Abstract
Exploitation of the heterosis of hybrid rice has shown great success in the improvement of rice yields. However, few genotypes exhibit strong restoration ability as effective restorers of cytoplasmic male sterility (CMS) in the development of hybrid rice. In this study, we developed a platform for the breeding by design of CMS restorer lines based on a library of chromosomal single segment substitution lines (SSSLs) in the Huajingxian74 (HJX74) genetic background. The target genes for breeding by design, Rf34 and Rf44, which are associated with a strong restoration ability, and gs3, gw8, Wxg1 and Alk, which are associated with good grain quality, were selected from the HJX74 SSSL library. Through pyramiding of the target genes, a restorer line, H121R, was developed. The H121R line was then improved regarding blast resistance by pyramiding of the qBLAST11 gene. Hence, a new restorer line with blast resistance, H131R, was developed. The platform involving the Rf34 and Rf44 restorer genes would be used for the continuous improvement of restorer lines through breeding by design in rice.
Keywords: cytoplasmic male sterility, restorer, single segment substitution line, breeding by design, hybrid rice
Introduction
Cytoplasmic male sterility (CMS) is the foundation for the exploitation of the heterosis of hybrid rice (Yuan and Tang 1999). The male sterility of CMS lines can be restored by nuclear-encoded restorer of fertility (Rf) genes in rice (Chen and Liu 2013). For gametophytic CMS, Chinsurah Boro II (indica) cytoplasm with Taichung 65 (japonica) nucleus (BT-CMS) is restored by the Rf1a or Rf1b gene on chromosome 10, and red-awned wild rice (Oryza rufipogon) cytoplasm with Liantangzao (indica) necleus (HL-CMS) is restored by Rf5, which is another Rf1a allele (Akagi et al. 2004, Hu et al. 2012, Kazama and Toriyama 2003, Komori et al. 2004, Liu et al. 2004, Wang et al. 2006). Sporophytic CMS, including wild abortive rice (Oryza rufipogon) cytoplasm with Eejiunan 1 (indica) nucleus (WA-CMS), dwarf abortive rice (Oryza rufipogon) cytoplasm with Xieqingzao (indica) nucleus (DA-CMS) and Yegong (indica landrace) cytoplasm with BII44-5 (indica) nucleus (YA-CMS) types, is restored by the Rf3 and Rf4 genes, located on chromosomes 1 and 10, respectively (Dai et al. 2015, Suresh et al. 2012, Yao et al. 1997, Zhang et al. 1997, 2002). Recently, the WA352 gene, which controls WA-CMS, and the Rf4 gene, which encodes a PPR protein, were cloned (Kazama and Toriyama 2014, Luo et al. 2013, Tang et al. 2014). CMS-based hybrid seed technology uses a three-line system that consists of a CMS line (A line), a maintainer (B line), and a restorer (R line) to produce F1 seeds. Since China pioneered hybrid rice production in the 1970s, the yield of hybrid rice has been increased by more than 20% compared with conventional varieties and now accounts for more than half of the annual rice planting area in China (Cheng et al. 2007, Huang et al. 2014). However, few genotypes exhibit a strong restoration ability as effective restorers for CMS in the development of hybrid rice (Sharma et al. 2012, Singh et al. 2012). Therefore, the development of elite restorers is an important aim of breeding programs for hybrid rice production.
Chromosomal single segment substitution lines (SSSLs), which carry a particular chromosomal segment from a donor in the genetic background of a recipient, eliminate background noise to a large extent and are powerful tools for the genetic analysis of quantitative trait loci (QTLs) (Ebitani et al. 2005, Kubo et al. 2002, Xi et al. 2006, Zhang et al. 2004). We have constructed a library of 1,123 SSSLs in rice using Huajingxian74 (HJX74), an elite indica variety from south China, as the recipient and 26 genetically diverse accessions collected worldwide as donors. Each SSSL in the library has only one chromosomal segment from a donor in the HJX74 genetic background (Zhang et al. 2004). These SSSLs have been employed to detect and clone QTLs for complex traits (He et al. 2005, Liu et al. 2010, Naeem et al. 2013, Wang et al. 2012, Zhang et al. 2012, Zhao et al. 2007), to assess allelic variations at loci of interest (Cai et al. 2013, Teng et al. 2012, Zeng et al. 2006), and to analyze gene by gene and gene by environment interactions (Chen et al. 2014, Liu et al. 2008, 2012, Zhu et al. 2015). To assess allelic variations at the Rf3 and Rf4 loci, 57 SSSLs carrying one of the loci in the substituted segments were selected from the library. Four alleles were identified at both loci in the set of SSSLs, which were designated Rf31, Rf32, Rf33 and Rf34 and Rf41, Rf42, Rf43 and Rf44, respectively, ranging from weak to strong in terms of their restoration ability. The HJX74 recipient harbors the Rf34 allele, with the strongest restoration ability, at the Rf3 locus, but a weaker allele at the Rf4 locus. One SSSL, W23-19-06-06-11, was found to carry the Rf44 allele, with the strongest restoration ability, in the substituted segment from the Lemont donor (Cai et al. 2013).
Peleman and van der Voort (2003) proposed the concept of ‘breeding by design’. This goal can be reached by following a three-step approach: mapping loci involved in all agronomically relevant traits, assessment of the allelic variation at those loci, and breeding by design. We proposed a strategy to practice breeding by design using the HJX74 SSSL library as a platform for rice breeding (Xi et al. 2006). The SSSL W14-18-6-10-1, which carries the purple pericarp gene Pb in the substituted segment on chromosome 4 from Lianjian33, was selected from the SSSL library. After comprehensive evaluation, the SSSL became a new variety designated Huaxiaohei1, which was released in 2005. Furthermore, a pyramiding line with three substituted segments in an HJX74 genetic background was designated Huabiao1 and released to farmers in 2009 (Dai et al. 2015). Because HJX74 is neither a CMS maintainer nor a CMS restorer, the HJX74 SSSL library cannot be directly used as a platform for the development of CMS lines and restorer lines. Recently, the platform was successfully improved to allow the development of CMS lines through breeding by design. Three isonuclear alloplasmic CMS lines with an HJX74 genetic background, designated W-H121A (WA type), D-H121A (DA type) and Y-H121A (YA type), and their maintainer lines were the first to be developed. The CMS lines were then improved through breeding by design to develop three new isonuclear alloplasmic CMS lines, designated D-H131A, W-H131A and Y-H131A, by pyramiding target genes from the HJX74 SSSL library (Dai et al. 2015). In this study, we developed a platform for the breeding by design of CMS restorer lines based on the HJX74 SSSL library. By using the Rf34 and Rf44 restorer genes, a restorer line, H121R with a strong restoration ability was developed in the HJX74 genetic background. The H121R line was then improved to develop an elite restorer line, H131R, with blast resistance. This work provides an example of conducting breeding by design of restorer lines based on the HJX74 SSSL library.
Materials and Methods
Plant materials
HXJ74, an elite variety that is planted widely in south China, is the recipient of the SSSLs and contains the Rf34 gene (Cai et al. 2013, Xi et al. 2006, Zhang et al. 2004). Three isonuclear alloplasmic CMS lines, W-H121A (with wild abortive cytoplasm), D-H121A (with dwarf wild abortive cytoplasm), and Y-H121A (with Yegong abortive cytoplasm), exhibit an HJX74 nuclear background, with the exception of non-functional rf3 and rf4 genes from a CMS line Xieqingzao A (Dai et al. 2015). Five SSSLs with an HXJ74 genetic background, W03-14-10-04-02, W08-15-08-28, W23-07-06-10-06, W23-07-06-05-02-02, and W23-19-06-06-11, were employed as the donors of target genes (Table 1).
Table 1.
Chromosomal substituted segments with target genes in the SSSLs
| SSSL | Donor | Chr. | Substitution segment | Interval of substituted segments (cM) | Target gene | Target trait |
|---|---|---|---|---|---|---|
| W08-15-08-28 | IR64 | 3 | RM2453-gs3-RM6146-RM3646-RM16 | 83.0–94.9 | gs3 | LG |
| W23-07-06-10-06 | Lemont | 6 | RM508-RM589-RM190(Wxg1)-RM204-RM402-Alk-RM539-RM541 | 1.4–68.5 | Wxg1 | MAAC |
| Alk | HGT | |||||
| W03-14-10-04-02 | Zhong4188 | 8 | RM256-RM5493-gw8-RM447 | 96.6–111.2 | gw8 | NG |
| W23-19-06-06-11 | Lemont | 10 | RM258-RM5373-Rf44-RM6100-RM25685 | 30.2–58.5 | Rf44 | SRF |
| W23-07-06-05-02-02 | Lemont | 11 | RM224-qBLAST-11-RM144 | 110.1–116.5 | qBLAST-11 | HRB |
LG, long grain; MAAC, medium apparent amylose content; HGT, high gelatinization temperature; NG, narrow grain; SRF, strong restoration of fertility; HRB, high resistance to blast.
All of the materials used in this study were grown at an experimental station of South China Agricultural University, Guangzhou (23°07′N, 113°15′E), China, in two cropping seasons from 2008 to 2014. The first cropping season was from late February to mid-July, and the second cropping season was from late July to mid-November. The seeds were sown in seed beds, and the seedlings were transplanted to fields. Seedlings were transplanted at a density of 16.7 cm × 32.4 cm, with one seedling per hill. The adopted field management procedures, including irrigation, fertilizer application and pest control, essentially followed normal agricultural practice.
Development of the restorer lines H121R and H131R
For development of the restorer line H121R, four SSSLs, W08-15-08-28 with the gs3 gene, W23-07-06-10-06 with the Wxg1 gene, W03-14-10-04-02 with the gw8 gene, and W23-19-06-06-11 with the Rf44 gene in their chromosomal substituted segments, were selected from the HXJ74 SSSL library (Table 1). W08-15-08-28 and W23-07-06-10-06 were first crossed, and 6 homozygous plants with gs3 and Wxg1 gene were selected from an F2 population of 100 plants by marker-assisted selection (MAS). The homozygotes with gs3 and Wxg1 gene were then crossed with the SSSL W03-14-10-04-02 with the gw8 gene, and the line H121 with gs3, Wxg1 and gw8 genes were obtained from the F3 population. To improve the fertility restoration, the H121 line was crossed with W23-19-06-06-11 with the Rf44 gene. Good quality of restorer line, H121R, was obtained, which carried the gs3, Wxg1 and gw8 genes from the H121 line and the Rf44 gene from W23-19-06-06-11 in the F3 population by MAS.
For improvement of blast resistance in the H121R restorer line, an SSSL, W23-07-06-05-02-02, was selected from the SSSL library. W23-07-06-05-02-02 carries the qBLAST-11 gene in their chromosomal substituted segments in the HXJ74 genetic background (Table 1). The H121R line was crossed with W23-07-06-05-02-02. The restorer line with blast resistance, H131R with the desirable homozygous alleles in all target genes out of about 1000 lines was obtained from the F3 population through MAS using the linked markers.
DNA extraction and marker assay
DNA was extracted from fresh young leaves using the CTAB method (Murray and Thompson 1980). Miniscale DNA extraction was carried out according to the procedure described by Zheng et al. (1995). The PCR profile used for amplification basically followed a previously described protocol (Cai et al. 2013). The PCR amplified products were analyzed via electrophoresis in 6% polyacrylamide denaturing gels and subjected to the silver staining procedure, as described by Li et al. (2006).
The target genes in the SSSLs were identified using SSR markers that are linkage with the genes (Table 1). The gs3 gene, conferring a long-grain trait (Fan et al. 2006), is located in the substituted segment of chromosome 3 in SSSL W08-15-08-28. The genes Wxg1, conferring a medium apparent amylose content (Teng et al. 2012), and Alk, conferring a high gelatinization temperature (Gao et al. 2003), are both located in the same substituted segment of chromosome 6 in W23-07-06-10-06. The gw8 gene, conferring a narrow-grain trait (Wang et al. 2012), is located in the substituted segment of chromosome 8 in W03-14-10-04-02. W23-19-06-06-11 carries the Rf44 gene, conferring a strong restoration ability, in the substituted segment of chromosome 10 (Cai et al. 2013). The qBLAST11 QTL, for resistance to blast, was mapped between the RM224 and RM144 markers in the substituted segment of chromosome 11 in W23-07-06-05-02-02 (Zhang et al. 2012) (Table 1).
Observation of pollen and spikelet fertility
The pollen and spikelet fertility of F1 hybrids in testcrosses were used to evaluate the fertility restoration ability of restorer lines. Mature anthers in spikelets were collected to determine pollen fertility. The pollen was stained with a 1% (m/v) I2-KI solution. The numbers of stainable and un-stainable pollen grains in each individual were counted under an optical microscope. Twenty plants were collected for pollen and spikelet fertility traits. All statistical analyses were performed using SPSS version 18 in IBM and GraphPad Prism 5 in GraphPad Software.
Assessment of agronomic traits and grain quality
Each of the traits was tested in 40 plants per line during the second cropping season in 2012. The grain traits of grain length, grain width and grain weight were evaluated as described previously (Fan et al. 2006). Tests of apparent amylose content (AAC) and gelatinization temperature (GT) which was indirectly estimated via alkali spreading value (ASV), were conducted following procedures described elsewhere (Tan et al. 1999, Teng et al. 2012). All statistical analyses were performed using IBM SPSS version 18.
Evaluation of resistance to blast
Blast inoculation and the evaluation of resistance were performed followed the methods described by Zhu et al. (2012). Three rows of seedlings, containing 20 plants each, were planted in a greenhouse in 30 cm × 20 cm × 5 cm trays. Inoculation of the lines was performed at the 3.5–4 leaf stage using a set of 10 blast isolates selected to show a diverse spectrum of virulence. A 20 ml spore suspension (105 spores/ml) was applied to each tray using an airbrush connected to a source of compressed air. Each isolate-host combination was assessed in three replications. After inoculation, the trays were maintained in the dark for 24 h at a relative humidity of 95–100% and a temperature of 25°C, after which they were transferred to a greenhouse where the ambient temperature was maintained at 25–28°C. Six days later, disease symptoms were evaluated on a standard scale of 0–9 based on the type and size of lesions, as described by the International Rice Research Institute (IRRI 1996). Rice plants exhibiting reactions with a score of 0–3 were considered resistant, and those showing reactions with a score of 4–9 were categorized as susceptible.
Results
Development of the H121R restorer line, with a strong restoration ability and good grain quality, in the HJX74 genetic background
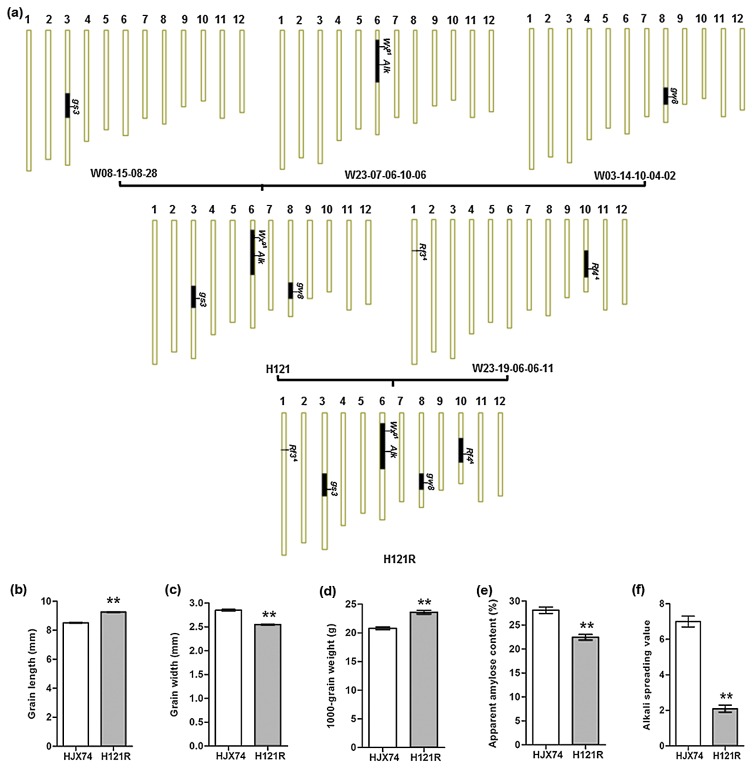
To improve the grain quality of HJX74 plants, four genes, gs3, gw8, Wxg1 and Alk, in three SSSLs were pyramided under the HJX74 genetic background. The pyramid line with the four genes (gs3, gw8, Wxg1 and Alk) was designated H121 (Fig. 1a). To improve the restoration ability of the H121 line, the SSSL W23-19-06-06-11 carrying Rf44 gene was then crossed with the H121 line. The pyramid line, carrying the gs3, gw8, Wxg1 and Alk genes from H121, the Rf44 gene from W23-19-06-06-11, and the Rf34 gene from the HJX74 genetic background, was designated H121R (Fig. 1a).
Fig. 1.
Development of the H121R restorer line. (a) Development of the H121R restorer line based on the HJX74 SSSL library. H121 was developed from the cross of W08-15-08-28/W23-07-06-10-06//W03-14-10-04-02. H121R was then developed from the cross of H121/W23-19-06-06-11. The vertical bars are a graphical representation of the chromosomes. Black regions represent substitute segments with target genes, and white regions represent the HJX74 genetic background. (b) Grain length. (c) Grain width. (d) 1000-grain weight. (e) Apparent amylase content. (f) Alkali spreading value. Error bars represent SD. **, significant at the 0.01 probability level by t test.
To evaluate the effect of breeding by design, the target traits controlled by the target genes in the H121R line were tested. As expected, the grain shape of H121R was long-slender, with a grain length of 9.4 mm and a grain width of 2.5 mm, whereas HJX74 presented a grain length of 8.6 mm and a grain width of 2.8 mm (Fig. 1b, 1c). The 1000-grain weight of H121R was 23.6 g, which was greater than the value of 20.8 g recorded in HJX74 (Fig. 1d). The AAC of H121R was 21.4%, which corresponded to the intermediate class and was significantly lower than the value of 28.5% obtained in HJX74 (Fig. 1e). Another trait related to eating and cooking, GT, was also greatly increased in H121R compared with HJX74 (Fig. 1f). However, there were no significant differences detected in the other main agronomic traits between H121R and HJX74, including the days to heading, plant height, number of panicles, panicle length, and filled grain number per panicle (Table 2).
Table 2.
Comparison of some agronomic traits in HJX74, H121R and H131R
| Trait | HJX74 | H121R | H131R |
|---|---|---|---|
| Plant height (cm) | 89.7 ± 1.5 | 90.3 ± 1.3 ns | 89.2 ± 0.8 ns |
| Heading date (d) | 77.5 ± 0.6 | 79.0 ± 1.5 ns | 78.2 ± 0.7 ns |
| No. of panicles | 7.7 ± 0.6 | 8.1 ± 1.6 ns | 7.9 ± 1.3 ns |
| Panicle length (cm) | 22.7 ± 1.7 | 23.3 ± 1.4 ns | 23.1 ± 1.6 ns |
| Filled grain number per panicle | 151.9 ± 4.7 | 143.7 ± 6.6 ns | 147.5 ± 5.3 ns |
“ns” indicates no significant difference from the control HJX74 at p < 0.05.
Improvement of the blast resistance of the H121R restorer line
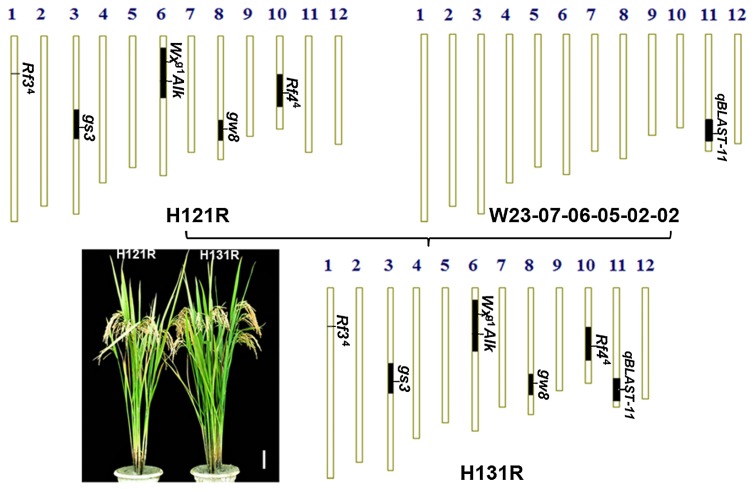
To improve the blast resistance of H121R, the SSSL W23-07-06-05-02-02 carrying blast resistance gene qBLAST11 was crossed with the H121R line. The obtained pyramid line, containing the gs3, gw8, Wxg1, Alk, and Rf44 genes from H121R, the qBLAST11 gene from W23-07-06-05-02-02, and the Rf34 gene from the HJX74 genetic background, was designated H131R (Fig. 2).
Fig. 2.
Improvement of the H121R restorer line regarding blast resistance. H131R was developed from the cross of H121R/W23-07-06-05-02-02. The vertical bars are a graphical representation of the chromosomes. Black regions represent substitute segments with target genes, and white regions represent the HJX74 genetic background.
To evaluate the reaction of the plants to blast in uniform blast nursery, HJX74, H121R and H131R were tested against 10 representative blast isolates collected in Guangdong province. The results indicated that the frequency of resistance in H131R was 100%, which was much higher than the frequency of 10.0% recorded in HJX74 and H121R (Table 3). However, there were no significant differences detected in the other main agronomic traits, including the days to heading, plant height, number of panicles, panicle length and filled grain number per panicle among H131R, H121R and HJX74 (Table 2).
Table 3.
Resistance to blast in HJX74, H121R, H131R and F1 hybrid of W-H121A/H131R
| Isolate | Disease score | |||
|---|---|---|---|---|
|
| ||||
| HJX74 | H121R | H131R | W-H121A/H131R | |
| 04-94 | 1(R) | 1(R) | 1(R) | 1(R) |
| Y98-66 | 9(S) | 9(S) | 1(R) | 1(R) |
| 97-322 | 9(S) | 7(S) | 1(R) | 1(R) |
| W06-2a | 8(S) | 9(S) | 1(R) | 1(R) |
| 93-286a | 9(S) | 8(S) | 1(R) | 1(R) |
| 07-4a | 7(S) | 7(S) | 1(R) | 2(R) |
| 06-141a | 9(S) | 8(S) | 1(R) | 1(R) |
| 93-203a | 5(S) | 5(S) | 1(R) | 1(R) |
| 00-193 | 9(S) | 8(S) | 1(R) | 1(R) |
| 00-173a | 7(S) | 8(S) | 1(R) | 1(R) |
| No. of infected isolates | 9 | 9 | 0 | 0 |
| Resistance frequency (%) | 10 | 10 | 100 | 100 |
| Resistance evaluation | Highly susceptible | Highly susceptible | Highly resistant | Highly resistant |
R or S is resistant or susceptible, respectively.
Restoration ability of the two restorer lines
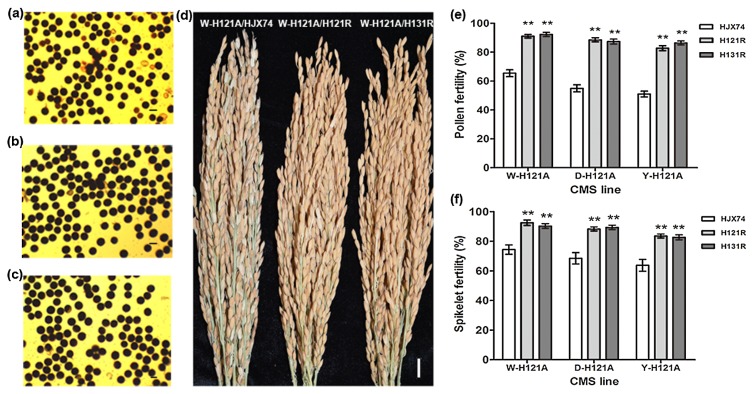
To assess the restoration ability of the restorer lines, H121R, H131R and HJX74 were test crossed with the W-H121A, D-H121A and Y-H121A CMS lines, respectively. The restoration ability of the restorer lines H121R and H131R was much stronger than that of HJX74 in terms of both pollen fertility and in spikelet fertility (Fig. 3a–3d). The pollen fertility and spikelet fertility of the F1 hybrids derived from H121R and the CMS lines W-H121A, D-H121A and Y-H121A were 95.4% and 93.8%; 91.9% and 91.4%; and 88.9% and 85.6%, respectively. Similarly, the pollen fertility and spikelet fertility of the F1 hybrids derived from H131R and the CMS lines W-H121A, D-H121A and Y-H121A were 94.3% and 95.8%; 92.5% and 90.7%; and 87.8% and 86.4%, respectively. The pollen fertility and spikelet fertility in the F1 hybrids derived from HJX74 and the CMS lines W-H121A, D-H121A and Y-H121A, which were used as controls, were only 68.5% and 72.7%; 57.3% and 66.9%; and 48.6% and 62.1%, respectively. These results indicate that the goal of breeding to achieve restoration of fertility in the restorer lines H121R and H131R was achieved through pyramiding of the target genes in the platform.
Fig. 3.
Restoration ability of the restorer lines H121R and H131R. (a) Pollen fertility in the F1 hybrid of W-H121A/HJX74. Scale bar, 50 μm. (b) Pollen fertility in the F1 hybrid of W-H121A/H121R. Scale bar, 50 μm. (c) Pollen fertility in the F1 hybrid of W-H121A/H131R. Scale bar, 50 μm. (d) Spikelet fertility in the F1 hybrids of W-H121A/HJX74, W-H121A/H121R, and W-H121A/H131R. Scale bar, 1 cm. (e) Pollen fertility of F1 hybrids from the crosses between the restorer lines HJX74, H121R and H131R and the CMS lines W-H121A, D-H121A, and Y-H121A, respectively. (f) Spikelet fertility of F1 hybrids from the crosses between the restorer lines HJX74, H121R and H131R and the CMS lines W-H121A, D-H121A, and Y-H121A, respectively. Error bars represent SD. **, significant at the 0.01 probability level by t test.
Discussion
Following the strategy of breeding by design proposed by Peleman and van der Voort (2003), we developed inbred varieties and CMS lines under the platform of HJX74 SSSL library in rice (Dai et al. 2015). In this study, the SSSL library was successfully improved with the Rf34 and Rf44 genes into a platform for developing restorer lines. These results indicate that the HJX74 SSSL library is a powerful platform for breeding by design in rice, not only for inbred varieties but also for CMS lines, maintainers and restorers in hybrid rice. As a platform for breeding by design, the HJX74 SSSL library has several advantages. First, HJX74, the recipient parent of the SSSL library, is an elite variety that has been widely planted in south China in the past decade. Second, the substituted segments in the SSSL library cover the entire rice genome, with more than 18 equivalents of the rice genome. Third, the substituted segments in the library come from 26 genetically diverse donors. Fourth, each of the SSSLs shares the same HJX74 genetic background, with only one substituted segment from a donor (Xi et al. 2006, Zhang et al. 2004). Therefore, the three-steps involved in breeding by design proposed by Peleman and van der Voort (2003) can be conducted using the platform of the HJX74 SSSL library (Dai et al. 2015).
Since China pioneered hybrid rice production in the 1970s, the sporophytic CMS system has been the most important system employed in hybrid rice development, both in China and around the world (Cheng et al. 2007, Yuan and Tang 1999). Sporophytic CMS, including the WA, DA and YA types, is controlled by the WA352 gene (Luo et al. 2013). The restoration of fertility in sporophytic CMS lines is controlled by two restorer genes, Rf3 on chromosome 1 and Rf4 on chromosome 10 (Cai et al. 2014, Tang et al. 2014, Xie et al. 2002, Yao et al. 1997, Zhang et al. 1997, 2002). CMS-based hybrid seed technology uses a three-line system consisting of a CMS line (A line), a maintainer line (B line), and a restorer line (R line). The A line exhibits male-sterile cytoplasm and two non-functional nuclear restorer genes, rf3 and rf4. The B line displays normal fertile cytoplasm but contains the same nuclear genome as the A line. The R line possesses two functional nuclear restorer genes, Rf3 and Rf4. Therefore, the three-line system is basically a CMS/Rf system. Understanding CMS and fertility restoration in the three-line system lays a foundation for breeding by design of the “three lines” in hybrid rice. In this study, the HJX74 SSSL library was successfully developed into a platform for developing restorer lines through breeding by design. Taken together with the previous development of CMS lines and maintainer lines using the platform of the HJX74 SSSL library (Dai et al. 2015), the HJX74 SSSL library has become a platform for breeding by design for the “three lines” in hybrid rice. Employing this platform, a series of “three lines” can be designed according to the goals regarding improvement. The available “three lines” developed on the platform can be then improved by the use of target genes selected from the HJX74 SSSL library. Thus, various new superior versions of the “three lines” can be developed easily and effectively under the SSSL platform. It is expected that the sustainable improvement of these “three lines” will facilitate hybrid rice development.
Acknowledgements
This work was supported by grants from the major program of transgenic new variety breeding of China (2014ZX08009-037B) and from the National Natural Science Foundation of China (91435207 and U1031002).
Literature Cited
- Akagi, H., Nakamura, A., Yokozeki-Misono, Y., Inagaki, A., Takahashi, H., Mori, K. and Fujimura, T. (2004) Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108: 1449–1457. [DOI] [PubMed] [Google Scholar]
- Cai, J., Liao, Q., Dai, Z., Zhu, H., Zeng, R., Zhang, Z. and Zhang, G. (2013) Allelic differentiations and effects of the Rf3 and Rf4 genes on fertility restoration in rice with wild abortive cytoplasmic male sterility. Biol. Plant. 57: 274–280. [Google Scholar]
- Cai, J., Dai, Z., Zhu, H., Zeng, R., Zhang, Z., Ma, T. and Zhang, G. (2014) Comparative analysis of fertility restoration genes for WA, Y, and DA cytoplasmic male sterility in rice. Biol. Plant. 58: 241–246. [Google Scholar]
- Chen, J., Li, X., Cheng, C., Wang, Y., Qin, M., Zhu, H., Zeng, R., Fu, X., Liu, Z. and Zhang, G. (2014) Characterization of epistatic interaction of QTLs LH8 and EH3 controlling heading date in rice. Sci. Rep. 4: 4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. and Liu, Y. (2013) Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65: 579–606. [DOI] [PubMed] [Google Scholar]
- Cheng, S., Zhuang, J., Fan, Y., Du, J. and Cao, L. (2007) Progress in research and development on hybrid rice: a super-domesticate in China. Ann. Bot. 100: 959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Z., Lu, Q., Luan, X., Cai, J., Zhu, H., Liu, Z., Zeng, R., Zhang, Z., Wang, S., Zheng, L.et al. (2015) Development of a platform for breeding by design of CMS lines based on an SSSL library in rice (Oryza sativa L.). Euphytica 205: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K. and Yano, M. (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed. Sci. 55: 65–73. [Google Scholar]
- Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., Li, X. and Zhang, Q. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Gao, Z., Zeng, D., Cui, X., Zhou, Y., Yan, M., Huang, D., Li, J. and Qian, Q. (2003) Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci. China, C, Life Sci. 46: 661–668. [DOI] [PubMed] [Google Scholar]
- He, F., Xi, Z., Zeng, R., Talukdar, A. and Zhang, G. (2005) Identification of QTLs for plant height and its components by using single segment substitution lines in rice (Oryza sativa). Rice Sci. 12: 151–156. [Google Scholar]
- Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., Huang, Q., Ji, Y., Qin, X., Wan, L.et al. (2012) The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRRI (1996) Standard evaluation system for rice, 4th edn International Rice Research Institute, Manila, Philippines: p. 52. [Google Scholar]
- Kazama, T. and Toriyama, K. (2003) A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544: 99–102. [DOI] [PubMed] [Google Scholar]
- Kazama, T. and Toriyama, K. (2014) A fertility restorer gene, Rf4, widely used for hybrid rice breeding encodes a pentatricopeptide repeat protein. Rice 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori, T., Ohta, S., Murai, N., Takakura, Y., Kuraya, Y., Suzuki, S., Hiei, Y., Imaseki, H. and Nitta, N. (2004) Map-based cloning of a fertility restorer gene Rf-1 in rice (Oryza sativa L.). Plant J. 37: 315–325. [DOI] [PubMed] [Google Scholar]
- Kubo, T., Aida, Y., Nakamura, K., Tsunematsu, H., Doi, K. and Yoshimura, A. (2002) Reciprocal chromosome segment substitution series derived from japonica and indica cross of rice (Oryza sativa L.). Breed. Sci. 52: 319–325. [Google Scholar]
- Li, W., Zeng, R., Zhang, Z., Ding, X. and Zhang, G. (2006) Fine mapping of locus S-b for F1 pollen sterility in rice (Oryza sativa L.). Chin. Sci. Bull. 51: 675–680. [Google Scholar]
- Liu, G., Zhang, Z., Zhu, H., Zhao, F., Ding, X., Zeng, R., Li, W. and Zhang, G. (2008) Detection of QTLs with additive effects and additive-by-environment interaction effects on panicle number in rice (Oryza sativa L.) with single-segment substitution lines. Theor. Appl. Genet. 116: 923–931. [DOI] [PubMed] [Google Scholar]
- Liu, G., Zhu, H., Liu, S., Zeng, R., Zhang, Z., Li, W., Ding, X., Zhao, F. and Zhang, G. (2010) Unconditional and conditional QTL mapping for the developmental behavior of tiller number in rice (Oryza sativa L.). Genetica 138: 885–893. [DOI] [PubMed] [Google Scholar]
- Liu, G., Zhu, H., Zhang, G., Li, L. and Ye, G. (2012) Dynamic analysis of QTLs on tiller number in rice (Oryza sativa L.) with single segment substitution lines. Theor. Appl. Genet. 125: 143–153. [DOI] [PubMed] [Google Scholar]
- Liu, X., Xu, X., Tan, Y., Li, S., Hu, J., Huang, J., Yang, D., Li, Y. and Zhu, Y. (2004) Inheritance and molecular mapping of two fertility-restoring loci for Honglian gametophytic cytoplasmic male sterility in rice (Oryza sativa L.). Mol. Genet. Genomics 271: 586–594. [DOI] [PubMed] [Google Scholar]
- Luo, D., Xu, H., Liu, Z., Guo, J., Li, H., Chen, L., Fang, C., Zhang, Q., Bai, M., Yao, N.et al. (2013) A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 45: 573–577. [DOI] [PubMed] [Google Scholar]
- Murray, M. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem, M., Freed, S. and Zhang, G. (2013) Molecular genetic studies of heading date gene OsMADS50 by using single segment substitution lines in rice. Int. J. Agric. Biol. 15: 631–639. [Google Scholar]
- Peleman, J.D. and van der Voort, J.R. (2003) Breeding by design. Trends Plant Sci. 8: 330–334. [DOI] [PubMed] [Google Scholar]
- Sharma, S., Singh, S., Nandan, R. and Kumar, M. (2012) Identification of restorers and maintainers for CMS lines of rice (Oryza sativa L.). Indian J. Plant Genet. Resour. 25: 186–188. [Google Scholar]
- Singh, U., Lal, J. and Kumar, H. (2012) Isolation and evaluation of restorers and examining possibility of developing new version of CMS lines for upland rainfed rice hybrids. Environ. Ecol. 30: 872–876. [Google Scholar]
- Suresh, P.B., Srikanth, B., Kishore, V.H., Rao, I.S., Vemireddy, L.R., Dharika, N., Sundaram, R.M., Ramesha, M.S., Rao, K.R.S.S., Viraktamath, B.C.et al. (2012) Fine mapping of Rf3 and Rf4 fertility restorer loci of WA-CMS of rice (Oryza sativa L.) and validation of the developed marker system for identification of restorer lines. Euphytica 187: 421–435. [Google Scholar]
- Tan, Y., Li, J., Yu, S., Xing, Y., Xu, C. and Zhang, Q. (1999) The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid ‘Shanyou 63’. Theor. Appl. Genet. 99: 642–648. [DOI] [PubMed] [Google Scholar]
- Tang, H., Luo, D., Zhou, D., Zhang, Q., Tian, D., Zheng, X., Chen, L. and Liu, Y. (2014) The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol. Plant 7: 1497–1500. [DOI] [PubMed] [Google Scholar]
- Teng, B., Zeng, R., Wang, Y., Liu, Z., Zhang, Z., Zhu, H., Ding, X., Li, W. and Zhang, G. (2012) Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol. Breed. 30: 583–595. [Google Scholar]
- Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., Zeng, R., Zhu, H., Dong, G., Qian, Q.et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44: 950–954. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Zou, Y., Li, X., Zhang, Q., Chen, L., Wu, H., Su, D., Chen, Y., Guo, J., Luo, D.et al. (2006) Cytoplasmic male sterility of rice with Boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18: 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, Z., He, F., Zeng, R., Zhang, Z., Ding, X., Li, W. and Zhang, G. (2006) Development of a wide population of chromosome single-segment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.). Genome 49: 476–484. [DOI] [PubMed] [Google Scholar]
- Xie, J., Zhuang, J., Fan, Y., Tu, G., Xia, Y. and Zheng, K. (2002) Mapping of fertility-restoring genes with main effects and epistatic effects for CMS-DA in rice. J. Genet. Genomics 29: 616–621. [PubMed] [Google Scholar]
- Yao, F., Xu, C., Yu, S., Li, J., Gao, Y., Li, X. and Zhang, Q. (1997) Mapping and genetic analysis of two fertility restorer loci in the wild-abortive cytoplasmic male sterility system of rice (Oryza sativa L.). Euphytica 98: 183–187. [Google Scholar]
- Yuan, L. and Tang, C. (1999) Retrospect, current status and prospect of hybrid rice. Rice Sci. 4: 3–6. [Google Scholar]
- Zeng, R., Zhang, Z., He, F., Xi, Z., Talukdar, A., Shi, J., Qin, L., Huang, C. and Zhang, G. (2006) Identification of multiple alleles at the Wx locus and development of single segment substitution lines for the alleles in rice. Rice Sci. 13: 9–14. [Google Scholar]
- Zhang, G., Lu, Y., Bharaj, T.S., Virmani, S.S. and Huang, N. (1997) Mapping of the Rf-3 nuclear fertility-restoring gene for WA cytoplasmic male sterility in rice using RAPD and RFLP markers. Theor. Appl. Genet. 94: 27–33. [DOI] [PubMed] [Google Scholar]
- Zhang, G., Zeng, R., Zhang, Z., Ding, X., Li, W., Liu, G., He, F., Tulukdar, A., Huang, C., Xi, Z.et al. (2004) The construction of a library of single segment substitution lines in rice (Oryza sativa L.). Rice Genet. Newsl. 21: 85–87. [Google Scholar]
- Zhang, Q., Liu, Y., Zhang, G. and Mei, M. (2002) Molecular mapping of the fertility restorer gene Rf-4 for WA cytoplasmic male sterility in rice. J. Genet. Genomics 29: 1001–1004. [PubMed] [Google Scholar]
- Zhang, Y., Yang, J., Shan, Z., Chen, S., Qiao, W., Zhu, X., Xie, Q., Zhu, H., Zhang, Z., Zeng, R.et al. (2012) Substitution mapping of QTLs for blast resistance with SSSLs in rice (Oryza sativa L.). Euphytica 184: 141–150. [Google Scholar]
- Zhao, F., Zhu, H., Ding, X., Zeng, R., Zhang, Z., Li, W. and Zhang, G. (2007) Detection of QTLs for important agronomic traits and analysis of their stabilities using SSSLs in rice. China Agric. Sci. 6: 769–778. [Google Scholar]
- Zheng, K., Subudhi, P., Domingo, J., Magpantay, G. and Huang, N. (1995) Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet. Newsl. 12: 255–258. [Google Scholar]
- Zhu, H., Liu, Z., Fu, X., Dai, Z., Wang, S., Zhang, G., Zeng, R. and Liu, G. (2015) Detection and characterization of epistasis between QTLs on plant height in rice using single segment substitution lines. Breed. Sci. 65: 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Chen, S., Yang, J., Zhou, S., Zeng, L., Han, J., Su, J., Wang, L. and Pan, Q. (2012) The identification of Pi50(t), a new member of the rice blast resistance Pi2/Pi9 multigene family. Theor. Appl. Genet. 124: 1295–1304. [DOI] [PubMed] [Google Scholar]