Abstract
Background:
The established clinical staging systems (Rai/Binet) of chronic lymphocytic leukemia (CLL) cannot accurately predict the appropriate treatment of patients in the earlier stages. In the past two decades, several prognostic factors have been identified to predict the outcome of patients with CLL, but only a few studies investigated more markers together. To predict the time to first treatment (TTFT) in patients of early stages, we evaluated the prognostic role of conventional markers as well as cytogenetic abnormalities and combined them together in a new prognostic scoring system, the CLL prognostic index (CLL-PI).
Methods:
Taking advantage of a population of 406 untreated Chinese patients with CLL at early and advanced stage of disease, we identified the strongest prognostic markers of TTFT and, subsequently, in a cohort of 173 patients who had complete data for all 3 variables, we integrated the data of traditional staging system, cytogenetic aberrations, and mutational status of immunoglobulin heavy chain variable region (IGHV) in CLL-PI. The median follow-up time was 45 months and the end point was TTFT.
Results:
The median TTFT was 38 months and the 5-year overall survival was 80%. According to univariate analysis, patients of advanced Rai stages (P < 0.001) or with 11q- (P = 0.002), 17p- (P < 0.001), unmutated IGHV (P < 0.001), negative 13q- (P = 0.007) and elevated lactate dehydrogenase levels (P = 0.001) tended to have a significantly shorter TTFT. And subsequently, based on multivariate Cox regression analysis, three independent factors for TTFT were identified: advanced clinical stage (P = 0.002), 17p- (P = 0.050) and unmutated IGHV (P = 0.049). Applying weighted grading of these independent factors, a CLL-PI was constructed based on regression parameters, which could categorize four different risk groups (low risk [score 0], intermediate low [score 1], intermediate high [score 2] and high risk [score 3–6]) with significantly different TTFT (median TTFT of not reached (NR), 65.0 months, 36.0 months and 19.0 months, respectively, P < 0.001).
Conclusions:
This study developed a weighted, integrated CLL-PI prognostic system of CLL patients which combines the critical genetic prognostic markers with traditional clinical stage. This novel modified PI system could be used to discriminate among groups and may help predict the TTFT and prognosis of patients with CLL.
Keywords: 17p Deletion, Chronic Lymphocytic Leukemia, Immunoglobulin Heavy Chain Variable Mutation, Prognostic Index, Time to First Treatment
Introduction
Patients with chronic lymphocytic leukemia (CLL), the most common leukemia in the Western world, exhibit heterogeneous clinical courses and survival rates;[1] some patients have an indolent disease and do not require treatment whereas others have an aggressive disease, requiring treatment at initial presentation. The traditional staging systems independently developed by Rai et al.[2] and Binet et al.[3] have been widely applied for prognostication for more than 30 years. Despite their utility, there is still a significant heterogeneity regarding prognosis in each stage category. Using only clinical features is not sufficient to predict the risk of progression from the diagnosis, particularly in patients with early-stage disease.[4]
During the past two decades, several studies have identified some clinical and biological markers to further stratify patients’ survival. Cytogenetic abnormalities, such as deletions in 13q (13q−), 11q (11q−), and 17p (17p−), are critical events in the pathogenesis of CLL and important predictors of disease progression and survival,[5] as detected by interphase fluorescence in situ hybridization (I-FISH). Immunoglobulin heavy chain variable region (IGHV) mutational status also plays a critical role, as patients with unmutated IGHV (U-IGHV) genes have a distinctly more malignant disease and much shorter survival than those with mutated IGHV (M-IGHV).[6]
However, only a few studies have combined different markers together. In patients with sole 13q−, more than 20% present with U-IGHV status, and these patients have lower overall survival (OS) than patients carrying M-IGHV;[7] similar results have been found in 11q− and 17p− patients.[8] These studies indicate that multiple prognostic factors should be taken under consideration to design treatment and predict clinical outcome, and thus a comprehensive CLL prognostic index (CLL-PI) is necessary.[9]
In addition, CLL is rare in Asians, and the characteristics of CLL patients among an Asian population may not be similar to those in a Caucasian population. The incidence rates of CLL are approximately 80% lower among Asians compared to Caucasians,[10] and the difference in incidence rates between Asian immigrants and Western locals is maintained in all age groups and over time,[11] which suggests that genetic factors outplay environmental factors to give lower CLL rates in Chinese.
In the present study, we took a single-institutional cohort of Chinese patients with CLL and evaluated the prognostic role of conventional biomarkers, as well as cytogenetic abnormalities, to identify the most valuable prognosticators regarding the time to first treatment (TTFT). Subsequently, we integrated data from the traditional staging system, cytogenetic lesions, and IGHV mutational status in a new prognostic scoring system, the CLL-PI. We propose a new method to better predict the TTFT of patients with CLL.
Methods
Patients
A total of 406 treatment-naive CLL patients who were patients at the Institute of Hematology and Blood Disease Hospital between July 2007 and January 2015 were included in this study. The diagnosis in each case was confirmed according to the World Health Organization classification.[12] Evidence of persistent lymphocytosis and a compatible immunophenotype were required for diagnosis. In all cases, an immunophenotypic analysis was performed by flow cytometry, including CD19, CD5, CD22, CD23, CD38, CD25, CD103, CD11c, FMC7, BCL2, CD10, CD20, and surface immunoglobulins κ and λ. All patients enrolled gave informed consent in accordance with requirements of the Declaration of Helsinki, and the research project was approved by the Institutional Ethics Review Boards. For this study, clinical follow-up data were available until the beginning of February 2015.
Fluorescence in situ hybridization and immunoglobulin heavy chain variable mutational analysis
I-FISH was performed on standard cytogenetic preparations as previously reported.[5,13,14] The CLL FISH panel included probes for the chromosome 12 centromere (CEP12), 13q14.3 (LSI RB1), 14q32 (LSI IGHC/IGHV), 17p13 (LSI TP53), 11q22 (LSI ATM), and LSI CCND1/IGH. A dual-color, dual-fusion translocation probe was used to exclude the possibility of mantle cell lymphoma in the case of t(14q32)-positive results. All probes were Vysis FISH products (Abbott Molecular, Abbott Park, IL, USA). Signal screening was carried out on at least 200 cells with well-delineated signals. The cutoff points for positive values (mean of normal control + 3 standard deviation) determined from samples of ten cytogenetically normal people were as follows: 7.5% for CEP 12, 20.0% for deletion of TP53, 6.5% for the deletion of RB1 and ATM, and 4.5% for IgH translocation and CCND1/IGH. IGHV mutation was performed as previously reported.[15,16] Sequence homology ≤98% from the corresponding germ line gene was considered M-IGHV, as opposed to U-IGHV.
Survival and statistical analysis
Categorical variables were compared by the Chi-square test, and continuous variables were compared by the Mann-Whitney test. The TTFT was defined as the time from the date of diagnosis to the date of the initiation of the first treatment or the death or last follow-up at which the patient was known to be untreated. The OS was defined as the time from the date of diagnosis to the date of death or last follow-up. Kaplan-Meier methodology and log-rank test were undertaken for survival analyses using the SPSS statistical software package (version 19.0, IBM, New York, USA). Cox regression models were used to estimate hazard ratios (HRs). All tests were two sided. An effect was considered statistically significant if the P ≤ 0.05.
Results
Clinical characteristics of chronic lymphocytic leukemia population
A total of 406 CLL patients constituted the population of the study. The patients’ characteristics are summarized in Table 1. The median follow-up time was 45 (2–288) months, median TTFT was 38 (95% confident interval [CI]: 29–47) months, and 3-year cumulative probability of receiving treatment was 51.0%; the median estimated OS was 128 (95% CI: 108–147) months, and the 5-year OS was 80% [Supplementary Figure 1a (595.2KB, tif) and 1b (595.2KB, tif) ].
Table 1.
Clinical and biologic characteristics of chronic lymphocytic leukemia population, cohort, and CLL-PI class of risk patients
| Characteristics | Population (n = 406) | Cohort (n = 173) | t/X2 | P | Low risk* (n = 22) | Intermediate low risk* (n = 54) | Intermediate high risk* (n = 43) | High risk* (n = 54) | F/X2 | P |
|---|---|---|---|---|---|---|---|---|---|---|
| Age at diagnosis (years), mean (range) | 58 (26–85) | 56 (26–85) | 1.043 | 1.0 | 61 (47–71) | 54 (36–72) | 57 (38–85) | 53 (26–80) | 3.639 | 0.297 |
| Gender, n (%) | ||||||||||
| Male | 266 (66) | 120 (69) | 0.700 | 0.403 | 13 (59) | 39 (72) | 34 (79) | 34 (63) | 4.160 | 0.245 |
| Female | 140 (34) | 53 (31) | 9 (41) | 15 (28) | 9 (21) | 20 (37) | ||||
| Rai, n (%) | ||||||||||
| Low risk (0) | 60 (15) | 27 (16) | 0.087 | 0.958 | 22 (100) | 0 | 4 (9) | 1 (2) | 219.927 | <0.001 |
| Intermediate risk (I–II) | 211 (52) | 88 (51) | 0 | 54 (56) | 0 | 34 (63) | ||||
| High risk (III–IV) | 135 (33) | 58 (34) | 0 | 0 | 39 (91) | 19 (35) | ||||
| Binet, n (%) | ||||||||||
| A | 157 (40) | 66 (39) | 1.389 | 0.499 | 22 (100) | 29 (56) | 4 (9) | 11 (21) | 144.437 | <0.001 |
| B | 104 (27) | 53 (31) | 0 | 23 (44) | 1 (2) | 29 (55) | ||||
| C | 131 (33) | 51 (30) | 0 | 0 | 38 (88) | 13 (24) | ||||
| Elevated LDH, n (%) | ||||||||||
| Yes | 91 (26) | 37 (25) | 0.063 | 0.802 | 2 (12) | 10 (22) | 7 (19) | 18 (35) | 5.515 | 0.138 |
| No | 265 (74) | 114 (75) | 15 (88) | 36 (78) | 30 (81) | 33 (65) | ||||
| Elevated β2-MG, n (%) | ||||||||||
| Yes | 99 (42) | 48 (43) | 0.001 | 0.976 | 3 (27) | 7 (20) | 18 (60) | 20 (54) | 14.077 | 0.003 |
| No | 135 (58) | 65 (58) | 8 (73) | 28 (80) | 12 (40) | 17 (46) | ||||
| B symptoms, n (%) | ||||||||||
| Yes | 90 (28) | 35 (23) | 0.969 | 0.325 | 1 (6) | 6 (13) | 10 (27) | 18 (35) | 9.770 | 0.021 |
| No | 236 (72) | 115 (77) | 16 (94) | 39 (87) | 27 (73) | 33 (65) | ||||
| Hepatomegaly, n (%) | ||||||||||
| Yes | 23 (7) | 10 (7) | 0.000 | 0.984 | 1 (6) | 2 (5) | 3 (8) | 4 (9) | 0.639 | 0.887 |
| No | 313 (93) | 135 (93) | 15 (94) | 42 (95) | 35 (92) | 43 (91) | ||||
| Splenomegaly, n (%) | ||||||||||
| Yes | 149 (43) | 63 (43) | 0.003 | 0.953 | 1 (6) | 20 (45) | 20 (53) | 22 (46) | 10.510 | 0.015 |
| No | 194 (57) | 83 (57) | 15 (94) | 24 (55) | 18 (47) | 26 (54) | ||||
| FISH, n (%) | ||||||||||
| 13q− | 102 (35) | 42 (32) | 1.577 | 0.904 | 11 (69) | 16 (43) | 14 (42) | 1 (2) | 54.676 | <0.001 |
| Normal | 75 (26) | 34 (26) | 4 (25) | 8 (22) | 7 (21) | 15 (33) | ||||
| +12 | 43 (15) | 21 (16) | 0 | 9 (24) | 6 (18) | 6 (13) | ||||
| 11q− | 34 (12) | 19 (14) | 1 (6) | 4 (11) | 5 (15) | 9 (20) | ||||
| 17p− | 39 (13) | 16 (12) | 0 | 0 | 1 (3) | 15 (33) | ||||
| IGHV, n (%) | ||||||||||
| M-IGHV | 131 (71) | 120 (69) | 0.089 | 0.765 | 22 (100) | 54 (100) | 40 (93) | 4 (7) | 142.439 | <0.001 |
| U-IGHV | 54 (29) | 53 (31) | 0 | 0 | 3 (7) | 50 (93) |
*This analysis was performed on cohort of 173 patients. LDH: Lactate dehydrogenase; β2-MG: β2-microglobulin; FISH: Fluorescence in situ hybridization; CLL-PI: Chronic lymphocytic leukemia prognostic index; IGHV: Immunoglobulin heavy chain variable region; M: Mutated; U: Unmutated.
Kaplan-Meier curves for TTFT and OS of the 406 patients with CLL. (a) Kaplan-Meier curves for TTFT of the enrolled patients. (b) Kaplan-Meier curves for OS of the patients.
Univariate analysis and multivariate analysis of time to first treatment
As a preliminary step to the development of a prognostic model, we assessed the independent prognostic value of traditional staging systems and the biologic variables such as FISH analysis, IGHV mutational status, CD38 levels, and some common clinical features.
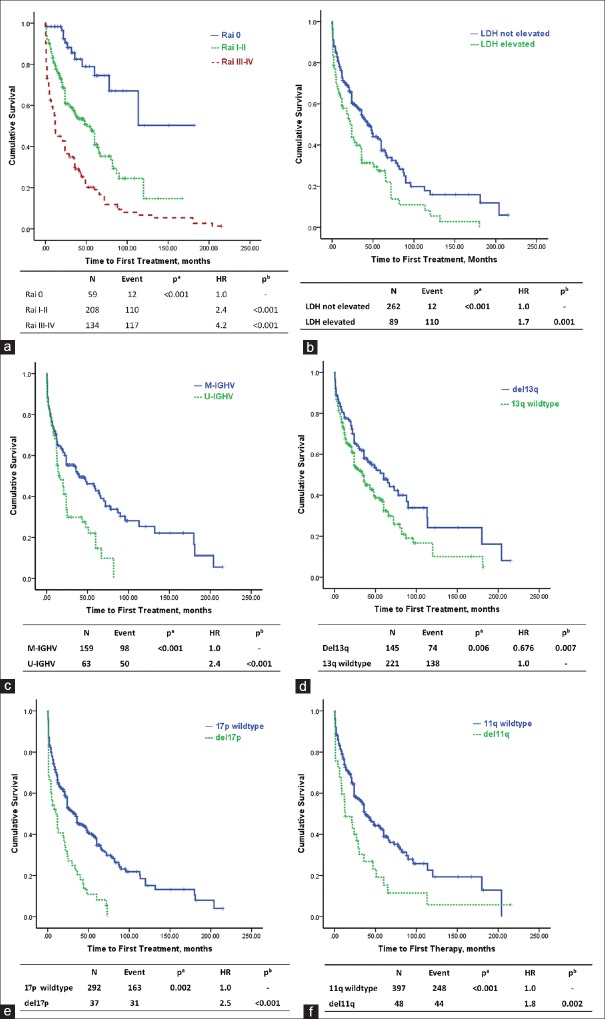
In our cohort, trisomy 12 failed to predict TTFT, and we thus did not consider it in further analyses. Other cytogenetic abnormalities detected by FISH, IGHV mutational status, and elevated levels of lactate dehydrogenase (LDH) were prognostic factors for treatment [Figure 1]. Specifically, patients with 11q−, 17p−, U-IGHV, CD38+, and elevated LDH levels tended to have a shorter TTFT.
Figure 1.
Kaplan-Meier curves for TTFT according to cytogenetic abnormalities, immunoglobulin heavy chain variable region (IGHV) mutational status and lactate dehydrogenase (LDH) level. Pa is the value calculated by log-rank test for differences in variables and Pb value is calculated by Cox multivariate analysis for differences in variables. (a) Kaplan-Meier curves for TTFT according to Rai staging system. (b) Kaplan-Meier curves for TTFT according to LDH. (c) Kaplan-Meier curves for TTFT according to IGHV mutational status. (d) Kaplan-Meier curves for TTFT according to 13q. (e) Kaplan-Meier curves for TTFT according to 17p. (f) Kaplan-Meier curves for TTFT according to 11q.
Multivariate Cox models were constructed to identify independent prognostic factors for predicting TTFT using LDH levels, the Rai staging system, 11q−, 13q−, 17p−, and IGHV mutational status. The model indicated that Rai risk groups, IGHV mutational status, and 17p− were independent factors to predict treatment [Table 2].
Table 2.
Multivariate Cox regression analysis for TTFT
| Items | β | P | HR | 95% CI |
|---|---|---|---|---|
| Rai staging system | 0.582 | 0.002 | 1.8 | 1.2–2.6 |
| U-IGHV | 0.525 | 0.049 | 1.7 | 1.0–2.9 |
| Elevated LDH | 0.110 | 0.690 | 1.1 | 0.7–1.9 |
| del11q | 0.341 | 0.286 | 1.4 | 0.8–2.6 |
| del17p >20% | 0.662 | 0.050 | 1.9 | 1.0–3.8 |
TTFT: Time to first treatment; HR: Hazard ratio; CI: Confidence interval; IGHV: Immunoglobulin heavy chain variable region; LDH: Lactate dehydrogenase; U: Unmutated.
Chronic lymphocytic leukemia prognostic index for time to first treatment
According to the β regression coefficients [Table 2] and the HR for each variable [Table 3], we developed a clinicobiological prognostic scoring system by assigning 1 point for Rai I–II stages and 2 points for Rai III–IV stages, the presence of an U-IGHV gene, or 17p deletion.
Table 3.
Score assignment of the prognostic factors
| Items | HR | 95% CI | P | Score |
|---|---|---|---|---|
| Rai | ||||
| 0 | 1.0 | – | – | 0 |
| I–II | 2.4 | 1.5–3.7 | <0.001 | 1 |
| III–IV | 4.2 | 2.7–6.6 | <0.001 | 2 |
| IGHV mutational status | ||||
| M-IGHV | 1.0 | – | – | 0 |
| U-IGHV | 2.4 | 1.6–3.6 | <0.001 | 2 |
| Del17p >20% | ||||
| Negative | 1.0 | – | – | 0 |
| Positive | 2.5 | 1.8–3.7 | <0.001 | 2 |
HR: Hazard ratio; CI: Confidence interval; IGHV: Immunoglobulin heavy chain variable region; M: Mutated; U: Unmutated.
Further analyses were performed on a cohort of 173 patients who had complete data for all 3 variables. The characteristics of this cohort of patients are summarized in Table 1. To assess the potential selection bias, we compared clinical and biologic features of the cohort (n = 173) with those of the whole population (n = 406) and did not find any significant difference between these two groups regarding clinical and biologic features [Table 1]. The score distribution of this CLL-PI in the cohort is shown in Figure 2a, and four prognostic groups were stratified, with Scores of 0, 1, 2, and 3–6.
Figure 2.
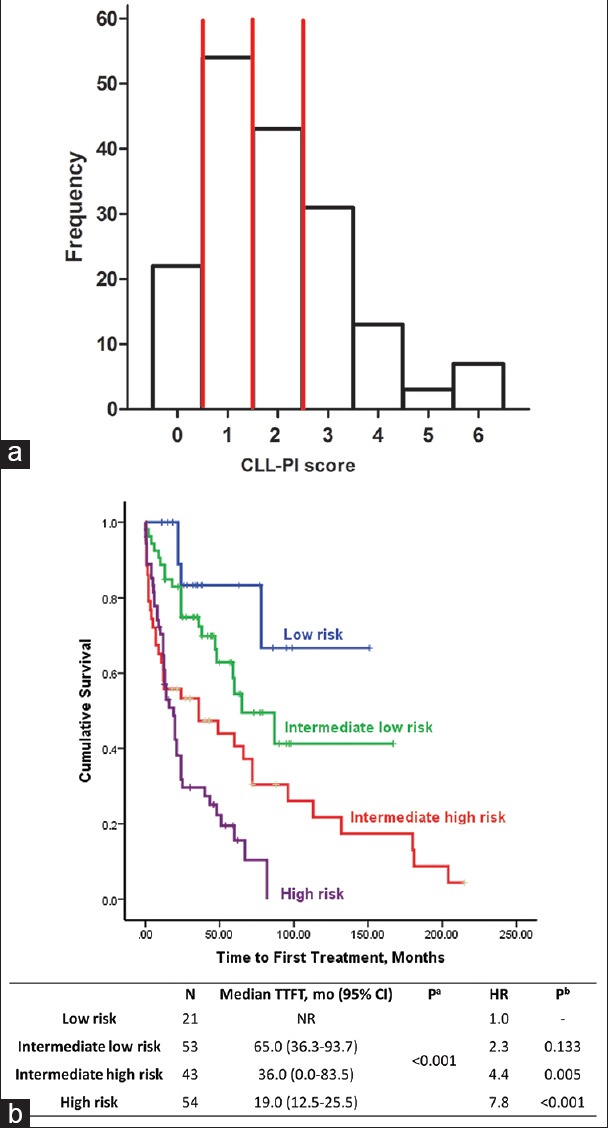
Chronic lymphocytic leukemia prognostic index. (a) Histogram of score point distribution according to chronic lymphocytic leukemia-prognostic index in the cohort of 173 patients. Vertical red lines show the positions of cut points splitting sample in four risk groups. (b) Kaplan-Meier curves for time to first treatment according to chronic lymphocytic leukemia-prognostic index.
Kaplan–Meier plots of the four risk groups are shown in Figure 2b. The 3-variable CLL-PI had good discriminatory power for TTFT in the cohort of 173 CLL patients. In particular, 22 of the patients (Score 0) were at low risk, 31% of patients (Score 1) were at intermediate-low risk, 25% of patients (Score 2) were at intermediate-high risk, and 31% of patients (Score 3–6) were at high risk. Estimated treatment-free rates in low-, intermediate-low-, intermediate-high-, and high-risk groups were 83%, 72%, 47%, and 30% at 3 years, respectively [Table 4]. To show the combination of predictive variables in each patient and in each group, we used a heat-map plot [Figure 3].
Table 4.
Cumulative probability of receiving treatment at 3 years from diagnosis for chronic lymphocytic leukemia population, cohort, Rai risk groups, IGHV/TP53 score, and CLL-PI
| Characteristics | n (%) | Median (months) | 3 years (%) | P* | HR | 95% CI | P† |
|---|---|---|---|---|---|---|---|
| Population | 406 | 38.0 | 51 | 0.443 | 1.0 | – | 0.451 |
| Cohort | 173 | 48.0 | 54 | 1.1 | 0.9–1.4 | ||
| Rai risk group | |||||||
| Low (0) | 59 (15) | NR | 83 | <0.001 | 1.0 | – | – |
| Intermediate (I–II) | 208 (52) | 51.0 | 56 | 2.4 | 1.5–3.7 | <0.001 | |
| High (III–IV) | 134 (33) | 12.0 | 30 | 4.2 | 2.7–6.6 | <0.001 | |
| Rai low risk | |||||||
| 0 | 21 (81) | NR | 83 | <0.001 | 1.0 | – | – |
| 2 | 4 (15) | 60.0 | 100 | 1.0 | 0.9–10.8 | 1.000 | |
| 4 | 1 (4) | 0.25 | 0 | 1.0 | 0–26873.8 | 1.000 | |
| Rai intermediate risk | 0.001 | ||||||
| 0 | 53 (61) | 65.0 | 72 | 1.0 | – | ||
| 2 | 31 (36) | 20.0 | 38 | 3.0 | 1.6–5.4 | <0.001 | |
| 4 | 3 (3) | NR | 67 | 2.0 | 0.3–14.9 | 0.511 | |
| Rai high risk | |||||||
| 0 | 39 (67) | 24.0 | 43 | 0.067 | 1.0 | – | – |
| 2 | 12 (21) | 12.0 | 25 | 1.7 | 0.8–3.5 | 0.155 | |
| 4 | 7 (12) | 12.0 | 0 | 2.5 | 1.0–5.9 | 0.040 | |
| CLL-PI class | |||||||
| Low | 22 (13) | NR | 83 | <0.001 | 1.0 | – | – |
| Intermediate low | 54 (31) | 65.0 | 72 | 2.3 | 0.8–6.6 | 0.133 | |
| Intermeidate high | 43 (25) | 36.0 | 47 | 4.4 | 1.6–12.6 | 0.005 | |
| High | 54 (31) | 19.0 | 30 | 7.8 | 2.8–21.8 | <0.001 |
*P value calculated by log-rank test for differences in variables; †P value calculated by Cox multivariate analysis for differences in variables. CI: Confidence interval; FISH: Fluorescence in situ; HR: Hazard ratio; CLL-PI: Chronic lymphocytic leukemia prognostic index; IGHV: Immunoglobulin heavy chain variable region.
Figure 3.
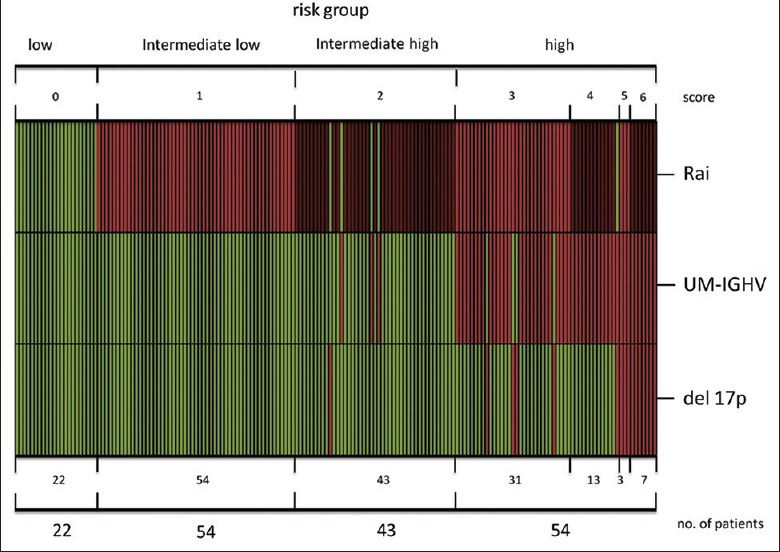
Heat-map of individual patient chronic lymphocytic leukemia-prognostic index scores. Columns refer to individual patients; rows refer to predictors.
Comparison of the chronic lymphocytic leukemia-PI system with the Rai staging system
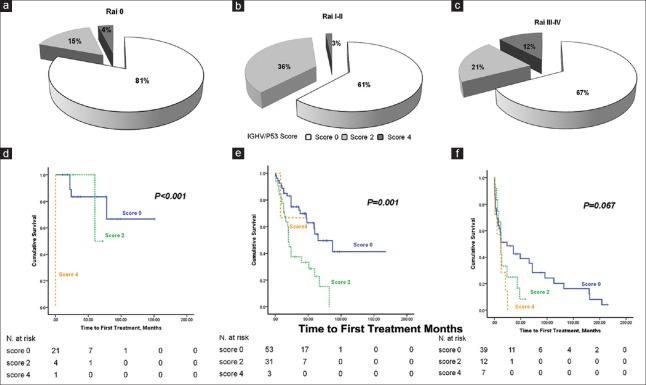
Compared with the Rai staging system, the CLL-PI integrated the prognostic value of IGHV mutational status and 17p deletion, so we further investigated the effect of IGHV mutational status and 17p– on different Rai classes of risk. Because the prognostic scores for U-IGHV and 17p– were both 2 points, the IGHV/17p score varied from 0 to 4, and the IGHV/17p score distribution within the Rai class of risk is shown in Table 4 and Figure 4a–4c. We observed that percentages of Score 4 patients significantly increased to 12.1% in the Rai high-risk (Rai III–IV) group. Then, we assessed the prognostic impact of the IGHV/17p score and found that it could predict the TTFT for the Rai low-risk [Table 4 and Figure 4d] and intermediate-risk [Table 4 and Figure 4e] groups and had marginal significance in predicting the TTFT for the Rai high-risk group [Table 4 and Figure 4f]. Table 4 also reports the median and 3-year TTFT for the above-mentioned groups.
Figure 4.
IGHV/17p Score and Survival Curves for Patient Subgroups. The IGHV/17p score distribution within (a) the Rai low-risk group, (b) intermediate-risk group and (c) high-risk group. Kaplan-Meier curves for TTFT according to IGHV/17p Score for (d) the Rai low-risk group, (e) intermediate-risk group and (f) high-risk group.
Due to the presence of U-IGHV and 17p−, the stratification of Rai risk groups was modified. Thus, in the Rai low-risk group, 4 patients (14.8%) were reclassified to the CLL-PI intermediate-high-risk group, and one patient (3.7%) was assigned to the CLL-PI high-risk group. In the Rai intermediate-risk group, 34 patients (38.6%) were reclassified to the CLL-PI high-risk group, and in the Rai high-risk group, 39 patients (67.2%) were reclassified to the CLL-PI intermediate-high-risk group [Table 1].
Discussion
CLL is described as exhibiting heterogeneous clinical courses; the disease can present in aggressive and indolent forms.[17] Traditional staging systems, such as those developed by Rai and Binet, are based on physical examination and simple laboratory parameters and are known as the basis for evaluation of prognosis in CLL. However, significant heterogeneity still remains within each category, and these systems are not robust enough to predict the risk of progression in patients at early stages.[4] They are still useful for estimating treatment in patients at late stages, and they use anemia and thrombocytopenia as markers for discriminating between groups, as well as indications for therapy. In our study, approximately 67.2% of patients belonging to a high-risk group under the Rai system (Rai III–IV) were reclassified to intermediate-high risk because patients with a stable level of hemoglobin or platelet count should be monitored without therapy unless they have evidence of progressive bone marrow failure, as manifested by worsening of anemia or thrombocytopenia.
In the past few decades, a number of new prognostic markers have been identified to discriminate between stable and progressive forms of the disease at the early-stage, such as serum-based markers,[18,19] IGHV gene mutational status,[6] flow cytometry-based markers,[20,21,22] and cytogenetic aberrations.[5] More recently, several novel gene mutations have been identified using next-generation sequencing approaches, including NOTCH1, MYD88, SF3B1, NRAS, KRAS, and others.[23,24] Some RNA-based markers have also been shown to be associated with clinical outcomes.[25,26]
Combining these markers in prognostic indices improves the accuracy of the TTFT or survival prediction.[27,28] Such prognostic models allow clinicians to apply the collective results of prognostic tests and to develop risk-adapted therapies. For patients with high risk of progress, earlier treatment may be necessary. Therefore, the aim of this study was to identify markers that have independent prognostic value among routine clinical assays, particularly in an Asian population. In our cohort, we demonstrated that apart from the Rai staging system, 17p deletion and IGHV mutational status were reliable independent predictors for TTFT. Subsequently, we defined the CLL-PI by combining only these three variables together and identifying four classes of risk for TTFT.
13q− has been shown to be the most favorable prognosticator when compared with 11q−, 17p−, and trisomy 12,[29] but in our cohort, 13q deletion was not associated with longer TTFT. Interestingly, it has been shown that CLL patients with a larger deletion of 13q have a shorter TTFT.[30] Trisomy 12 has been connected with a good response to treatment; thus, no association has been found with TTFT.[29] Further, the deletion of 11q is associated with younger age at diagnosis and an inferior outcome.[5] According to our analyses, patients harboring 11q− received treatment earlier in the univariate analysis, but 11q− failed to predict treatment independently in multivariate analyses.
This analysis has several strengths. First, the population was unselected and untreated at diagnosis, and the follow-up was consistent. Second, this analysis incorporates both a traditional staging system and novel genetic abnormalities to identify markers independently associated with TTFT. Third, this analysis highlights the importance of understanding the real significance of several prognostic factors taken together. Parameters analyzed in this index are objective and readily available in laboratory assays; subjective features, such as the size of lymph nodes, are not included, unlike in other studies.[28] Moreover, this study was performed in an Asian cohort and of predictive value in Asian patients with CLL since the epidemiologic and genetic characteristics between the Caucasians and Asians are quite different.[11,23]
Nevertheless, our study also presents several limitations. First, it was a single-center study of a referral population, and the patients included in this study had a higher proportion of advanced cases compared with general-practice CLL. Second, the CLL-PI has not been evaluated in an independent validation population, preferably in the context of large, randomized clinical trials. Moreover, these newly discovered novel gene mutations, such as NOTCH1, SF3B1, and BIRC3,[23] have been reported to be associated with disease progression and may have critical role in predicting the TTFT,[23,31] but they are not routinely evaluated in our clinical laboratory and were therefore not included in the development of this prognostic index.
In summary, we developed a weighted, integrated CLL-PI which combines the most important genetic prognostic markers (IGHV mutation status, 17p deletion) with traditional clinical staging. This newly modified PI could be used to discriminate among groups and may help predict the TTFT and prognosis of patients with CLL.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
This work was supported by grants from the National Natural Science Foundation of China (No. 81370632, 81200395), the National Science and Technology Supporting Program (No. 2014BAI09B12), the Fundamental Application and Advanced Technology Research Program of Tianjin (No. 15JCYBJC27900), and the National Public Health Grand Research Foundation (No. 201202017).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yi Cui
References
- 1.Dighiero G. Unsolved issues in CLL biology and management. Leukemia. 2003;17:2385–91. doi: 10.1038/sj.leu.2403154. doi:10.1038/sj.leu.2403154. [DOI] [PubMed] [Google Scholar]
- 2.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–34. doi: 10.1182/blood-2016-08-737650. [DOI] [PubMed] [Google Scholar]
- 3.Binet JL, Auquier A, Dighiero G, Chastang C, Piguet H, Goasguen J, et al. Anew prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48:198–206. doi: 10.1002/1097-0142(19810701)48:1<198::aid-cncr2820480131>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 4.Parker TL, Strout MP. Chronic lymphocytic leukemia: Prognostic factors and impact on treatment. Discov Med. 2011;11:115–23. [PubMed] [Google Scholar]
- 5.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. doi:10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 6.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 7.Kröber A, Seiler T, Benner A, Bullinger L, Brückle E, Lichter P, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–6. [PubMed] [Google Scholar]
- 8.Gladstone DE, Blackford A, Cho E, Swinnen L, Kasamon Y, Gocke CD, et al. The importance of IGHV mutational status in del(11q) and del(17p) chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2012;12:132–7. doi: 10.1016/j.clml.2011.12.005. doi:10.1016/j.clml.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenquist R, Cortese D, Bhoi S, Mansouri L, Gunnarsson R. Prognostic markers and their clinical applicability in chronic lymphocytic leukemia: Where do we stand?Leuk Lymphoma. 2013;54:2351–64. doi: 10.3109/10428194.2013.783913. doi:10.3109/10428194.2013.783913. [DOI] [PubMed] [Google Scholar]
- 10.Dores GM, Anderson WF, Curtis RE, Landgren O, Ostroumova E, Bluhm EC, et al. Chronic lymphocytic leukaemia and small lymphocytic lymphoma: Overview of the descriptive epidemiology. Br J Haematol. 2007;139:809–19. doi: 10.1111/j.1365-2141.2007.06856.x. doi:10.1111/j.1365-2141.2007.06856.x. [DOI] [PubMed] [Google Scholar]
- 11.Mak V, Ip D, Mang O, Dalal C, Huang S, Gerrie A, et al. Preservation of lower incidence of chronic lymphocytic leukemia in Chinese residents in British Columbia: A 26-year survey from 1983 to 2008. Leuk Lymphoma. 2014;55:824–7. doi: 10.3109/10428194.2013.827785. doi:10.3109/10428194.2013.827785. [DOI] [PubMed] [Google Scholar]
- 12.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, et al. The World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues. Report of the Clinical Advisory Committee Meeting, Airlie House, Virginia, November, 1997. Ann Oncol. 1999;10:1419–32. doi: 10.1023/a:1008375931236. [DOI] [PubMed] [Google Scholar]
- 13.An G, Xu Y, Shi L, Shizhen Z, Deng S, Xie Z, et al. Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica. 2014;99:353–9. doi: 10.3324/haematol.2013.088211. doi:10.3324/haematol.2013.088211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An G, Li Z, Tai YT, Acharya C, Li Q, Qin X, et al. The impact of clone size on the prognostic value of chromosome aberrations by fluorescence in situ hybridization in multiple myeloma. Clin Cancer Res. 2015;21:2148–56. doi: 10.1158/1078-0432.CCR-14-2576. doi:10.1158/1078-0432.CCR-14-2576. [DOI] [PubMed] [Google Scholar]
- 15.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 16.Terrin L, Trentin L, Degan M, Corradini I, Bertorelle R, Carli P, et al. Telomerase expression in B-cell chronic lymphocytic leukemia predicts survival and delineates subgroups of patients with the same igVH mutation status and different outcome. Leukemia. 2007;21:965–72. doi: 10.1038/sj.leu.2404607. doi:10.1038/sj.leu.2404607. [DOI] [PubMed] [Google Scholar]
- 17.MacIntyre DA, Jiménez B, Lewintre EJ, Martín CR, Schäfer H, Ballesteros CG, et al. Serum metabolome analysis by 1H-NMR reveals differences between chronic lymphocytic leukaemia molecular subgroups. Leukemia. 2010;24:788–97. doi: 10.1038/leu.2009.295. doi:10.1038/leu.2009.295. [DOI] [PubMed] [Google Scholar]
- 18.Lee JS, Dixon DO, Kantarjian HM, Keating MJ, Talpaz M. Prognosis of chronic lymphocytic leukemia: A multivariate regression analysis of 325 untreated patients. Blood. 1987;69:929–36. [PubMed] [Google Scholar]
- 19.Hallek M, Wanders L, Ostwald M, Busch R, Senekowitsch R, Stern S, et al. Serum beta(2)-microglobulin and serum thymidine kinase are independent predictors of progression-free survival in chronic lymphocytic leukemia and immunocytoma. Leuk Lymphoma. 1996;22:439–47. doi: 10.3109/10428199609054782. doi:10.3109/10428199609054782. [DOI] [PubMed] [Google Scholar]
- 20.Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–75. doi: 10.1056/NEJMoa023143. doi:10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 21.Hamblin TJ, Orchard JA, Ibbotson RE, Davis Z, Thomas PW, Stevenson FK, et al. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002;99:1023–9. doi: 10.1182/blood.v99.3.1023. doi:10.1182/blood.V99.3.1023. [DOI] [PubMed] [Google Scholar]
- 22.Rassenti LZ, Jain S, Keating MJ, Wierda WG, Grever MR, Byrd JC, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–30. doi: 10.1182/blood-2007-05-092882. doi:10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121:1403–12. doi: 10.1182/blood-2012-09-458265. doi:10.1182/blood-2012-09-458265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. doi:10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–47. doi: 10.1084/jem.194.11.1639. doi:10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. AmicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. doi:10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 27.Visentin A, Facco M, Frezzato F, Castelli M, Trimarco V, Martini V, et al. Integrated CLL Scoring System, a new and simple index to predict time to treatment and overall survival in patients with chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2015;15:612–20. doi: 10.1016/j.clml.2015.06.001. e1-5. doi:10.1016/j.clml.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Wierda WG, O'Brien S, Wang X, Faderl S, Ferrajoli A, Do KA, et al. Multivariable model for time to first treatment in patients with chronic lymphocytic leukemia. J Clin Oncol. 2011;29:4088–95. doi: 10.1200/JCO.2010.33.9002. doi:10.1200/JCO.2010.33.9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunnarsson R, Mansouri L, Isaksson A, Göransson H, Cahill N, Jansson M, et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica. 2011;96:1161–9. doi: 10.3324/haematol.2010.039768. doi:10.3324/haematol.2010.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68:1012–21. doi: 10.1158/0008-5472.CAN-07-3105. doi:10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 31.Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: Association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–8. doi: 10.1182/blood-2011-08-373159. doi:10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan-Meier curves for TTFT and OS of the 406 patients with CLL. (a) Kaplan-Meier curves for TTFT of the enrolled patients. (b) Kaplan-Meier curves for OS of the patients.