Abstract
The erect panicle model super-rice can rationally transform the solar energy into accumulated organic matter (biomass) and increase grain yield. The phenotype of erect panicle architecture controlled by DENSE AND ERECT PANICLE 1 (DEP1) has been used in rice breeding for nearly a century owing to its high-yield, lodging tolerance with strong stem, reasonable population structure and high nitrogen use efficiency. DEP1 is a G protein γ subunit that is involved in the regulation of erect panicle, number of grains per panicle, nitrogen uptake, and stress-tolerance through the G protein signal pathway. Here we review the development of erect panicle rice varieties, DEP1 alleles and regulatory network, and its physiological and morphological functions. Additionally, the further increasing the yield potential of erect-panicle super-rice, and the development of molecular designing breeding for indica-japonica hybrid rice with the dep1 gene are also prospected.
Keywords: rice, erect panicle, heterotrimeric G proteins, DEP1, high-yield rice
Introduction
The past century has witnessed two major breakthroughs in rice breeding. The first is the green revolution of the 1960s in which the development of semi-dwarf lines greatly enhanced the rice yield (Peng et al. 1999). The second breakthrough is the application of heterosis in hybrid rice, which allowed a continuous increase in rice yield in the 1970s (Virmani et al. 1982, Yuan 1998b). Although these two breakthroughs dramatically improved the rice yield, the imminent food crisis that humanity will face in the next 50 years has forced breeders and scientists to search for next breakthrough. To achieve this goal, national and international rice breeding programs have emphasized the improvement of crop productivity by selecting for superior grain yield components and an ideal plant architecture (Huang et al. 2009, Khush 1999, Peng et al. 2008, Yuan 1998a). Five super rice ideal plant types for different geographical zones in China have been proposed by the Chinese Ministry of Agriculture, which included the erect panicle model that has dominated the northern japonica zone (Qian et al. 2016). The erect panicle architecture is conferred by a heterotrimeric G protein gene DENSE AND ERECT PANICLE 1 (DEP1), which significantly increases the rice production (Huang et al. 2009, Wang et al. 2009). Although more than 34 several increasing yield genes have recently been identified in rice (Fan et al. 2006, Ikeda et al. 2013, Komatsu et al. 2003, Li et al. 2011, Oikawa and Kyozuka 2009, Shomura et al. 2008, Song et al. 2007), only DEP1 has been extensively studied and widely used in rice breeding (Huang et al. 2009, Wang et al. 2009, Zhou et al. 2009).
The objectives of this review are to provide an overview of the DEP1 function in relation to single plant grain yield and population production, as well as to build a mechanistic model that represents the functions of the DEP1 gene and its associated pathways. We also discuss the implications of these findings in advancing our knowledge base and in improving rice production.
The history of the erect panicle type
In 1924, ‘Balilla’ was first variety in Italy that was reported to have an erect panicle architecture. This variety was derived from a Chinese japonica variety ‘Chinese Originario’, which was introduced from Japan in 1902, and was subsequently used in the generation of a new variety, ‘Ardito’, and later released as ‘Balilla’. Ten rice varieties were introduced into China, including ‘Balilla’ from Italy in 1958, and a number of varieties were bred using ‘Balilla’ as a parent (Xu et al. 1995). The profound significance of erect panicle architecture was not appreciated until the first widely expanded commercial erect panicle type variety ‘Liaojing 5’ was released in 1976. ‘Liaojing 5’ not only has high yield potential but also has preferable integrated traits, such as lodging resistance and a well-developed vascular system, thereby replacing the predominant Japanese japonica in northern China. Shortly afterwards, a number of high-yield japonica rice strains with erect panicle were released as commercial varieties (Yang 1984), with several of these meeting the super rice criteria of the Chinese Ministry of Agriculture, including ‘Shennong 265’ and ‘Qinzhonglang2’ (Huang et al. 2009, Sun et al. 2014). As the erect panicle type varieties began to occupy a predominant place among japonica rice in northern China (Chen et al. 2001), scientists and breeders began to pay close attention to the erect panicle architecture. Xu et al. (1995) earlier reported that the erect panicle trait was controlled by a single dominant gene using the posterity of the cross between ‘Liaojing 5’ and ‘Toyonishiki’ (a popular japonica variety). In 2007, a major quantitative trait locus (QTL) that conferred the erect panicle was mapped between DNA markers RM5652 and H90 on chromosome 9 (Yan et al. 2007). In 2009, the gene for erect panicle was identified by three independent research teams as Os09g0441900 and was subsequently named DEP1/EP/qPE9-1 (Huang et al. 2009, Wang et al. 2009, Zhou et al. 2009). Afterwards, a major rice NUE QTL (qNGR9) was cloned from the japonica variety ‘Qianzhongliang 2’, which is largely unresponsive to nitrogen fertilization during the vegetative growth period. A subsequent experiment demonstrated that qNGR9 is synonymous with DEP1 (Sun et al. 2014).
Function of DEP1/EP/qPE9-1
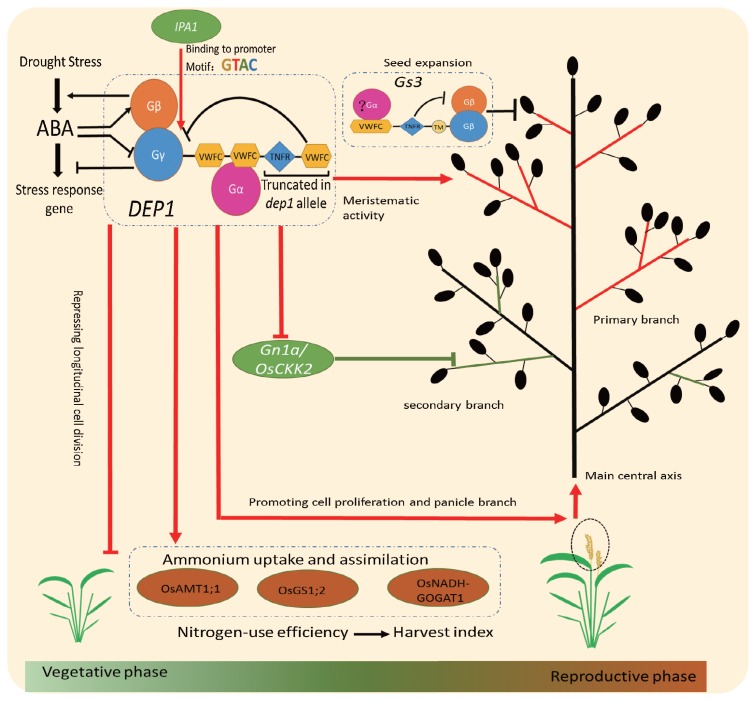
The DEP1 locus is pleiotropic for erect panicle, number of grains per panicle, nitrogen uptake and metabolism, ABA response and drought tolerance (Huang et al. 2009, Sun et al. 2014, Zhang et al. 2015). DEP1 has a modular arrangement with a conventional plant-specific Gγ subunit protein domain at its N-terminus, followed by two von Willebrand factor type C (VWFC) domains, a tumor necrosis factor receptor (TNFR)/nerve growth factor receptor (NGFR) family cysteine-rich domain, and a VWFC domain at the C-terminus. G protein signaling participates in various kinds of growth and developmental processes in animals and plants (Temple and Jones 2007). Heterotrimeric G proteins consisting of three subunits, namely, Gα, Gβ, and Gγ (New and Wong 1998). The rice genome contains a single Gα (RGA1) and a single Gβ (RGB1) subunits, two canonical Gγ subunits (RGG1, RGG2), and three non-canonical Gγ subunits (GS3, OsGGC2 and DEP1) (Botella 2012). The non-canonical Gγ subunits, which do not occur in mammals, have a conventional Gγ protein like domain at its N-terminus and a cysteine-rich domain at its C-terminus. A 637-bp stretch in the 5th exon of the dominate DEP1 locus (dep1 allele) is replaced by a 12-bp sequence, thereby eliminating the cysteine-rich domain, but not the Gγ domain. Plant with a dep1 genetic background that were subjected to RNA interference developed curved panicles and showed a lower number of grains per panicle, whereas those with a DEP1 genetic background exhibited no obvious changes (Huang et al. 2009, Taguchi-Shiobara et al. 2011) (Table 1). Similarly, the dep1-overexpressing plants presented a dwarf and short panicle phenotype, whereas DEP1-overexpressing plants showed no visible changes (Huang et al. 2009, Taguchi-Shiobara et al. 2011) (Table 1). These experiment evidence demonstrated that dep1 allele is a gain-of-function mutation. A self-inhibition model was constructed to explain the gain-of function mechanism (Xu et al. 2016). The observations that were derived from the DEP1 mutation and another non-canonical Gγ subunit GS3 showed that the cysteine-rich domain at the C-terminus has an inhibitory effect on the Gγ protein like domain at N-terminus (Botella 2012, Mao et al. 2010, Xu et al. 2016). Replacement of the dep1 allele resulted in the elimination of the inhibition of the cysteine-rich domain, thereby resulting in an increase of signaling by the Gβγ dimer. The increased Gβγ dimer signal induced two contrasting effects on plant architecture. First, longitudinal cell division and plant height during the vegetative growth period were repressed. Second, cell proliferation and panicle branching during the reproductive stage, which increases meristematic activity and thereby resulting in reduced inflorescence internode lengths (Huang et al. 2009, Sun et al. 2014). These opposing functions of the dep1 allele lead to an erect panicle architecture, well developed vascular bundles, an increased number of grains per panicle, and consequently, an increase in the grain yield (Huang et al. 2009, Xu et al. 2015) (Fig. 1). Because the cysteine-rich region suppressed the Gβγ dimer signal, variation in the cysteine-rich region caused varying degrees of erect panicle architecture and also changed the dominance, semi-dominance and recessiveness of the alleles (Huang et al. 2009, Sun et al. 2014, Taguchi-Shiobara et al. 2011, Wang et al. 2009, Zhou et al. 2009). Several studies have proven that the truncated DEP1 protein (dep1 allele) increases the number of grains per panicle and subsequently enhance the yield per plant (Huang et al. 2009, Wang et al. 2009, Xu et al. 2014). Recently, the genome editing lines using CRISPR/Cas9 constructs to target DEP1 in exon 5 also confirmed the function of the truncated DEP1 protein (Li et al. 2016). A genetic diversity analysis indicated that a G/C SNP at the promoter region results in a core sequence shift (TGGGCC) for a sit II transcriptional regulatory element, and is significantly associated with the number of primary and secondary branches, as well as number of grains per panicle (Zhao et al. 2016). However, Zhou et al. (2009) showed that the dep1/qpe9-1 allele is responsible for the development of an erect panicle architecture and a decrease in the number of grains per panicle, which may due to the specific genetic background or cultivation conditions used in the experiment (Zhou et al. 2009) (Table 1).
Table 1.
Phenotypes of transgenosis lines of DEP1 and of NIL lines with different genetic backgrounds
| Vector | Receptor | Phenotype | References |
|---|---|---|---|
| RNAi-DEP1 | Shao313 (dep1) | Curved panicles and fewer grains | Huang et al. (2009) |
| pDEP1:dep1 | Shao314 (DEP1) | Erect panicle and increased number of grains | |
| pActin:dep1 | Nipponbare (DEP1) | Dwarfed with erect panicles | |
| pActin:DEP1 | Nipponbare (DEP1) | No change | Taguchi-Shiobara et al. (2011) |
| pActin:Dn1-1 | Nipponbare (DEP1) | Dwarfed with erect panicles | |
| pActin:DN1 | Nipponbare (DEP1) | No change | |
| RNAi-DN1 | Nipponbare (DEP1) | No change | |
| Crispr/Cas9:DEP1 | Zhonghua 11 (DEP1) | Dwarfed with erect panicles | Li et al. (2016) |
| Genetic background | Subspecies | Grain number | Primary branch | Secondary branch | Yield-increasinga | References | |||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| DEP1 | dep1 | DEP1 | dep1 | DEP1 | dep1 | ||||
| Toyonishiki | japonica | 108 | 146 | 11 | 13 | 16 | 25 | +10.16% | Wang et al. (2009) |
| Shao314 | japonica | 130 | 251 | 11 | 16 | 18 | 49 | +40.90% | Huang et al. (2009) |
| ZF802 | indica | 60 | 290 | – | – | – | – | – | Huang et al. (2009) |
| R6547 | indica | 303 | 288 | 17 | 17 | 62 | 57 | –17.20% | Zhou et al. (2009) |
| RIL-ipa1 | japonica | – | – | 15 | 15 | 63 | 60 | n.s. | Lu et al. (2013) |
| Zhonghua 11 | japonica | 104 | 150 | – | – | – | – | – | Li et al. (2016) |
“–” indicates the missing data; “n.s.” means no significant change.
The increase of yield per plant caused by the dep1 allele.
Fig. 1.
The DEP1 network, including the structural characteristics of DEP1 and GS3, the different functions in vegetative and reproductive phases, and upstream and downstream genes. Details are described in the text.
The Gγ protein like domain of DEP1 strongly interacts with RGB1, and the VWFC domain at the C-terminus may participate in association between Gγ and Gα. The dep1 allele inhibits nitrogen responses through interactions with the Gβ and Gα subunits, and reduced Gα or enhanced Gβ activity inhibits nitrogen responses (Sun et al. 2014). A recent study showed that high ABA concentrations decrease the transcription level of DEP1 and increase the transcription level of RGB1. Moreover, DEP1 negatively regulates the ABA response via suppressing the expression of ABA and stress response related gene, whereas RGB1 positively regulates ABA biosynthesis through upregulating NCED gene expression (Zhang et al. 2015). Thus, DEP1 is involved in ABA response and drought tolerance, and modulated by RGB1 (Fig. 1). DEP1 is also involved in conferring cadmium tolerance to yeast cells and plants, and its cysteine-rich region influences this activity, whereas the Gγ domain might not be involved (Kunihiro et al. 2013).
The DEP1 Regulation Network
Plant and panicle architectures are complex traits that are determined by several factors. Although our understanding is limited, the available data allowed us to construct a simple network that summarizes how DEP1 affects plant and panicle architectures (Fig. 1). IPA1, which confers the ideal rice plant architecture with reduced tiller numbers and increased grain numbers per panicle, directly binds to the DEP1 promoter at different sites, and the GTAC core motifs are responsible for IPA1 binding (Jiao et al. 2010, Lu et al. 2013, Miura et al. 2010). RIL lines, which are derived from a cross between ‘Ri22’, an ipa1-carrying cultivar (Jiao et al. 2010) and ‘Liaogeng5’, an erect panicle variety, demonstrated that IPA1 functions as a positive regulator of DEP1 for plant height and panicle length in rice (Lu et al. 2013). A study performed using the mixed genetic background of the F2 populations showed that the effects of DEP1 associated with IPA1 binding sites might be obscured or disrupted by different pathways (Xu et al. 2014). Thus, a complex relationship may exist between DEP1 and IPA1. IPA1 also apparently induces two contrasting effects on plant architecture. A higher IPA1 expression level at the vegetative stage represses shoot branching, thereby leading to a decrease in panicle number. However, a higher IPA1 expression level in young panicles promotes branching, which increases the number of grains per panicle (Jiao et al. 2010). The transition from the vegetative to reproductive phase is marked by the transformation of the shoot apical meristem into an inflorescence meristem. Factors that trigger this functional transition between DEP1 and IPA1 in the vegetative and reproductive phases is worth further study. Xu et al. (2014) demonstrated that IPA1 and DEP1 have contrasting effects on most yield components, and thus the identification of a balance between IPA1 and DEP1 may be another key point that would facilitate in yield improvement.
Gn1a, the first major QTL of yield-related traits is located downstream of DEP1 (Huang et al. 2009). Gn1a encodes a cytokinin oxidase/dehydrogenase, which regulates the meristematic activity, panicle branching and grain number through its effects on cytokinin levels (Ashikari et al. 2005). The dep1 allele down-regulated the expression of Gn1a (Huang et al. 2009). A NIL-Gn1a line had the same number of primary branches as the control line but developed a higher number of secondary branches (Ashikari et al. 2005). This finding suggests that dep1 genetically controls the number of primary and secondary branches at the panicle top, whereas Gn1a regulates the number of secondary branches at the panicle base (Fig. 1). The restorer line 93-11, which harbors the dep1 and gn1a allele, exhibited a remarkable yield performance, reaching as high as 12 t ha−1 (Liu et al. 2012). LAX, FZP and RCN1 are involved in axillary meristem initiation and development, and DEP1 rarely affects these genes (Komatsu et al. 2003, Nakagawa et al. 2002, Oikawa and Kyozuka 2009).
GS3, another non-canonical Gγ subunit, is a homolog of DEP1 and was identified a grain size QTL under different genetic backgrounds (Fan et al. 2006, Mao et al. 2010, Takano-Kai et al. 2009). GS3 has a similar isoform as DEP1, where it comprises a Gγ subunit domain, a transmembrane domain, a TNFR/NGFR family cysteine-rich domain, and a VWFC domain at the C-terminus. The Gγ domain functions as a negative regulator of grain size and organ size, and the C-terminal TNFR/NGFR domain plays an inhibitory role to the Gγ subunit domain (Mao et al. 2010). Although no experimental evidence as demonstrated the interactions between DEP1 and GS3, these provably function using the same pathway. These may be manipulated simultaneously because these are both Gγ subunits, and they confer signals through a Gβγ dimer. Erect panicle varieties show a common disadvantage of low 1,000 grain weights, and GS3 regulates the grain size and weight. Thus, the identification of a factor that simultaneously regulates DEP1 and GS3 may improve the yield of erect panicle varieties. Although a number of studies have demonstrated the relationship among DEP1, IPA1 and Gn1a (Huang et al. 2009, Lu et al. 2013, Xu et al. 2014), there is a need to elucidate the relationship between DEP1 and GS3.
DEP1 regulates nitrogen uptake and metabolism by affecting genes that are associated with ammonium uptake and assimilation (such as OsAMT1;1, OsGS1;2 and OsNADH-GOGAT1) (Huang et al. 2009). In dep1 allelic plants, these genes were up-regulated under low nitrogen conditions. The dep1 allelic plants have higher glutamine synthase activities and accumulate higher amounts of internal nitrogen than the DEP1 allelic plants, regardless of limited nitrogen supply (Sun et al. 2014). DEP1 also regulates ABA responses by affecting the expression of ABA- and stress inducible genes, such as OsABI5, OsRAB16A, OsLIP9, OsNAC5 and OsNAC6 (Zhang et al. 2015).
Homologs of DEP1 and other related genes
The homologs of DEP1 regulate organ size and shape in other species. AGG3, a Gγ subunit in Arabidopsis that has a 22.5% amino acid sequence identity with DEP1, regulates petal growth by influencing the cell proliferation period (Chakravorty et al. 2011, Li et al. 2012). Several homologs of DEP1 with truncated C-terminal deletions are gain-of-function mutants in barley, bread wheat and its diploid wild progenitor Triticumurartu (Huang et al. 2009). Because the Gγ subunits participate in the regulation of organ size in Arabidopsis, and the truncated of DEP1 homologs are also occur in some cereals, the use of DEP1 homologs in other crops may contribute to solving the global food crisis.
DEP2, which is allelic to EP2 (Os07g0616000), was identified from mutations of ‘Zhonghua 11’ and ‘Nipponbare’ (Abe et al. 2010, Li et al. 2010). DEP3 ( Os06g0677000) was identified from a mutation in an elite Korean japonica cultivar (Qiao et al. 2011), and both dep3 and the dep2/ep2 alleles are recessive alleles that cause a dense and erect panicle architecture. Experimental evidence did not indicate whether DEP2/EP2 or DEP3 is involved in the G protein complex or affects the nitrogen-use efficiency in rice. Further functional characterization of DEP2/EP2 and DEP3 may provide a better understanding of the erect panicle mechanism and improve the genetic diversity of erect panicle germplasm.
The effects of DEP1 on the morphology and physiology in rice breeding
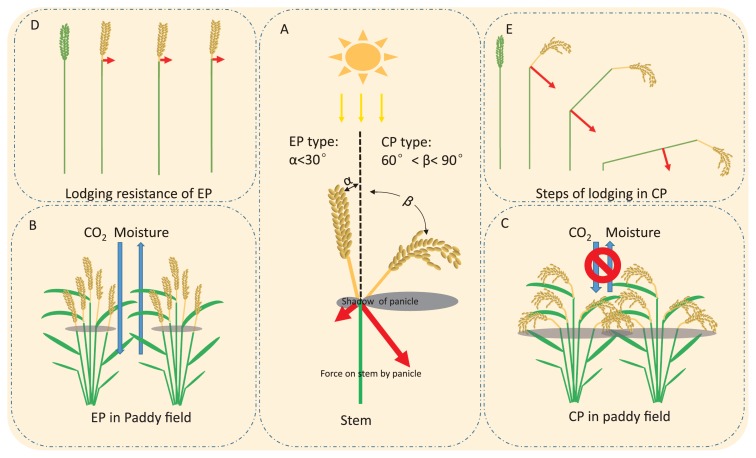
The advantages of the dep1 allele not only involve its high meristematic activity and the high nitrogen-use efficiency, but also its improved population canopy structure that is caused by erect panicles (Fig. 2). Before the molecular function of DEP1 was elucidated, breeders and scientists focused on increasing rice yield by inducing morphological changes that result in erect panicles. The photosynthetic ability of panicles was negligible, but the panicles’ shadows may obstruct the light, thereby preventing it from reaching the leaves. Xu et al. (1990) conducted a light intensity investigation by comparing the light intensity in the canopy before and after cutting the panicles, and the results showed that the erect panicle plants were less affected by panicle shadows than the curved panicle plants (Fig. 2A). Moreover, the erect panicle architecture may improve the circulation of CO2 and moisture (Xu et al. 1995) (Fig. 2). A population structure analysis based on field investigation showed that the subsequent opening of the plant canopy through erect panicle architecture allows a 10% increase in sunlight penetration compared to curved panicle varieties (Xu et al. 1996). In addition, the canopy temperature of erect type varieties was about 2°C higher than that of curved panicle type varieties during the midday (Xu et al. 1996). Wind speed within the canopy is also much faster with erect panicle varieties than with curved panicle varieties, thereby resulting in a 6% decrease in canopy humidity using erect panicle varieties compared to that with curved panicle varieties (Xu et al. 1996). The relatively low level of humidity may be beneficial to improving tolerance to plant diseases and insect pests, such as sheath blight tolerance. In the curved panicle variety, gravity bends the panicle neck, thereby forcing the upper internode, and ultimately inducing the entire plant to lodge. The erect panicle varieties showed strong lodging resistance (Xu et al. 2004b) (Fig. 2). Well development vascular bundles and large diameter stems contribute to lodging resistance, and the erect panicle leads to a smaller panicle neck angle compared to that of the curved type, which decreases the force on the panicle neck and stem, and consequently improves the resistance to lodging (Xu et al. 2004b).
Fig. 2.
The effects of DEP1 on morphology. (A) The panicle shadow on a leaf at noon and the force of the panicle on the stem between erect and curve panicle plants. (B, C) The erect panicle improves the circulation of oxygen and moisture compared to that in the curved panicle. (D, E) The erect panicle decreases the force on the panicle neck and stem, and consequently improves the resistance to lodging compared to that in the curved panicle. EP and CP represent erect panicle architecture and curved panicle architecture, respectively.
Application of DEP1 in rice production
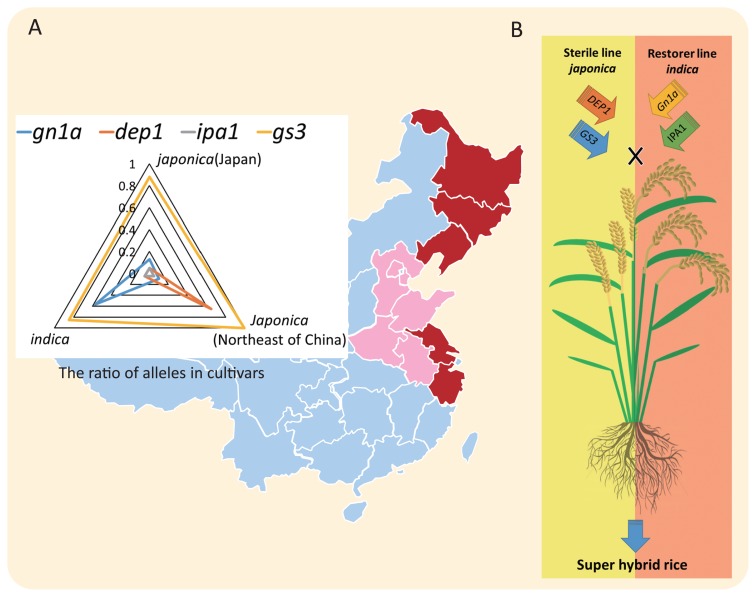
Unlike most other yield-related genes that were identified through molecular research, DEP1 has been extensively used in rice breeding (Qian et al. 2016). The 3-years average yield of an erect panicle variety, ‘Shennong 606’, has reached 12.34 t ha−1 (Xu et al. 2004a). An allelic investigation of 78 japonica rice cultivars that were widely cultivated in northeast China between 1963 and 2008 showed that after the release of ‘Liaojing 5’, the application of the dep1 allele continued to increase in japonica varieties in northern of China (Zhao et al. 2016). More than 70% cultivars in Liaoning Province carry the dep1 allele, and more than 50% of the dep1 carrying cultivars are propagated in northeast of China. At the downstream region of the Yangtze River, nearly all the japonica varieties have erect panicle architecture (Xu et al. 2007) (Fig. 3A). The northeast area and downstream region of the Yangtze River represents over 70% of entire japonica rice cultivated area in China. Moreover, the cultivation area for erect panicle varieties has annually increased in the japonica zone (Fig. 3A). For other yield related genes, the long grain type allele of GS3 has historically predominated in northern China (Sun et al. 2012). A high proportion of the dep1 and gn1a alleles have been detected in japonica (northeast of china) and in indica (Fig. 3A), indicating that dep1 and gn1a contribute the rice production improvement in japonica and indica rice production, respectively. Both high yielding alleles dep1 and gn1a do exist in Japanese japonica varieties (Sun et al. 2012), which are generally low yielding. We also observed that even though the ipa1 allele showed an unequaled phenotype for rice production, the ipa1 allele rarely existed in both indica and japonica rice varieties (Sun et al. 2012).
Fig. 3.
The application of DEP1 in rice breeding. (A) Distribution map of erect panicle varieties carrying the dep1 allele. Regions of large-scale cultivation of erect panicle varieties are indicated in dark red, the area that has generated erect panicle varieties are shown in pink. The top left radar chart showe the allele ratio of gn1a, dep1, ipa1, and gs3 in japonica (separated into japonica in Japan and japonica in northeast of China) and indica varieties (Sun et al. 2012). (B) A model for breeding super hybrid rice by rational genetic design using DEP1 and related genes. The japonica sterile plant with the desired alleles of DEP1 and GS3, and the indica restorer plant carrying the favorable alleles of IPA1 and Gn1a. Thus, the F1 hybrids may exhibit an optimized yield components.
The bloom of genome information from the global rice germplasm and next generation sequence may facilitate in the breeding of an elite super rice variety using indica-japonica heterosis. The DEP1 also has a potential to contribute to the hybrid super rice breeding by rational design. In the F2 population derived from the cross between ‘Liaojing 5’ and ‘Ri22’, heterozygous plants showed better yield performance compared to plants that were homozygous at both DEP1 and IPA1 loci (Xu et al. 2014). Crossing a japonica sterile line with a desired allele of DEP1 and GS3 with an indica restorer line that carried a favorable allele of Gn1a and IPA1 is expected to show an optimal yield components (Liu et al. 2012) (Fig. 3B). The F1 plant with these elite alleles will exhibit an erect panicle architecture, large grains, increase in number of grains at the both top and bottom of the panicle, moderate plant height, and a balanced relationship between the number of grains per panicle and the number of panicles (Chen 2013).
Conclusions and Prospective
The Gγ subunit protein, which is encoded by DEP1 gene, contributes to an increase in grain number, a high nitrogenuse efficiency, and improved drought tolerance. The erect panicle architecture also lead to an improvement in canopy structure and lodging resistance, which in turn benefits rice production (Huang et al. 2009, Xu et al. 1990, 2004b). Thus, DEP1 can increase the rice yield, enhance the tolerance to environmental changes, and relieve the pressure of environmental pollution such as excessive application of nitrogenous fertilizers. These elite traits persuade us that DEP1 may be the key factor that drives improved rice production breakthrough. However, although the findings of the present study represent significant progress in the study of DEP1 functions, several important pieces of information have yet to be elucidated. As the G protein works very upstream of growth and developmental processes, and Gγ subunits affects stomatal movement and lateral roots in Arabidopsis (Trusov et al. 2007), DEP1 probably affects other crucial growth processes in rice. As illustrated in Fig. 1, details on the pathways and the network underlying the DEP1 functions have gradually been uncovered, and the identification of other genes involved in these processes is essential for the characterization of the regulatory network. DEP1 may regulate cytokinin production through Gn1a, as well as the responses to ABA, but whether auxin, strigolactone, brassinolide or other hormones are involved in the DEP1 network have yet to be established. The introduction of the dep1 allele in japonica breeding makes its absence in indica varieties difficult to understand. We believe the indica breeding has primarily focused on the heterosis of the F1 plants in the past decades, while japonica breeding has built ideal plant and panicle architectures to achieve higher yields. As the dep1 allele showed superior yield components under the indica genetic background compared to that of the japonica background (Xu et al. 2015), we believe that the combination of the heterosis in F1 plants and the dep1 allele may improve indica yield. And rational design of ideal architecture using defined genes, such as DEP1 and its related genes, will exploit strong indica-japonica heterosis along with predetermined specific morphological changes and functional characters. However, erect panicle is not perfect, it showed a low setting rate at the bottom of panicle and are considered to be only mediocre eating quality. Future breeding projects on DEP1 utilization should address these inadequacies. Because the erect panicle type has a specific morphology and nitrogen response traits, a matching cultivation method that includes the density of plant and the amount of fertilizer, should be developed. Four step identification process for rice breeding were proposed as followings: (i) identification of genes that control traits, (ii) assessment of the characteristics of the particular architecture, (iii) establishment of the population structure, and (iv) population structure that makes the maximum use of the solar energy in given ecological conditions to achieve the yield limits (Zhang et al. 2008). The ability to readily incorporate yield improve genes into rice varieties will become increasingly important. Thus, a system for the application of yield-related genes that have been identified through molecular research investigations to rice breeding schemes to maximize yield should be established in the near future.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 31430062, 31501284 and 31371587) and a class general financial grant from the China Postdoctoral Science Foundation (Grant No. 2014M560211, Postdoctoral No. 142541).
Literature Cited
- Abe, Y., Mieda, K., Ando, T., Kono, I., Yano, M., Kitano, H. and Iwasaki, Y. (2010) The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet. Syst. 85: 327–339. [DOI] [PubMed] [Google Scholar]
- Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., Angeles, E.R., Qian, Q., Kitano, H. and Matsuoka, M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Botella, J.R. (2012) Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 17: 563–568. [DOI] [PubMed] [Google Scholar]
- Chakravorty, D., Trusov, Y., Zhang, W., Acharya, B.R., Sheahan, M.B., McCurdy, D.W., Assmann, S.M. and Botella, J.R. (2011) An atypical heterotrimeric G-protein gamma-subunit is involved in guard cell K(+)-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67: 840–851. [DOI] [PubMed] [Google Scholar]
- Chen, W., Xu, Z., Zhang, W., Zhang, L. and Yang, S. (2001) Creation of new plant type and breeding rice for super high yield. Acta Agron. Sin. 27: 665–674. [Google Scholar]
- Chen, Z.J. (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 14: 471–482. [DOI] [PubMed] [Google Scholar]
- Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., Li, X. and Zhang, Q. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Huang, X., Qian, Q., Liu, Z., Sun, H., He, S., Luo, D., Xia, G., Chu, C., Li, J. and Fu, X. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497. [DOI] [PubMed] [Google Scholar]
- Ikeda, M., Miura, K., Aya, K., Kitano, H. and Matsuoka, M. (2013) Genes offering the potential for designing yield-related traits in rice. Curr. Opin. Plant Biol. 16: 213–220. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., Dong, G., Zeng, D., Lu, Z. and Zhu, X. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Khush, G.S. (1999) Green revolution: preparing for the 21st century. Genome 42: 646–655. [PubMed] [Google Scholar]
- Komatsu, K., Maekawa, M., Ujiie, S., Satake, Y., Furutani, I., Okamoto, H., Shimamoto, K. and Kyozuka, J. (2003) LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100: 11765–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro, S., Saito, T., Matsuda, T., Inoue, M., Kuramata, M., Taguchi-Shiobara, F., Youssefian, S., Berberich, T. and Kusano, T. (2013) Rice DEP1, encoding a highly cysteine-rich G protein gamma subunit, confers cadmium tolerance on yeast cells and plants. J. Exp. Bot. 64: 4517–4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Liu, W., Tang, J., Chen, J., Tong, H., Hu, B., Li, C., Fang, J., Chen, M. and Chu, C. (2010) Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20: 838–849. [DOI] [PubMed] [Google Scholar]
- Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., Pan, X., Lin, Q., Luo, W., Wu, G. and Li, H. (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Liu, Y., Zheng, L., Chen, L., Li, N., Corke, F., Lu, Y., Fu, X., Zhu, Z., Bevan, M.W.et al. (2012) The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New phytol. 194: 690–703. [DOI] [PubMed] [Google Scholar]
- Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., Shao, D., Xu, C., Li, X., Xiao, J.et al. (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Liu, J., Tao, H., Shi, S., Ye, W., Qian, Q. and Guo, L. (2012) Genetics and breeding improvement for panicle type in rice. Chinese J. Rice Sci. 26: 227–234. [Google Scholar]
- Lu, Z., Yu, H., Xiong, G., Wang, J., Jiao, Y., Liu, G., Jing, Y., Meng, X., Hu, X. and Qian, Q. (2013) Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., Li, X. and Zhang, Q. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 107: 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K., Ikeda, M., Matsubara, A., Song, X.-J., Ito, M., Asano, K., Matsuoka, M., Kitano, H. and Ashikari, M. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Nakagawa, M., Shimamoto, K. and Kyozuka, J. (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29: 743–750. [DOI] [PubMed] [Google Scholar]
- New, D.C. and Wong, J.T. (1998) The evidence for G-protein-coupled receptors and heterotrimeric G proteins in protozoa and ancestral metazoa. Biol. Signals Recept. 7: 98–108. [DOI] [PubMed] [Google Scholar]
- Oikawa, T. and Kyozuka, J. (2009) Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S., Cassman, K.G., Virmani, S., Sheehy, J. and Khush, G. (1999) Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci. 39: 1552–1559. [Google Scholar]
- Peng, S., Khush, G.S., Virk, P., Tang, Q. and Zou, Y. (2008) Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 108: 32–38. [Google Scholar]
- Qian, Q., Guo, L., Smith, S.M. and Li, J. (2016) Breeding high-yield superior-quality hybrid super-rice by rational design. Nat. Sci. Rev. 3: 283–294. [Google Scholar]
- Qiao, Y., Piao, R., Shi, J., Lee, S.-I., Jiang, W., Kim, B.-K., Lee, J., Han, L., Ma, W. and Koh, H.-J. (2011) Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 122: 1439–1449. [DOI] [PubMed] [Google Scholar]
- Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S. and Yano, M. (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Song, X.-J., Huang, W., Shi, M., Zhu, M.-Z. and Lin, H.-X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S., Zhang, C., Ma, Y., Liu, Q., Huang, X. and Yuan, Q. (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46: 652–656. [DOI] [PubMed] [Google Scholar]
- Sun, J., Liu, D., Wang, J.-Y., Ma, D.-R., Tang, L., Gao, H., Xu, Z.-J. and Chen, W.-F. (2012) The contribution of intersubspecific hybridization to the breeding of super-high-yielding japonica rice in northeast China. Theor. Appl. Genet. 125: 1149–1157. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara, F., Kawagoe, Y., Kato, H., Onodera, H., Tagiri, A., Hara, N., Miyao, A., Hirochika, H., Kitano, H., Yano, M.et al. (2011) A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets. Breed. Sci. 61: 17–25. [Google Scholar]
- Takano-Kai, N., Jiang, H., Kubo, T., Sweeney, M., Matsumoto, T., Kanamori, H., Padhukasahasram, B., Bustamante, C., Yoshimura, A., Doi, K.et al. (2009) Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple, B.R. and Jones, A.M. (2007) The plant heterotrimeric G-protein complex. Annu. Rev. Plant Biol. 58: 249–266. [DOI] [PubMed] [Google Scholar]
- Trusov, Y., Rookes, J.E., Tilbrook, K., Chakravorty, D., Mason, M.G., Anderson, D., Chen, J.G., Jones, A.M. and Botella, J.R. (2007) Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 19: 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virmani, S., Aquino, R. and Khush, G. (1982) Heterosis breeding in rice (Oryza sativa L.). Theor. Appl. Genet. 63: 373–380. [DOI] [PubMed] [Google Scholar]
- Wang, J., Nakazaki, T., Chen, S., Chen, W., Saito, H., Tsukiyama, T., Okumoto, Y., Xu, Z. and Tanisaka, T. (2009) Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 119: 85–91. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Xu, N., Xu, H., Tang, L., Liu, J., Sun, J. and Wang, J. (2014) Breeding value estimation of the application of IPA1 and DEP1 to improvement of Oryza sativa L. ssp. japonica in early hybrid generations. Mol. Breed. 34: 1933–1942. [Google Scholar]
- Xu, Q., Liu, T., Bi, W., Wang, Y., Xu, H., Tang, L., Sun, J. and Xu, Z. (2015) Different effects of DEP1 on vascular bundle- and panicle-related traits under indica and japonica genetic backgrounds. Mol. Breed. 35: 1–10. [Google Scholar]
- Xu, Q., Zhao, M., Wu, K., Fu, X. and Liu, Q. (2016) Emerging insights into heterotrimeric G protein signaling in plants. J. Genet. Genomics 43: 495–502. [DOI] [PubMed] [Google Scholar]
- Xu, Z., Chen, W., Zhang, L. and Yang, S. (1990) Comparative study on light distribution in rice canopies with different panicle types. Sci. Agric. Sin. 13: 10–16. [Google Scholar]
- Xu, Z., Chen, W., Zhang, L. and Zhang, C. (1995) The heredity of the erect panicle character and relation with other characters in rice. Journal of Shenyang Agricultural University 26: 1–7. [Google Scholar]
- Xu, Z., Chen, W., Zhou, H., Zhang, L. and Yang, S. (1996) Physiological and ecological characteristics of rice with erect panicle and prospects of their utilization. Science Bulletin: 1642–1648. [Google Scholar]
- Xu, Z., Chen, W., Zhang, W., Zhou, S., Liu, L., Zhang, L. and Yang, S. (2004a) New plant type breeding for super-high yielding northern japonica rice. Scientia Agricultura Sinica 37: 1407–1413. [Google Scholar]
- Xu, Z., Zhang, S., Zhou, S. and Liu, L. (2004b) Primary analysis of relationship between rice panicle type and lodging resistance. Plant Physiology Communications 40: 561–563. [Google Scholar]
- Xu, Z., Chen, W., Han, Y., Shao, G., Zhang, W. and Ma, D. (2007) Classification of panicle type and its relationship with grain yield and quality of rice in Liaoning Province. Acta Agron. Sin. 33: 1411–1418. [Google Scholar]
- Yan, C.J., Zhou, J.H., Yan, S., Chen, F., Yeboah, M., Tang, S.Z., Liang, G.H. and Gu, M.H. (2007) Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (Oryza sativa L.). Theor. Appl. Genet. 115: 1093–1100. [DOI] [PubMed] [Google Scholar]
- Yang, S. (1984) The primary discussion on the theories and methods of the ideotype breeding of rice. Acta Agron. Sin. 17: 6–13. [Google Scholar]
- Yuan, L. (1998a) Hybrid rice breeding for super high yield. 21st century: 10. [Google Scholar]
- Yuan, L.P. (1998b) Hybrid rice breeding in China. Advances in Hybrid Rice Technology. Philippines: International Rice Research Institute: 27–33. [Google Scholar]
- Zhang, D.-P., Zhou, Y., Yin, J.-F., Yan, X.-J., Lin, S., Xu, W.-F., Baluška, F., Wang, Y.-P., Xia, Y.-J. and Liang, G.-h. (2015) Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J. Exp. Bot. 66: 6371–6384. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Li, J., Xue, Y., Han, B. and Deng, X.W. (2008) Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol. Plant 1: 715–719. [DOI] [PubMed] [Google Scholar]
- Zhao, M., Sun, J., Xiao, Z., Cheng, F., Xu, H., Tang, L., Chen, W., Xu, Z. and Xu, Q. (2016) Variations in DENSE AND ERECT PANICLE 1 (DEP1) contribute to the diversity of the panicle trait in high-yielding japonica rice varieties in northern China. Breed. Sci. 66: 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Zhu, J., Li, Z., Yi, C., Liu, J., Zhang, H., Tang, S., Gu, M. and Liang, G. (2009) Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]