Abstract
Sulfur to alpha carbon thioether-containing peptides (sactipeptides) are ribosomally synthesized post-translationally modified peptides (RiPPs) with bacteriocidal activities. The thioether crosslink, which is required for biological activity, is installed by a member of the radical S-adenosyl-l-methionine (SAM) superfamily in the peptide substrate. Herein we show that the radical SAM enzyme, SkfB, utilizes the 5′-deoxyadenosyl radical generated from the reductive cleavage of SAM to abstract a hydrogen atom from the α-carbon of the amino acid at position 12 in the substrate, SkfA, to initiate the installation of a thioether crosslink. The insights from this work are applicable to all radical SAM sactipeptide maturases.
Graphical abstract
Peptide derived secondary metabolites are produced by microorganisms via one of two synthetic pathways, non-ribosomal peptide synthetases (NRPS) or post-ribosomal peptide synthesis (PRPS).1–3 While the biosynthesis of NRPS peptides, such as vancomycin, has been extensively studied, the biosynthetic strategies for PRPS metabolites are a subject of considerable recent interest. By contrast to NRPS peptides, which are modified both during and after synthesis, PRPS peptides are extensively modified after translation by the ribosome. The natural products produced by the PRPS mechanism are referred to as ribosomally synthesized post-translationally modified peptides (RiPPs).3 The wide distribution of these natural products suggests that they provide a selective advantage to the producing organism in complex biological niches by acting as signaling molecules, cofactors, or bacteriocidal molecules.3
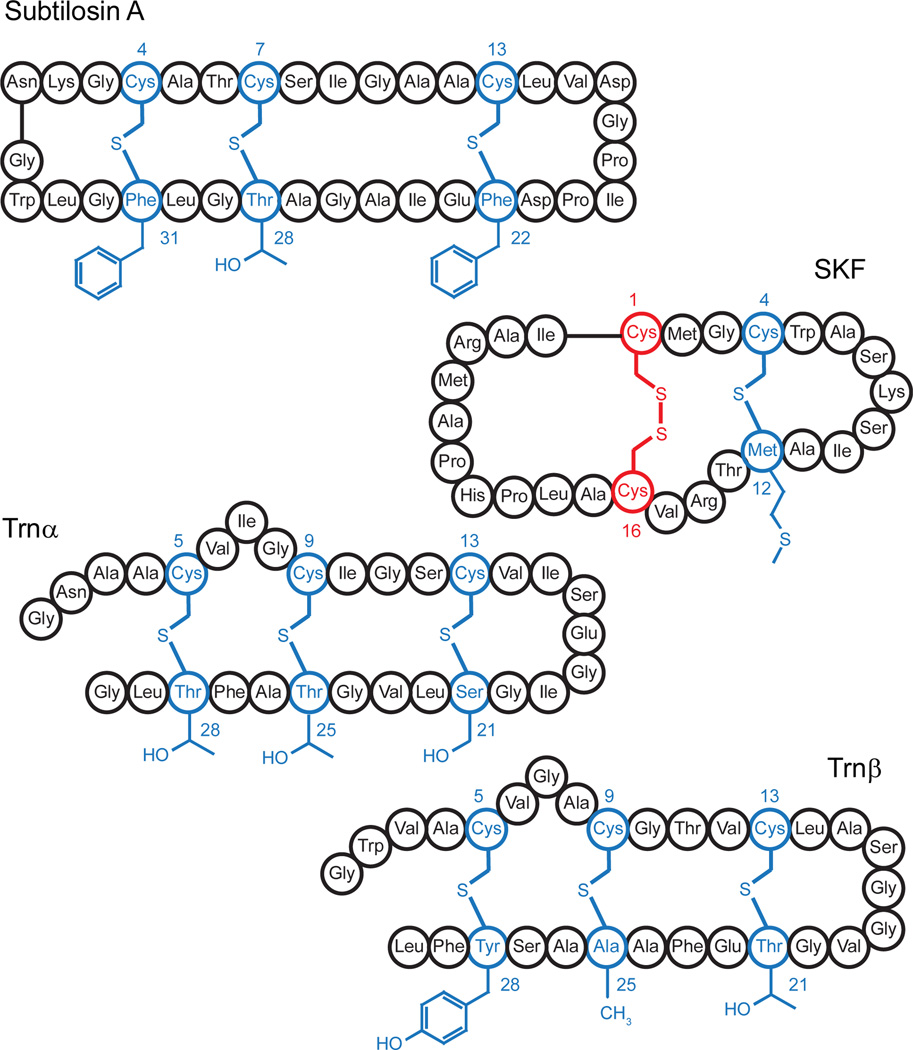
Many distinct subclasses of RiPP natural products have been identified and classified on the basis of the structures of the mature metabolites.3 One of the best studied classes of RiPPs are the lanthipeptides, which contain intramolecular thioether crosslinks. These crosslinks are installed between the sulfur atom of a cysteine and the beta carbon of either a dehydrated serine or threonine in a redox neutral mechanism. Another class of metabolites that contain thioether crosslinks is the sulfur to alpha carbon thioether containing peptides, sactipeptides (Fig. 1).3–7 One of the distinguishing features between the lanthipeptide and sactipeptide metabolites is that the sactipeptides contain one or more intramolecular thioether linkages between a Cys thiol side chain and the alpha carbon of an amino acid in the peptide. The sactipeptide natural products include thurincin H, thuricin CD, subtilosin A, and sporulation killing factor (SKF), which are shown in Fig. 1. In the limited number of sactipeptides characterized to date, the amino acids Ala, Met, Phe, Ser, Thr, and Tyr have been observed in the acceptor positions.
Figure 1.
Schematic of the sactipeptides subtilosin A, sporulation killing factor (SKF), and the two peptides, Trnα and Trnβ. The sulfur to α-carbon thioether crosslinks are highlighted in blue. The sidechains of the accepting residues remain unchanged.
Similar to the lanthipeptides, the sactipeptides are generated as precursor peptides that contain a N-terminal leader sequence important for the recognition by the maturase enzymes.3 The maturation of the precursor peptide begins with the installation of the thioether crosslink(s). Recent investigations on the maturation of subtilosin A, thurincin H, six cysteines in forty-five (SCIFF), and SKF peptides have shown that the thioether is installed in the precursor peptide by a radical S-adenosyl-l-methionine (SAM) enzyme, which is encoded by an orf clustered with the orf encoding for the peptide.8–11 Members of the radical SAM superfamily are primarily identified by the presence of a CxxxCxxC motif.12 The three Cys thiolates in this motif coordinate three iron atoms of a [4Fe-4S] cluster, while the fourth iron is coordinated by the α-amino and α-carboxylate moieties of SAM.13,14 The [4Fe-4S] cluster rests in the +2 oxidation state and it mediates the reductive cleavage of SAM to commonly afford the 5′-deoxyadenosyl radical (dAdo•) upon reduction to the +1 oxidation state. Radical SAM enzymes use this strong oxidant to abstract a hydrogen atom from the substrate to initiate catalysis.
The sactipeptide maturase radical SAM enzymes AlbA, ThnB, Tte1186, and SkfB have been shown to all reductively cleave SAM to generate the dAdo•, which is subsequently used to catalyze the formation of one or more thioether crosslinks in their respective peptide substrates.8–11 Site-directed variants lacking the [4Fe-4S] cluster are unable to reductively cleave SAM and, more importantly, unable catalyze the formation of the thioether crosslink(s).
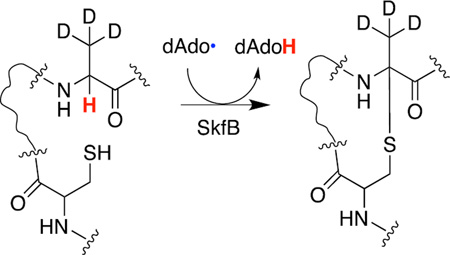
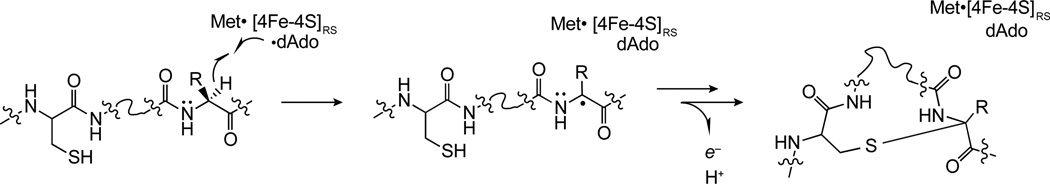
In contrast to redox neutral mechanism of thioether crosslink formation in the lanthipeptides, the mechanism of thioether installation in sactipeptides entails a two electron oxidation of the peptide. To date, multiple mechanisms for peptide thioether crosslink formation have been proposed.8,11 While the mechanisms differ in the details of thioether bond formation, all of them posulate that the dAdo• abstracts the hydrogen atom from the Cα position of the acceptor amino acid (see Scheme 1). Recently, it was shown that AlbA uses the dAdo• to abstract a H-atom from Phe31 in the maturation of the subtilosin A precursor peptide, SboA.15 However, the investigators used a uniformly deuterated Phe in the investigation and one cannot rule out the dAdo• abstracting a H-atom from the Cβ position of the acceptor amino acid. In this report we provide the first biochemical evidence that the radical SAM enzyme SkfB uses the dAdo• to abstract the H-atom from the Cα of the acceptor amino acid in the SKF precursor peptide SkfA.
Scheme 1.
A proposed mechanism of thioether crosslink formation in sactipeptides. The dAdo• abstracts the Cα hydrogen atom from the acceptor residue to initiate catalysis to generate the captodatively stabilized radical intermediate.
SkfB installs the thioether crosslink between Cys4 and Met12 in the mature SKF product (Fig. 1).9,16 Additionally, SkfB tolerates a variety of sidechains at position 12 of the substrate, SkfA, including A, L, S, T, N, F, and Y.9 We took advantage of the acceptor site promiscuity to pinpoint the site of H-atom abstraction. Fmoc protected L-Ala, both uniformly deuterated (Fmoc-[U-D4]A) and containing three deuterium at the β-methyl moiety (Fmoc-[β-D3]A), are commercially available (Cambridge Isotope Laboratories). Therefore, we synthesized two M12A SkfA variants which were labeled with [β-D3]A or [U-D4]A at position 12 only. After synthesis, [β-D3]A– and [U-D4]A–SkfA were purified by HPLC and determined to be at least 97% pure, as judged by UHPLC-MS (see Fig S1 and S2 respectively). We reasoned that if the H-atom is derived from the Cα at position 12, then we should observe deuterium transfer to 5′-deoxyadenosine (dAdo) with [U-D4]A– but not [β-D3]A–SkfA. Moreover, as a corollary to this, we would observe retention of three deuterium atoms in the final product regardless of whether the substrate contained [U-D4]A or [β-D3]A at position 12.
We first demonstrated that synthetic wild-type SkfA is a substrate for SkfB. To this end, wild-type SkfA was synthesized and purified in the same manner as the deuterium containing peptides. Purified wild-type SkfA (≥93% pure, Fig S3) was incubated with SkfB and SAM with reducing equivalents supplied from NADPH via the recombinant Bacillus subtilis flavodoxin (YkuN) and Escherichia coli ferredoxin (flavodoxin):NADP+ reductase (Fpr). Previous studies with 7-carboxy-7-deazaguanine synthase have shown that use of the native reducing system may help maximize enzymatic activity in radical SAM enzymes.17 After 3 h, an aliquot of the reaction was quenched with acid and the dAdo was analyzed by UHPLC-MS. The remaining portion of the reaction was treated with iodoacetamide to alkylate the free Cys thiolates in the peptide then acid quenched. Both dAdo and the peptide from each quenched aliquot were analyzed by UHPLC-MS using an LTQ OrbiTrapXL mass spectrometer. The exquisite resolution of the instrument has been used previously to monitor deuterium incorporation into dAdo.18
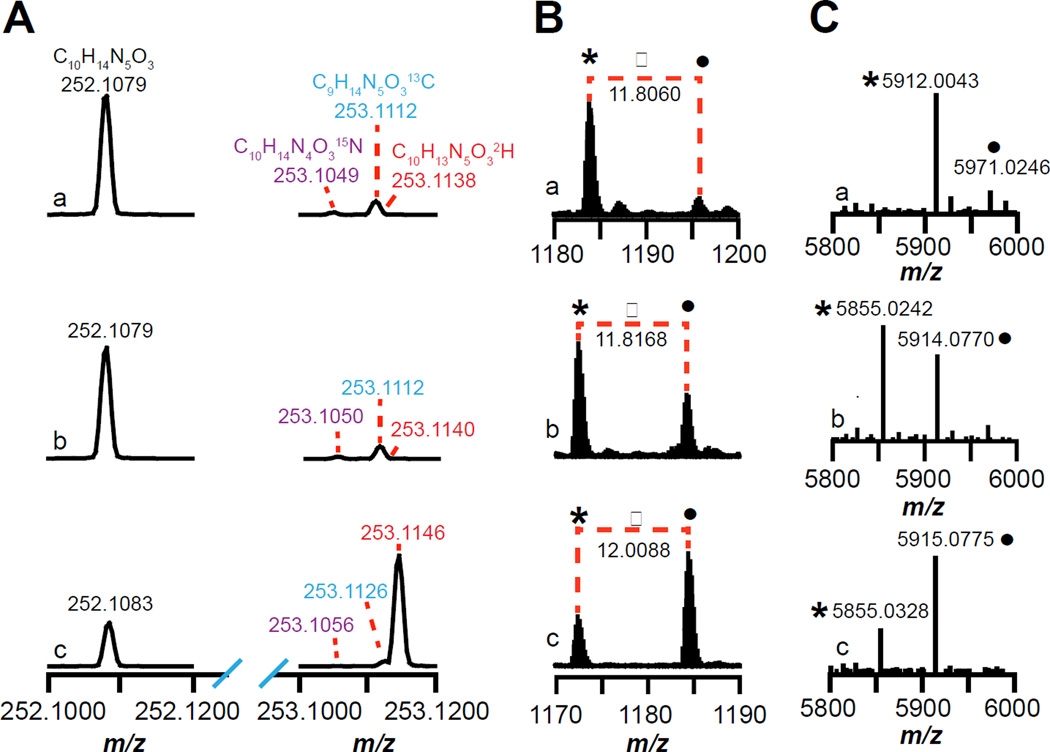
The results of the incubations are shown in Fig. 2. The m/z of 252.1079 in Fig 2Aa corresponds to the [M+H]+ for dAdo (C10H14N5O3, theoretical m/z = 252.1097, 7.1 ppm error) showing that SkfB produces dAdo in the presence of the natural abundance synthetic wild-type SkfA peptide. The 15N (m/z of 253.1049), 13C (m/z of 253.1112), and 2H (m/z of 253.1138) isotopic peaks of the +1 m/z envelope are resolved and are 1.8% (theoretical = 1.8%), 10.9% (theoretical = 10.8%), and 0.3% (theoretical = 0.1%) relative to the corresponding molecular ion peak (m/z of 252.1079).
Figure 2.
Mass spectra of (A) dAdo or (B and C) SkfA isolated from reactions containing (a) wild-type, (b) M12[β-D3]A–, or (c) M12[U-D4]A–SkfA. (A) The mass spectra show the monoisotopic (m/z = 252.1079 – 252.1083) and the corresponding 15N (m/z = 253.1049 – 253.1056), 13C (m/z = 253.1112 – 253.1126), and 2H isotope peaks (m/z = 253.1138 – 253.1146) of dAdo. (B) Mass spectra of SkfA peptides zooming in on the +5 charge state (see Fig S4a–c for the full mass spectra). The peak indicated by • corresponds to SkfA with three carbamidomethylated cysteines, whereas the * corresponds to SkfA containing one thioether crosslink and two carbamidomethylated cysteines. (C) The deconvoluted mass spectra generated using the Xtract software (Thermo Fisher) from the full mass spectra shown in Fig S4a–c.
We next incubated M12[β-D3]A– or M12[U-D4]A–SkfA variants with SkfB in the same manner as described above for wild-type SkfA and examined the deuterium content of dAdo isolated from the reactions containing the M12[β-D3]A– or M12[U-D4]A–SkfA variants (Fig. 2Ab and 2Ac). The dAdo from the reaction containing the M12[β-D3]A–SkfA shows an isotopic distribution that is identical to that of the natural abundance dAdo. However, dAdo formed in the presence of the M12[U-D4]A–SkfA shows significant enrichment in the deuterium isotope (m/z = 253.1146) relative to the molecular ion peak at m/z of 252.1083. The transfer of deuterium into dAdo with the M12[U-D4]A– and not with the M12[β-D3]A–SkfA supports the role of the dAdo• in abstracting a H-atom directly from the Cα of the substrate. The mass spectrum corresponding to dAdo shown in Fig 2Ac shows a mixture of natural abundance dAdo (m/z = 252.1083) and dAdo enriched with deuterium (m/z = 253.1146). The natural abundance deuterium contained in dAdo (m/z = 252.1083) corresponds to 0.1% of the molecular ion peak, which contributes a negligible amount to the species with m/z of 253.1146. Therefore, 72% of the dAdo is generated upon abstraction of the α-deuterium atom from the M12[U-D4]A–SkfA. The remaining 28% of the dAdo generated in the reaction is likely due to SkfB catalyzing the reductive cleavage of SAM uncoupled to catalysis.
We next examined the mass spectrum of the SkfA peptide from these reactions to determine if the dAdo formation correlates with thioether crosslink formation. While thioether crosslink formation in the peptide is nearly complete for wild-type SkfA, we can readily observe multiple charge states corresponding to both the unmodified and modified substrate (Table S1, Fig S4a). In the foregoing discussion, we will focus on the +5 charge state envelopes for clarity. The mass envelope corresponding to the unmodified wild-type SkfA with three carbamidomethyl groups (•) and the single thioether-containing wild-type SkfA with two carbamidomethyl groups (*) are 11.8060 amu apart, corresponding to a difference of 59.0300 amu (11.8060 amu × 5) (Fig 2Ba). The observed mass shift is consistent with the lighter species (*) lacking one carbamidomethyl group (57.0215 amu) and two hydrogen atoms (2.0156 amu) relative to the intact substrate. The corresponding deconvoluted spectrum (Fig 2Ca) shows a peak at 5971.0246 for the unmodified wild-type SkfA, which is triply alkylated by iodoacetamide (theoretical m/z 5971.0505, 4.3 ppm error). The peak at 5912.0043 corresponds to the product peak, which is consistent with the presence of a single thioether crosslink and doubly carbamidomethylated Cys sidechains (theoretical m/z 5912.0134, 1.5 ppm error).
Incubation of either the M12[β-D3]A– or M12[U-D4]A– SkfA led to a mixture of unmodified substrate and singly modified product (Fig 2Bb and 2Bc, Fig S4 b–c, see Table S2 and S3 for all of the charge states). With the M12[β-D3]A–SkfA peptide, the difference between the unmodified (•) and modified (*) peptide for the +5 charge state is 11.8168 amu (Fig 2Bb), corresponding to a difference of 59.0840 amu (11.8168 amu × 5). This is accounted for with the lighter species (*) lacking one carbamidomethyl group (57.0215 amu) and two hydrogen atoms (2.0156 amu) relative to the intact substrate. The corresponding deconvoluted spectrum (Fig 2Cb) shows a peak at 5914.0770 for the unmodified M12[β-D3]A–SkfA, which is triply alkylated by iodoacetamide (theoretical m/z 5914.0660, 1.9 ppm error). The peak at 5855.0242 corresponds to the product peak, which is consistent with the presence of a single thioether crosslink and two carbamidomethylated Cys sidechains (theoretical m/z 5855.0288, 0.8 ppm error). Moreover, the mass of the peptide product is consistent with retention of all of the starting deuterium atoms.
With the M12[U-D4]A–SkfA the difference between the unmodified (•) and modified (*) peptide for the +5 charge state is 12.0088 amu (Fig 2Bc), corresponding to a difference of 60.0440 amu (12.0088 amu × 5). This difference is accounted for with the lighter species (*) lacking one carbamidomethyl group (57.0215 amu), one hydrogen atom (1.0078 amu), and one deuterium atom (2.0141 amu). The corresponding deconvoluted spectrum (Fig 2Cc) shows a peak at 5915.0775 for the unmodified M12[U-D4]A–SkfA, which is triply alkylated by iodoacetamide and contains four deuterium atoms (theoretical m/z 5915.0722, 0.9 ppm error). The peak at 5855.0328 corresponds to the product peak, which is consistent with the presence of a single thioether crosslink, two carbamidomethylated Cys sidechains and retention of only three deuterium atoms (theoretical m/z 5855.0288, 0.7 ppm error). Collectively, these data show that formation of the thioether crosslink in SkfA is coupled with the loss of a hydrogen atom from the Cα of the peptide at position 12.
Prior to this investigation all of the proposed mechanisms for thioether crosslink formation begin with the dAdo• abstracting the Cα hydrogen atom from the acceptor residue in the peptide (Scheme 1). While deuterium transfer from the peptide substrate to dAdo has been shown previously in a SboA analog during reaction with AlbA15, the results presented in this investigation are the first to unambiguously localize the site of abstraction to the Cα hydrogen atom at position 12 of SkfA during catalysis to install the thioether crosslink in the peptide. The abstraction of the H-atom from the Cα of the acceptor site will be a general feature of sactipeptide forming maturases.
Supplementary Material
Acknowledgments
Funding Sources
The work reported in this publication was supported by National Institutes of General Medical Sciences of the National Institutes of Health grant R01 GM72623 awarded to VB. The content is solely the responsibility of the authors and does not necessarily represent the official views for the National Institutes of Health.
We would like to thank Prof. Hazel Holden for providing the pET28JT vector and the vector encoding the TEV protease.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website in a PDF. The supporting information contains the materials and methods used in the investigation, Table S1 – S3, and Figures S1 – S4.
No competing financial interests have been declared.
REFERENCES
- 1.Finking R, Marahiel MA. Ann Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 2.Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 3.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babasaki K, Takao T, Shimonishi Y, Kurahashi K. J Biochemistry. 1985;98:585–603. doi: 10.1093/oxfordjournals.jbchem.a135315. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Pastor JE. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 6.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Proc Natl Acad Sci U S A. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H, Churey JJ, Worobo RW. FEMS Microbiol Lett. 2009;299:205–213. doi: 10.1111/j.1574-6968.2009.01749.x. [DOI] [PubMed] [Google Scholar]
- 8.Flühe L, Knappe TA, Gattner MJ, Schafer A, Burghaus O, Linne U, Marahiel MA. Nat Chem Biol. 2012;8:350–357. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- 9.Flühe L, Burghaus O, Wieckowski BM, Giessen TW, Linne U, Marahiel MA. J Am Chem Soc. 2013;135:959–962. doi: 10.1021/ja310542g. [DOI] [PubMed] [Google Scholar]
- 10.Wieckowski BM, Hegemann JD, Mielcarek A, Boss L, Burghaus O, Marahiel MA. FEBS Lett. 2015;589:1802–1806. doi: 10.1016/j.febslet.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 11.Bruender NA, Wilcoxen J, Britt RD, Bandarian V. Biochemistry. 2016;55:2122–2134. doi: 10.1021/acs.biochem.6b00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 14.Chen D, Walsby C, Hoffman BM, Frey PA. J Am Chem Soc. 2003;125:11788–11789. doi: 10.1021/ja036120z. [DOI] [PubMed] [Google Scholar]
- 15.Benjdia A, Guillot B, Lefranc H, Vaudry J, Leprince J, Berteau O. Chem Comm. 2016;52:6249–6252. doi: 10.1039/c6cc01317a. [DOI] [PubMed] [Google Scholar]
- 16.Liu W-T, Yang Y-L, Xu Y, Lamsa A, Haste NM, Yang JY, Ng J, Gonzalez D, Ellermeier CD, Straight PD, Pevzner PA, Pogliano J, Nizet V, Pogliano K, Dorrestein PC. Proc Natl Acad Sci U S A. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruender NA, Young AP, Bandarian V. Biochemistry. 2015;54:2903–2910. doi: 10.1021/acs.biochem.5b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young AP, Bandarian V. Biochemistry. 2015;54:3569–3572. doi: 10.1021/acs.biochem.5b00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.