |
One of the fundamental challenges in single-cell analysis is the amplification of a small amount of initial nucleic acid material to reach the detection threshold level. Recently, significant technical advances in whole-genome amplification (WGA) have been achieved to overcome this challenge for single-cell genome analysis. Based on the protocols used for WGA, there are three main categories of single-cell techniques. GenomePlex PCR [32] uses degenerative-oligonuceotide PCR to amplify DNA from single cells. The method achieves low physical coverage, but the amplification is uniform across the genome. It is therefore suitable for copy number profiling in single cancer cells [32]. Another popular single-cell WGA method, Multiple Displacement Amplification (MDA), uses Phi29 and random primers to amplify DNA in a linear process through multiple displacement mechanisms [38,39]. This approach generates long DNA products, and achieves high-coverage amplification, and therefore is suitable for detection of point mutations at base-pair resolution. The MDA protocol was first used in single-cell exome sequencing studies to uncover the genetic landscape of cancer cells [38,39], and subsequently coupled with a microfluidic system to amplify genomes from single human sperms [40]. Multiple annealing and looping-based amplification cycles (MALBAC) [41,42] is a new WGA method that uses quasi-linear pre-amplification to reduce the bias associated with nonlinear amplification. In MALBAC, single-stranded amplicons generated through strand-displacement are used as templates to produce full amplicons, then full amplicons form looped DNA to avoid exponential amplification. This approach achieves high-coverage and uniform amplification, and enables genome-wide detection of both single nucleotide polymorphisms (SNPs) and copy number variations (CNVs) of a single cell. The method has been applied to single SW480 cancer cells [41] as well as human oocytes [42].
Another frontier with significant progress is single-cell transcriptomic analysis. Although there are more copies of mRNA than DNA in single cells, this application faces its own difficulties in quantification of different RNA species. In order to amplify the limited amount of mRNA in single cells, several approaches have emerged. The Poly-A tailing method uses terminal transferase to add anchoring sequences on the 3’ of synthesized cDNA, so that each cDNA has two primer binding sites for PCR amplification. The method was used in the first single-cell microarray study [43] and in the first single-cell mRNA-seq study [17]. Sequence specific amplification (SSA) uses multiplexed RT-PCR to amplify hundreds of specific targets in single cells. This method has a simple one-step protocol, but is limited to analyzing only a small number of genes [11,13]. The Smart-seq amplification method is a widely used approach for full-length mRNA analysis of single cells [16,21,44]. The method uses template-switching based protocol to anchor primer binding site on the 3’ cDNA. The cDNA is then amplified by PCR and sequenced by Illumina sequencers. Smart-seq has high coverage across transcripts, and enables identification of SNPs as well as different transcript isoforms. One limitation of Smart-seq is that the efficiency for template-switching is low and thus it has difficulty in profiling lowly-expressed mRNAs. CEL-seq adds T7 promoters to the cDNA and utilizes in vitro transcription (IVT) to amplify mRNA. The method also shows robust efficiency and sensitivity for single-cell transcriptomic profiling [15,45]. By coupling IVT with degenerative PCR based approach, the recently published DR-seq method even achieves integrated genome and transcriptome sequencing at the same time from the same cell [46].
For all the aforementioned single-cell transcriptomic methods, a common drawback is the need to handle each single cell samples independently, which limits the throughput of the analysis and also may inadvertently introduce human error. Very recent breakthroughs solve these problems by high-throughput molecular barcoding of single cells in microwells or microdroplets before sequencing library generation [47–49]. The CytoSeq platform randomly deposits single cells and transcript barcoding probes into an array of picoliter wells before cell lysis and reverse transcription; any selection of genes can be amplified and analyzed from the barcoded cDNAs [47]. The Drop-seq and inDrop strategies, however, separate thousands of single cells into aqueous droplets, associate a different barcode to each cell’s RNAs, and sequence them all together [48,49]. These massively parallel barcoding strategies have significantly increased the throughput of single-cell transcriptomic analysis.
The broad applications of single-cell genomic/transcriptomic analysis in the biomedical field have also been supported by the rapid development of microfluidic devices. Microfluidic devices help to automate the distribution, processing, and analysis of biological materials, and have significantly increased the measurement throughput. Microfluidic devices have been used as the basis for various single-cell technologies, such as the single-cell capture and amplification platforms [44,49], as well as high-throughput single-cell qPCR analysis [13]. As single-cell analysis protocols are highly sensitive to technical errors induced by manual processing, the accurate control provided by the microfluidic devices is a significant advantage. Microfluidic devices also improve the sensitivity of single-cell assays by confining the reaction volume and increasing the local concentration.
In comparison to the progress made in assaying nucleic acids, single-cell proteomic analysis is much more challenging because, unlike DNA or RNA sequences, it is not possible to amplify protein sequences using current technologies. Standard immunofluorescence methods have been routinely used to analyze four markers at single-cell level. Now, highly multiplexed fluorescence microscopic allows analysis of up to 60 proteins in tissue specimens [50]. Notably, the development of mass cytometry has dramatically increased the multiplexity of cytometry-based analysis by labeling antibodies with isotopes [24]. This innovation resolves the problem of spectral overlap that is common in normal flow cytometry. It is now possible to measure more than 40 parameters in a large number of single cells in a short period of time.
The methods discussed above require isolation of cells from their in situ environment. Recently in situ methods have been developed to preserve spatial information [51]. By computational integration of single-cell RNA-seq data with in situ RNA patterns, one can accurately infer cellular localization within complex patterned tissues [52–56]. Similarly, mass cytometry can be coupled with immunohistochemical data to obtain highly multiplexed proteomic information at subcellular resolution [57]. Another method, called multiplexed ion beam imaging (MIBI), uses secondary ion mass spectrometry to image antibodies tagged with isotopically pure elemental metal reporters [58]. Taken together, these technologies have greatly facilitated the systematic analysis of gene and protein expression variability at the single-cell resolution.
Computational methods for analyzing single-cell data
With the technological breakthroughs that have generated large amounts of high-throughput single-cell data, the development of novel computational tools has become an integral part of the analysis. Single-cell technologies present a number of challenges that cannot be addressed by traditional computational methods. First, each cell is typically measured only once and thus there are no technical replicates in the strict sense. Second, the amount of starting material is subject to strong stochastic variation. Still at an early stage, a number of computational methods have been developed to address these issues (Figure 2).
Figure 2. A typical flow chart for single-cell data analysis.
Representative methods are mentioned. See the main text for detailed description.
Preprocessing and quantification are the first steps of any large-scale data analysis. The purpose of these steps is to convert raw data to quantitative biological information. Significant effort is paid to estimation and removal of systematic biases due to technical variability. A major issue in single-cell analysis is that technical variation is always confounded with biological variation. One simple approach to estimate technical variability compared the pooled sample with bulk RNA-seq experiments [59]. More precise calibration can be achieved by adding spike-in RNA to the library as control, followed by building an error model based on the variation of the spike-in RNA [60,61]. Methods that account for single-cell specific noise, such as dropout events and amplification biases, can also help to separate technical and biological variability of individual genes [62]. Recently, scLVM (single-cell latent variable model) [63] was developed to account for the confounding effects of cell cycle on modulating differentiation and gene expression profiles. Another approach to estimating reproducibility is to divide the RNA material from a single cell into two equal fractions which are then analyzed independently [20]. A recent review article [64] has provided a detailed survey of the computational methods and, in particular, the normalization steps for single-cell RNA-seq data from counts to expression values with or without unique molecular identifiers. For single-cell qPCR data, normalization to an endogenous control is not usually recommended due to biological variation and transcriptional noise exhibited by single cells [65]. It was shown that normalizing by the median Ct (threshold cycle) reduces variability in single-cell qPCR data [65]. For mass cytometry data, technical variations can be corrected with bead standards [66]. Normalization methods for CNV detection based on channel, genome composition, and recurrent genome artifact corrections have also been developed [67]. A review of the computational approaches to correct for biases in the WGA procedure and accurately determine copy number profiles has been outlined before [68].
The high-dimensionality of single-cell data provides a challenge for visualization. Several dimensionality reduction approaches are available to map the data points into a lower dimensional space while maintaining the single-cell resolution. The conventional principal component analysis (PCA) has been used to visualize single-cell data in different contexts [13,24,69]. Despite its success, this method relies on a linear assumption, and thus cannot fully capture the nonlinear relationships inherent in many single-cell datasets. This limitation can potentially be overcome by using a wide variety of non-linear methods [27,30,70–73], although the performance of each method is likely to be context dependent. The t-distributed stochastic neighbor embedding (t-SNE) [72,73] preserves both the global layout and local structure of the high-dimensional data by converting the Euclidean distances between each pair of data points into heavy-tailed conditional probabilities. A distributed implementation of the t-SNE algorithm, called viSNE [27], has been employed to visualize single-cell mass cytometry data. Another approach based on the Gaussian process latent variable model generates a smooth mapping from the latent space to the original data space [71]. This method was extended to a probabilistic PCA approach to account for censoring effect due to undetected transcripts [70]. More recently, a dimensionality reduction approach based on diffusion maps was adapted to identify and visualize the hematopoietic developmental progression in mouse embryo [30]. In this approach, the cells are related to each other through a gradual but stochastic, diffusion-like process. All these methods can help interrogate the relationships among different cell types in a lower dimensional space.
Unsupervised clustering is a widely used approach to group samples with similar properties, which can be used for identifying previously unknown subpopulations from single-cell data. Besides the classical clustering methods, a number of approaches have recently been developed to analyze single-cell data. For example, ACCENSE (automatic classification of cellular expression by nonlinear stochastic embedding) [74] combines the t-SNE algorithm with density-based partitioning without the need to pre-specify the number of target clusters. Another recent effort in this direction is SNN-Cliq [75], which achieves clustering of single-cell transcriptomic data by a graph theory-based algorithm. For low-dimensional single-cell expression data emerging from qPCR or FACS, the multiresolution correlation analysis (MCA) [76] can be useful in identifying subpopulations based on local pairwise gene correlations. In an effort to improve traditional manual gating in flow cytometry data, Citrus [77] was developed, which identifies stratifying subpopulations of cells whose abundance or behavior is correlated with a known endpoint of interest.
Having identified the cell subpopulations, one can identify the sets of genes that best discriminate these subpopulations. In addition to the standard differential expression tools for bulk experiments, new methods have been developed to address the specific challenges in single-cell data analysis. For example, SCDE (single-cell differential expression) uses a Bayesian approach that accounts for the likelihood of dropout events in single-cell RNA-seq data [62]. In another approach, MIMOSA (mixture models for single-cell assays) employs a mixture model where information is shared across subjects through exchangeable priors allowing to increase the power of detecting true differences [78].
Although clustering approaches reveal the underlying group structure within the data, they cannot provide information on the lineage relationships between different developmental stages. One method along this direction is SCUBA (single-cell clustering using bifurcation analysis) [28], which first infers the cellular hierarchy using dynamic clustering and then models gene expression dynamics using bifurcation theory. However, the application of SCUBA requires temporal information, which is often difficult to obtain experimentally. Computational methods have been developed to infer temporal information from snapshot single-cell data, including using principal curve analysis [79], and graph-model based algorithms such as Monocle [29] and Wanderlust [80]. The inferred temporal information can then be used as input to identify bifurcation events [28]. However, it is challenging to accurately infer temporal information if the bifurcation structure is complex. A related approach is SPADE (spanning-tree progression analysis of density-normalized events) [24,81], which infers cell lineages without assigning temporal order. In this case, additional biological knowledge is needed to interpret the resulting tree structure. Similar approaches have been developed to infer clonal structure using single-cell genomic data [82].
Network modeling can provide mechanistic insights into the coordination of gene activities and help to understand the overall dynamics of the system. Efforts are underway to apply network-modeling approaches to single-cell data. A simple but popular approach is to construct networks based on co-expression. A variant of this approach, called weighted gene co-expression network analysis (WGCNA) [83,84], uses a soft threshold for modeling co-expression and also identifies network modules (i.e., genes with coordinated activities). Co-expression networks have been applied to single-cell analysis of the mammalian embryonic development [19], hematopoiesis [14], neural stem cells [85], and leukemia [33,86]. Although network analysis provided novel insights in these studies, existing methods are applicable only if the sample size is sufficiently large and therefore are not directly applicable for studying the networks associated with rare cell types. In addition, correcting for latent confounding factors in single-cell data can help to reduce false positive links in these networks [63,87]. Co-expression networks can also be integrated with other types of data, such as ChIP-seq data, to estimate the underlying gene regulatory network [10]. Most network models are only a static representation of the system and do not explicitly consider the underlying gene expression dynamics. Building self-contained dynamic network models is challenging, although there are some approaches, such as Boolean networks, that have been applied to study stem cell differentiation processes [30,88].
Taken together, these data analysis methods have greatly enabled researchers to systematically extract quantitative information from the single-cell data, thereby playing an important role in applying single-cell technologies to investigate biomedical problems.
Applications in cancer genomics
Genome instability is a hallmark of cancer. Spatial and temporal knowledge of the cancer genome will have a significant impact on identifying cancer subtypes and developing patient-specific treatment strategies. A notable application of single-cell genome sequencing is in inferring tumor evolution paths. For example, single-cell genome sequencing applied to two human breast cancer cases suggested that tumors grow by punctuated clonal expansions with few persistent intermediates [32]. Another study on a thrombocythemia patient uncovered the likely monoclonal origin of this neoplasm [38]. Compared to the widely used bulk sequencing methods, single-cell cancer genome analysis has the advantage of characterizing intra-tumor cellular heterogeneity. For example, it has been used to map the intra-tumoral genetic landscape in kidney cancer [39], colorectal cancer [31], and leukemia [82]. Another intensely researched area is the detection and sequencing of circulating tumor cells (CTCs) for either understanding the metastatic process or early tumor detection. For example, a study on reproducibility of CNV patterns in CTCs of lung cancer patients suggested that CNVs at certain genomic loci are selected for the metastasis of cancer [89]. A recent whole-exome sequencing of CTCs provided insights into the mutational landscape of metastatic prostate cancer [90]. Recently, a high-coverage, whole-genome/exome single-cell sequencing method (nuc-seq) was developed and applied to breast cancer data wherein a large number of subclonal and de novo mutations were found, suggesting that point mutations evolved gradually over long periods of time [91]. In another study, by using single-cell whole-exome sequencing in multiple myeloma, it was demonstrated that the disease develops through a branching and parallel evolutionary pattern, where two divergent clones independently acquired the same convergent phenotype [92].
Single-cell transcriptomic advances in cancer research are also notable. For example, by using single-cell qPCR analysis in human colon cancer, it was found that multi-lineage differentiation represents a key source of in vivo functional and phenotypic cancer cell heterogeneity [13]. The Smart-seq method was used for profiling full length mRNA from single cells wherein by analyzing CTCs from melanomas, distinct gene expression signatures as well as alternative splicing events specific to the disease were identified [16]. A recent single-cell qPCR analysis of a mouse model of acute myeloid leukemia identified two subpopulations of leukemic cells, each characterized by distinct co-expression networks [33]. Another study using single-cell RNA-seq analysis in five primary glioblastomas (GBs) revealed that current GB classifiers are variably expressed across single cells within a tumor, suggesting that single-cell data can capture the true diversity of transcriptional subtypes within a tumor that cannot be detected by population-level data alone [23].
Single-cell proteomic approaches from flow cytometry, to mass cytometry, and multiplexed imaging, have also made great contributions to cancer research [27,57,58,93,94]. For example, an application of the viSNE approach to mass cytometry data on healthy and leukemic bone marrow samples showed that while the maps of healthy samples overlap, the leukemic samples from different patients form distinct populations from healthy bone marrow and from each other [27]. Moreover, integration of mass cytometry with multiplexed imaging techniques on breast cancer samples revealed substantial tumor microenvironment heterogeneity [57,58].
All these examples demonstrate that single-cell technologies provide a powerful approach to study the diversity and evolution of single cancer cells, which can ultimately be applied to the clinic from early detection to identifying therapeutic strategies for cancer patients.
Challenges and prospects
Single-cell analysis is still a new field, and there remain a number of significant challenges ahead (Box 2). A major goal for technological development is to improve the throughput and accuracy of the assays while reducing the cost. Promising results have been obtained by recent development of several approaches such as massive barcoding, microwells, and microdroplets [47–49]. Most technologies for single-cell analysis require the destruction of cells, and thus the temporal information is lost during the process. Along these lines, live-cell imaging technologies have generated exciting results [95]. Similarly, isolating single cells from a tissue results in loss of information about the spatial context, imposing a barrier for investigating the role of microenvironmental factors in gene regulation and cell-fate decisions. This issue is especially problematic for studying tumor progression, which is known to depend heavily on its interaction with the microenvironment. In this direction, several promising approaches have been developed as discussed above [51–56]. Similarly, methods for single-cell epigenomic profiling are still underdeveloped, although some promising strides have been made [96–100]. Further development in this area would aid to dissect the role of DNA methylation heterogeneity in cancer cells. Ideally, this would involve the measurement of gene expression, chromatin states, and DNA methylation states in a single cell, in order to elucidate the precise regulatory mechanisms at the single-cell resolution. However, such an integrated approach will require applying multiple measurement platforms on the same molecule without alteration of its property, a task that seems to be daunting if not impossible.
Computational method development is an integral component of every new technology. However, single-cell analysis presents unique challenges that require not only incremental changes but also revolutionary breakthroughs. Each analysis pipeline begins with extracting the signal from raw data, which requires the identification and correction of systematic technical noise in order to properly calibrate different samples and batches. For single-cell analysis, it remains challenging to distinguish technical variations from biological variations [60–63]. Rare cell types, by definition, consist of a small cell-population and may be detectable only in a relatively large cell population. It is difficult to distinguish them from technical artifacts as traditional clustering algorithms, which favor robustness, tend to identify large subpopulations. Another challenge is to distinguish transient variations, such as the ones caused by stochastic noise or regular cell-cycle variation, from those that are essential for cell identity. Integration analysis of gene expression data with chromatin states and transcription factor binding information has been very useful for understanding the underlying gene regulatory networks. However, as it remains difficult to map chromatin states and transcription binding at single-cell resolution, it is important, but challenging, to develop novel computational methods to integrate single-cell gene expression data with population level datasets. Perhaps most importantly, substantial efforts are required to improve single-cell technologies and computational methods in order to have direct implications in clinical decision-making.
Despite these challenges, single-cell analysis undoubtedly presents tremendous opportunities. Taken together, applications of single-cell analysis will greatly enhance the power of systematic characterization of cancer heterogeneity and lead to mechanistic insights into cancer progression, which ultimately will aid the development of novel therapeutic strategies, help us better understand the mechanisms of drug resistance, and lead to improvement of clinical outcomes.
Supplementary Material
Acknowledgments
This work was supported in part by a Claudia Adams Barr Award to G.-C.Y.
References
- 1.Bedard PL, et al. Tumour heterogeneity in the clinic. Nature. 2013;501:355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 3.de Vargas Roditi L, Claassen M. Computational and experimental single cell biology techniques for the definition of cell type heterogeneity, interplay and intracellular dynamics. Curr Opin Biotechnol. 2014;34C:9–15. doi: 10.1016/j.copbio.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Di Palma S, Bodenmiller B. Unraveling cell populations in tumors by single-cell mass cytometry. Curr Opin Biotechnol. 2015;31:122–129. doi: 10.1016/j.copbio.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Junker JP, van Oudenaarden A. Every cell is special: genome-wide studies add a new dimension to single-cell biology. Cell. 2014;157:8–11. doi: 10.1016/j.cell.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452. doi: 10.1186/s13059-014-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsioris K, et al. A new toolbox for assessing single cells. Annu Rev Chem Biomol Eng. 2014;5:455–477. doi: 10.1146/annurev-chembioeng-060713-035958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholas CR, et al. A method for single-cell sorting and expansion of genetically modified human embryonic stem cells. Stem Cells Dev. 2007;16:109–117. doi: 10.1089/scd.2006.0059. [DOI] [PubMed] [Google Scholar]
- 9.Thorsen T, et al. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 10.Guo G, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13:492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moignard V, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 18.Tang F, et al. Development and applications of single-cell transcriptome analysis. Nat Methods. 2011;8:S6–S11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Z, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng Q, et al. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 21.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behbehani GK, et al. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amir el AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marco E, et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci U S A. 2014;111:E5643–E5650. doi: 10.1073/pnas.1408993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moignard V, et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015;33:269–276. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu C, et al. Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res. 2014;24:701–712. doi: 10.1038/cr.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saadatpour A, et al. Characterizing heterogeneity in leukemic cells using single-cell gene expression analysis. Genome Biol. 2014;15:525. doi: 10.1186/s13059-014-0525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fulwyler MJ. Electronic separation of biological cells by volume. Science. 1965;150:910–911. doi: 10.1126/science.150.3698.910. [DOI] [PubMed] [Google Scholar]
- 35.Maamar H, et al. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raj A, et al. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 39.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, et al. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zong C, et al. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 43.Kurimoto K, et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimshony T, et al. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Dey SS, et al. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan HC, et al. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 48.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerdes MJ, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crosetto N, et al. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 52.Achim K, et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33:503–509. doi: 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- 53.Chen KH, et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lubeck E, et al. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satija R, et al. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 58.Angelo M, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marinov GK, et al. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 61.Grun D, et al. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 62.Kharchenko PV, et al. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 64.Stegle O, et al. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 65.Livak KJ, et al. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods. 2013;59:71–79. doi: 10.1016/j.ymeth.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng J, et al. Single-cell copy number variation detection. Genome Biol. 2011;12:R80. doi: 10.1186/gb-2011-12-8-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baslan T, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stahlberg A, et al. Defining cell populations with single-cell gene expression profiling: correlations and identification of astrocyte subpopulations. Nucleic Acids Res. 2011;39:e24. doi: 10.1093/nar/gkq1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buettner F, et al. Probabilistic PCA of censored data: accounting for uncertainties in the visualization of high-throughput single-cell qPCR data. Bioinformatics. 2014;30:1867–1875. doi: 10.1093/bioinformatics/btu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buettner F, Theis FJ. A novel approach for resolving differences in single-cell gene expression patterns from zygote to blastocyst. Bioinformatics. 2012;28:i626–i632. doi: 10.1093/bioinformatics/bts385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Maaten L. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014;15:3221–3245. [Google Scholar]
- 73.van der Maaten LJP, Hinton GE. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 74.Shekhar K, et al. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proc Natl Acad Sci U S A. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu C, Su Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics. 2015;31:1974–1980. doi: 10.1093/bioinformatics/btv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feigelman J, et al. MCA: Multiresolution Correlation Analysis, a graphical tool for subpopulation identification in single-cell gene expression data. BMC Bioinformatics. 2014;15:240. doi: 10.1186/1471-2105-15-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bruggner RV, et al. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014;111:E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finak G, et al. Mixture models for single-cell assays with applications to vaccine studies. Biostatistics. 2014;15:87–101. doi: 10.1093/biostatistics/kxt024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hastie T, Stuetzle W. Principal curves. J Am Stat Assoc. 1989;84:502–516. [Google Scholar]
- 80.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gawad C, et al. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci U S A. 2014;111:17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 84.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo Y, et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161:1175–1186. doi: 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kouno T, et al. Temporal dynamics and transcriptional control using single-cell gene expression analysis. Genome Biol. 2013;14:R118. doi: 10.1186/gb-2013-14-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McDavid A, et al. Modeling bi-modality improves characterization of cell cycle on gene expression in single cells. PLoS Comput Biol. 2014;10:e1003696. doi: 10.1371/journal.pcbi.1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu H, et al. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput Biol. 2014;10:e1003777. doi: 10.1371/journal.pcbi.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ni X, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lohr JG, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Melchor L, et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28:1705–1715. doi: 10.1038/leu.2014.13. [DOI] [PubMed] [Google Scholar]
- 93.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat Biotechnol. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 94.DiGiuseppe JA, et al. Detection of minimal residual disease in B lymphoblastic leukemia using viSNE. Cytometry B Clin Cytom. 2015 doi: 10.1002/cyto.b.21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Etzrodt M, et al. Quantitative single-cell approaches to stem cell research. Cell Stem Cell. 2014;15:546–558. doi: 10.1016/j.stem.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 96.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo H, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 99.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 100.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015 doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.