Abstract
Streptococcus thermophilus ACA-DC 2 is a newly sequenced strain isolated from traditional Greek yogurt. Among the 14 fully sequenced strains of S. thermophilus currently deposited in the NCBI database, the ACA-DC 2 strain has the smallest chromosome, containing 1,731,838 bp. The annotation of its genome revealed the presence of 1,850 genes, including 1,556 protein-coding genes, 70 RNA genes and 224 potential pseudogenes. A large number of pseudogenes were identified. This was also accompanied by the absence of pathogenic features suggesting evolution of strain ACA-DC 2 through genome decay processes, most probably due to adaptation to the milk ecosystem. Analysis revealed the existence of one complete lactose-galactose operon, several proteolytic enzymes, one exopolysaccharide cluster, stress response genes and four putative antimicrobial peptides. Interestingly, one CRISPR-cas system and one orphan CRISPR, both carrying only one spacer, were predicted indicating low activity or inactivation of the cas proteins. Nevertheless, four putative restriction-modification systems were determined that may compensate any deficiencies of the CRISPR-cas system. Furthermore, whole genome phylogeny indicated three distinct clades within S. thermophilus. Comparative analysis among selected strains representative for each clade, including strain ACA-DC 2, revealed a high degree of conservation at the genomic scale, but also strain specific regions. Unique genes and genomic islands of strain ACA-DC 2 contained a number of genes potentially acquired through horizontal gene transfer events, that could be related to important technological properties for dairy starters. Our study suggests genomic traits in strain ACA-DC 2 compatible to the production of dairy fermented foods.
Keywords: Extended genome report, Streptococcus thermophilus, Yogurt, Horizontal gene transfer, CRISPR, Stress genes
Introduction
The use of microorganisms in food fermentations is the means for converting perishable and frequently inedible raw materials into safe, shelf-stable and nutritionally upgraded foods [1]. The economic importance of starter cultures for the food industry has led to the continuous search for the discovery of new microorganisms with important technological characteristics. In many cases it has been proven that traditionally fermented foods represent a natural reservoir of undiscovered microbial strains for possible diverse food applications [2, 3].
Streptococcus thermophilus is among the species commonly used in the dairy industry, mainly in the fermentation of yogurt and several cheese varieties, contributing to the desirable organoleptic characteristics of the final product [4, 5]. It is the sole species considered GRAS within the Streptococcus genus, which includes mostly pathogens and opportunistic pathogens [6]. Due to the industrial significance of the species, a plethora of studies has been conducted for a number of strains, revealing information about their diverse technological features [7, 8]. Furthermore, during the last 15 years, the advance of high-throughput sequencing techniques along with the development of novel bioinformatics tools facilitated the analysis of complete genome sequences, providing information for the overall genetic content of S. thermophilus [9–12]. These studies have demonstrated that S. thermophilus strains have been adapted to the milk environment through extensive reductive evolution as indicated by the large number of pseudogenes found in all strains. Adaptation to the milk environment is also supported by the loss of genes related to carbohydrate metabolism and virulence.
In this study, we present the analysis of the complete genome sequence of S. thermophilus ACA-DC 2. The genomic insights acquired could be proven useful for the exploitation of the specific strain in the production of fermented dairy products.
Organism information
Classification and features
Streptococcus thermophilus ACA-DC 2 is classified within the order Lactobacillales of the class Bacilli. It is a non-sporulating, Gram-positive bacterium with coccus-shaped cells (Fig. 1). The strain was isolated from traditional Greek yogurt manufactured through backslopping [13, 14]. Its optimum growth takes place in M17 medium at 42 oC under microaerophilic conditions within 24 h. Information about the classification and the features of S. thermophilus ACA-DC 2 is summarized in Table 1. The phylogenetic analysis was based on 16S rRNA gene sequences and places S. thermophilus ACA-DC 2 in the distinct cluster formed by the S. thermophilus strains and within the salivarius group, as shown in Fig. 2.
Fig. 1.
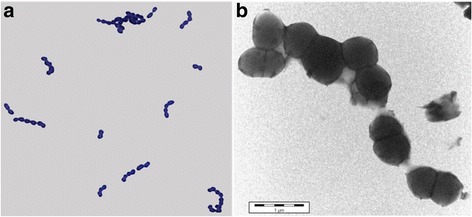
Photomicrographs of S. thermophilus ACA-DC 2. The images were obtained with (a) optical microscopy at magnification 1000x for Gram stained cells and (b) transmission electron microscopy of stained cells with 10% (w/v) PTA. Scale bar in (b) corresponds to 1 μm
Table 1.
Classification and general features of S. thermophilus strain ACA-DC 2 according to the MIGS recommendations [39]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [40] | |
| Phylum Firmicutes | TAS [41, 42] | ||
| Class Bacilli | TAS [43, 44] | ||
| Order Lactobacillales | TAS [44, 45] | ||
| Family Streptococcacceae | TAS [46–48] | ||
| Genus Streptococcus | TAS [47, 49, 50] | ||
| Species Streptococcus thermophilus | TAS [47, 51, 52] | ||
| Strain: ACA-DC 2 | TAS (this study) | ||
| Gram stain | Positive | IDA | |
| Cell shape | Coccus | IDA | |
| Motility | Non-motile | IDA | |
| Sporulation | Non-sporulating | NAS | |
| Temperature range | 30–50 °C | TAS [51] | |
| Optimum temperature | 42 °C | TAS [53] | |
| pH range; Optimum | 5–7; 6.5 | TAS [53] | |
| Carbon source | lactose; saccharose; d-glucose; galactose | IDA | |
| MIGS-6 | Habitat | Yogurt | TAS [13, 14] |
| MIGS-6.3 | Salinity | 2% NaCl (w/v) | TAS [51] |
| MIGS-22 | Oxygen requirement | Microaerophilic | TAS [51] |
| MIGS-15 | Biotic relationship | Free-living | NAS |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Greece | TAS [13, 14] |
| MIGS-5 | Sample collection | 1988 | NAS |
| MIGS-4.1 | Latitude | Unknown | |
| MIGS-4.2 | Longitude | Unknown | |
| MIGS-4.4 | Altitude | Unknown |
aEvidence codes - IDA inferred from direct assay, TAS traceable author statement (i.e., a direct report exists in the literature), NAS non-traceable author statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [54]
Fig. 2.
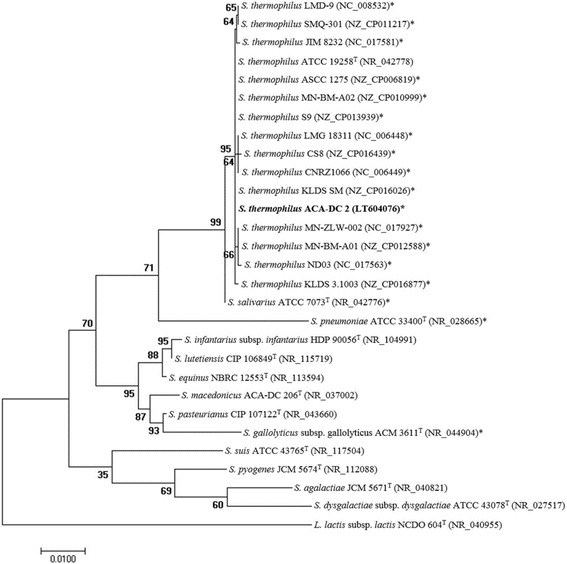
Phylogenetic tree highlighting the position of S. thermophilus ACA-DC 2 relative to other Streptococcus species. The tree was constructed based on 16S rRNA gene sequences. GenBank accession numbers are presented in parentheses and type strains are indicated with a superscript T (type strains = T). Strains with complete genome sequence are marked with an asterisk. 16S rRNA gene sequences were aligned using MUSCLE [55]. The phylogenetic tree was built by the Maximum Likelihood method within MEGA7 software [56] using the Tamura-Nei substitution model [57]. Lactococcus lactis subsp. lactis NCDO 604T served as the outgroup. Bootstrap values derived after 1,000 replicates. The scale bar indicates an estimated 0.01 nucleotide change per nucleotide position
Genome sequencing information
Genome project history
S. thermophilus ACA-DC 2 is deposited in the ACA-DC culture collection of the Laboratory of Dairy Research, Agricultural University of Athens, Athens, Greece. The strain was selected for sequencing in order to obtain information about its technological and probiotic potential, having as basic aim its application as a starter culture in the production of dairy fermented foods. The project was carried out in 2015 and the genome was sequenced, fully assembled and annotated. The genome sequencing project was registered in the European Nucleotide Archive database under accession number LT604076. The summary of the project information is shown in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Illumina genomic Nextera XT library; PacBio 10 kb genomic library |
| MIGS 29 | Sequencing platforms | Illumina HiSeq2500; PacBio RSII |
| MIGS 31.2 | Fold coverage | 636x |
| MIGS 30 | Assemblers | ABySS v1.5.1; BLASR; SSPACE v1.0; GapFiller v1.10 |
| MIGS 32 | Gene calling method | Prodigal; MeteGeneAnnotator; FGENESB |
| Locus Tag | STACADC2 | |
| Genbank ID | LT604076 | |
| GenBank Date of Release | 29-Jul-2016 | |
| GOLD ID | NA | |
| BIOPROJECT | PRJEB14916 | |
| MIGS 13 | Source Material Identifier | ACA-DC 2 |
| Project relevance | Dairy isolate |
Growth conditions and genomic DNA preparation
S. thermophilus ACA-DC 2 was grown in M17 medium (Biokar Diagnostics, Beauvais, France). For the isolation of the genomic DNA, 2 ml from an overnight culture incubated at 42 °C were used and the extraction procedure was performed according to the protocol of Pitcher et al. [15]. The purity and the concentration of the extracted DNA were measured with a UV-Vis spectrophotometer (Q5000, Quawell, San Jose, USA) while its integrity was evaluated electrophoretically in a 0.8% agarose gel.
Genome sequencing and assembly
Whole-genome sequencing was performed using the Illumina HiSeq2500 and the PacBio RSII platforms at BaseClear service laboratory for DNA-research (Leiden, The Netherlands). Paired-end sequence reads were generated using the Illumina HiSeq2500 system. FASTQ sequence files were obtained using the Illumina Casava pipeline v1.8.3. Initial quality assessment was based on data passing the Illumina Chastity filtering. Subsequently, reads containing adapters and/or PhiX control signal were removed using an in-house filtering protocol. The second quality assessment was based on the remaining reads using the FASTQC quality control tool v0.10.0 resulting in 4,403,680 reads.
The data collected from the PacBio RSII instrument were processed and filtered using the SMRT Analysis software suite. The Continuous Long Read data were filtered by Read-length (>50), Subread-length (>50) and Read quality (>0.75) resulting in 117,020 reads.
The quality of the Illumina FASTQ sequences was enhanced by trimming off low-quality bases using the program bbduk, which is part of the BBMap suite v34.46. The quality-filtered sequence reads were puzzled into a number of contig sequences. The analysis was performed using ABySS v1.5.1. The contigs were linked and placed into super-scaffolds based on the alignment of the PacBio CLR reads with BLASR [16]. The alignment was further used to estimate the orientation, order and distance between the contigs by the SSPACE-LongRead scaffolder v1.0 [17]. The gapped regions within the super-scaffolds were closed in an automated manner using GapFiller v1.10 [18]. The method takes advantage of the insert size between the Illumina paired-end reads. The assembly resulted in one circular chromosome of 1,731,838 bp.
Genome annotation
Prediction of genes was carried out with the online programs Prodigal [19], MetaGeneAnnotator [20] and FGENESB [21], for comparison and verification of the obtained results. Genome annotation was performed using RAST v2.0 [22]. Annotation anomalies, including pseudogenes, were identified using GenePRIMP [23]. All data acquired were combined and subjected to manual curation. The WebMGA server [24] and the EggNog v4.5 [25] were used for COG annotation, the Phobius web server was used for the identification of genes with transmembrane helices and genes with signal peptides [26] and the Pfam database was used for the identification of genes with Pfam domains [27]. Potential pathogenic features were identified using the MP3 tool [28]. The CRISPRs, the restriction-modification systems and the putative antimicrobial peptides were predicted using the CRISPRFinder web tool [29], the REBASE database [30] and BAGEL3 [31], respectively. The KODON software (Applied Maths NV, Sint-Martens-Latem, Belgium) was utilized for the visualization of synteny among the CRISPR regions of ACA-DC 2 and LMD-9 strains. The EDGAR server [32] was used for whole genome phylogeny and Venn diagram analysis. Circoletto [33] was employed for whole genome alignment among S. thermophilus strains. Finally, the genomic islands were identified through the IslandViewer 3 web-based resource [34].
Genome properties
The complete genome of S. thermophilus ACA-DC 2 consists of one circular chromosome containing 1,731,838 bp. The average GC content of the chromosome is 39.2%. A total of 1,850 genes were predicted after manual curation, including 1,556 protein-coding genes, 70 RNAs (56 tRNAs and 14 rRNAs) and 224 potential pseudogenes (Table 3). A circular map of the genome was generated using the CGView comparison tool [35] as shown in Fig. 3. Function was assigned to 1,182 genes (63.89%), while 1,318 genes (71.24%) had one or more conserved Pfam domains. The distribution of protein-coding genes into COG functional category is shown in Table 4. The analysis revealed that approximately 28.5% of the protein-coding genes do not have any described function.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 1,731,838 | 100.00 |
| DNA coding (bp) | 1,356,670 | 78.34 |
| DNA G + C (bp) | 679,104 | 39.21 |
| DNA scaffolds | 1 | 100.00 |
| Total genes | 1,850 | 100.00 |
| Protein coding genes | 1,556 | 84.11 |
| RNA genes | 70 | 3.78 |
| Pseudo genes | 224 | 12.11 |
| Genes in internal clusters | NA | NA |
| Genes with function prediction | 1,182 | 63.89 |
| Genes assigned to COGs | 1,327 | 71.73 |
| Genes with Pfam domains | 1,318 | 71.24 |
| Genes with signal peptides | 127 | 6.86 |
| Genes with transmembrane helices | 339 | 18.32 |
| CRISPR repeats | 2 |
Fig. 3.
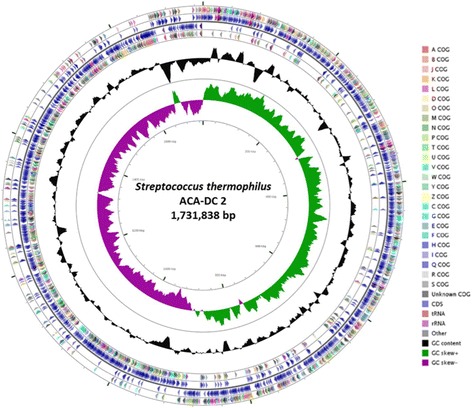
Circular map of S. thermophilus ACA-DC 2 genome features generated with the CGview tool. From periphery to center: Protein coding genes on forward strand colored by COG category assignment; Genes on forward strand; Protein coding genes on reverse strand colored by COG category assignment; Genes on reverse strand; GC content; GC skew; Genome region in kbp
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | %age | Description |
|---|---|---|---|
| J | 146 | 9.38 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.00 | RNA processing and modification |
| K | 89 | 5.72 | Transcription |
| L | 136 | 8.74 | Replication, recombination and repair |
| B | 0 | 0.00 | Chromatin structure and dynamics |
| D | 16 | 1.03 | Cell cycle control, Cell division, chromosome partitioning |
| V | 39 | 2.51 | Defense mechanisms |
| T | 43 | 2.76 | Signal transduction mechanisms |
| M | 80 | 5.14 | Cell wall/membrane biogenesis |
| N | 3 | 0.19 | Cell motility |
| U | 20 | 1.29 | Intracellular trafficking and secretion |
| O | 55 | 3.53 | Posttranslational modification, protein turnover, chaperones |
| C | 40 | 2.57 | Energy production and conversion |
| G | 66 | 4.24 | Carbohydrate transport and metabolism |
| E | 160 | 10.28 | Amino acid transport and metabolism |
| F | 67 | 4.31 | Nucleotide transport and metabolism |
| H | 49 | 3.15 | Coenzyme transport and metabolism |
| I | 33 | 2.12 | Lipid transport and metabolism |
| P | 67 | 4.31 | Inorganic ion transport and metabolism |
| Q | 13 | 0.84 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 63 | 4.05 | General function prediction only |
| S | 215 | 13.82 | Function unknown |
| - | 229 | 14.72 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
Main genome sequence characteristics
The genome of S. thermophilus ACA-DC 2 is the smallest one described to date among the fully sequenced strains of the species deposited in NCBI and it is approximately 200 kbp smaller than the larger described genome. The majority of potential pseudogenes encode hypothetical proteins, transposases and proteins involved in carbohydrate transport and metabolism. Analysis of the genome for virulence factors with the MP3 tool revealed a number of hits (data not shown). Detailed inspection of these hits with EDGAR demonstrated that several such genes are conserved among S. thermophilus strains indicating that it is rather unlikely to be related to pathogenicity, given the safe history of the species. The high percentage of pseudogenes along with the absence of typical virulence factors for streptococci suggest that the ACA-DC 2 strain evolved through genome decay during the adaptation to the rich in nutrients dairy environment [9, 11].
S. thermophilus ACA-DC 2 carries a complete lactose-galactose operon containing the galR, galK, galT, galE, galM, lacS and lacZ genes (STACADC2_1195-1189) and it is able to ferment lactose and galactose, the latter in a fairly slow rate (data not shown). It has been reported that fermentation of galactose is limited among the strains of S. thermophilus [11]. As mentioned above, several genes responsible for the transport and degradation of sugars, such as fructose, maltose and trehalose, have been identified as pseudogenes in the genome of ACA-DC 2, further supporting the specialization of the bacterium to catabolize lactose.
The proteolytic system of S. thermophilus ACA-DC 2 consists of several genes encoding aminopeptidases, such as pepA (STACADC2_1626), pepC (STACADC2_0202), pepF (STACADC2_0406), pepM (STACADC2_1333), pepN (STACADC2_0892), pepO (STACADC2_1656), pepP (STACADC2_1520), pepQ (STACADC2_0572), pepS (STACADC2_0058), pepT (STACADC2_0971), pepV (STACADC2_0960), and pepX (STACADC2_1446), one oligopeptide opp ABC transporter (STACADC2_1229-1233), four polar amino acid ABC transporters (STACADC2_0780-0782, STACADC2_0992-0995, STACADC2_1355-1358, STACADC2_1431-1433), two symporters for branched-chain amino acids (STACADC2_0872, STACADC2_1160), and two glutamine ABC transporters (STACADC2_0547-0548, STACADC2_1281-1282). Strain ACA-DC 2 lacks a cell wall-associated proteinase (PrtS). Although this gene may be important for optimal growth of S. thermophilus in milk, its absence is of minor significance when co-cultured with a proteolytic Lactobacillus delbrueckii subsp. bulgaricus strain, since the release of peptides by the latter is beneficial for the growth of S. thermophilus [10, 11].
Similar to other dairy bacteria, S. thermophilus ACA-DC 2 is able to synthesize exopolysaccharides (EPS) that may confer improved viscosity and texture to yogurt [4]. The EPS cluster is flanked by a deoD gene encoding a purine nucleoside phosphorylase (STACADC2_0949) and a pseudogene originally encoding a beta-glucosidase. Four of these genes, namely epsA (STACADC2_0948), epsB (STACADC2_0947), epsC (STACADC2_0946) and epsD (STACADC2_0945) are implicated in the regulation, polymerization, chain length and export of the EPS and are conserved among several S. thermophilus strains [36].
The genome analysis of strain ACA-DC 2 revealed a number of genes known to be responsive to unfavorable conditions prevailing during industrial applications. Among them we identified conserved heat shock genes like grpE, dnaK, dnaJ (STACADC2_0105-0107) and groES, groEL (STACADC2_0179-0180), genes encoding Clp proteases (STACADC2_0071, STACADC2_0315, STACADC2_0526, STACADC2_0544, STACADC2_1391), a proton translocating F0F1-ATPase system (STACADC2_0430-0437) and a P-type calcium pump ATPase (STACADC2_0983). The strain also harbors genes related to oxidative stress, namely a Mn-superoxide dismutase (STACADC2_0657), a glutathione reductase (STACADC2_0362), two thioredoxins (STACADC2_1043, STACADC2_1624), two thioredoxin reductases (STACADC2_1208, STACADC2_1429), a NADH oxidase (STACADC2_1113) and two sulfoxide reductases (STACADC2_1408, STACADC2_1159). Furthermore, the genome carries four putative antimicrobial peptides that need further investigation (STACADC2_0091, STACADC2_1453, STACADC2_1458 and STACADC2_1709).
Two candidate CRISPRs were detected in the chromosome of strain ACA-DC 2. Intriguingly, both CRISPRs contained only one spacer. One CRISPR was found surrounded by cas proteins (STACADC2_0849-0856) while the other was orphan. The CRISPR-cas system of strain ACA-DC 2 exhibited the same organization and high degree of identity to that described previously for strain LMD-9 (Fig. 4) [37]. The two CRISPR-cas systems differed mainly in the csm6 gene, which in the case of strain ACA-DC 2 is a potential pseudogene as well as in csm2 gene that seems to be distinct in the two strains. S. thermophilus LMD-9 carries three CRISPR-cas systems and the system that is similar to that of ACA-DC 2 carries the lowest number (three) of spacers. Combined these findings could indicate low activity or even inactivation of the entire CRISPR-cas system in strain ACA-DC 2. Another possibility that cannot be excluded concerns low exposure of strain ACA-DC 2 to foreign DNA. Of course, any deficiency in the activity of the CRISPR-cas system may be compensated by restriction-modification (RM) systems. Strain ACA-DC 2 carries four putative RM systems according to the REBASE database (data not shown) belonging to RM types I (STACADC2_0642, STACADC2_0645, STACADC2_0648), II (STACADC2_0597-0598), III (STACADC2_0788-0789) and IV (STACADC2_0626).
Fig. 4.
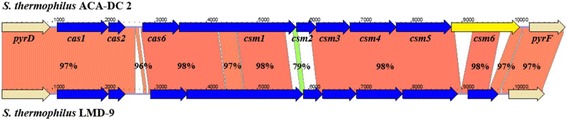
Synteny plot of the CRISPR loci between S. thermophilus strains ACA-DC 2 and LMD-9. The synteny of the two regions was calculated by the KODON software. In both strains the cas genes are denoted in blue. Gene csm6 in strain ACA-DC 2 is a potential pseudogene and it is denoted in yellow. The pyrD and pyrF genes colored in beige define the upstream and downstream limits of the CRISPR loci. Percentages displayed in the ribbon areas correspond to the % identity among the nucleotide sequences
Comparative genomic analysis, strain specific genomic features and genomic islands
Resolution of phylogenetic trees based on 16S rRNA gene sequences is limited due to high sequence identity, especially for strains of the same species. For this reason, we also performed whole genome phylogeny as implemented in EDGAR, using all available complete genomes of S. thermophilus. The phylogenetic tree produced revealed that S. thermophilus strains could be clustered in two distinct branches, the second of which could be also split in two sub-branches (Fig. 5). Strain ACA-DC 2 formed one of the branches along with strains CNRZ1066, LMG 18311, S9 and CS8. We chose strains ACA-DC 2, JIM 8232 and KLDS 3.1003 as representatives of each branch for comparative genomic analysis (Fig. 5). Whole genome alignments revealed extensive regions of high identity (>98%) among the genomes. However, regions of lower identity (between 80 and 98%) as well as strain specific regions were also identified. Using Venn diagram analysis as implemented in EDGAR, we determined a core genome of 1,303 genes among the three genomes as well as 137, 185 and 236 unique genes for strains ACA-DC 2, KLDS 3.1003 and JIM 8232, respectively.
Fig. 5.
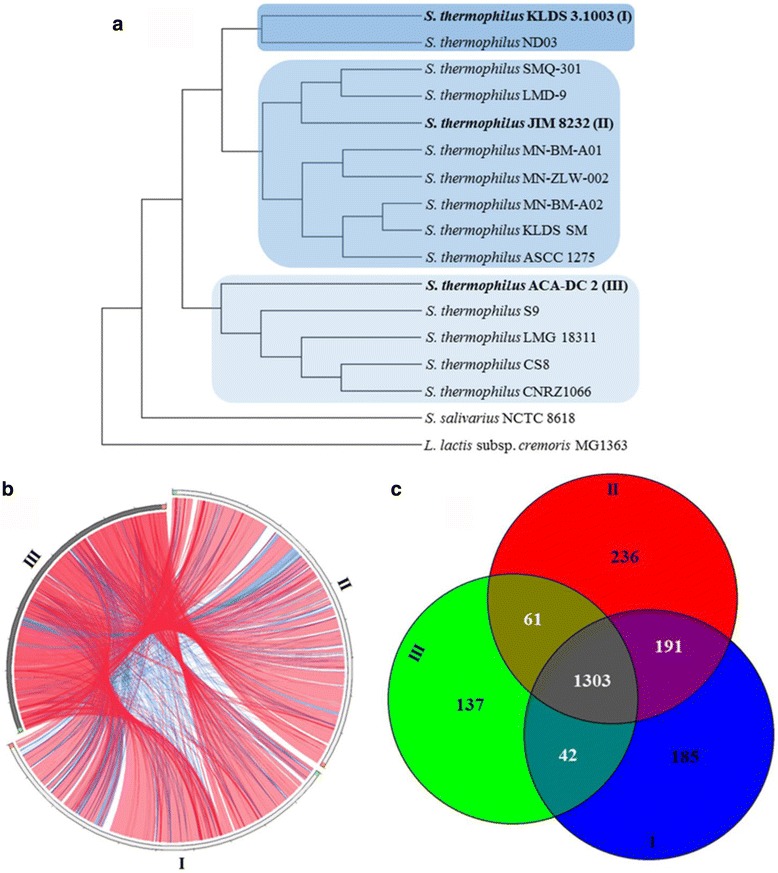
Comparative genomics of S. thermophilus strains. a Whole genome phylogeny of S. thermophilus strains with complete available genome sequences. The phylogenetic tree was calculated in EDGAR and it is presented as a cladogram ignoring branch length. The strains S. salivarius NCTC 8616 and Lactococcus lactis subsp. cremoris MG1363 were used as outgroups. Colored boxes indicate the three distinct S. thermophilus branches identified. Strains in each colored branch designated in bold were used for further comparative analysis. b Whole genome alignments of S. thermophilus strains KLDS 3.1003, JIM 8232 and ACA-DC 2 using Circoletto. Red and blue ribbons correspond to regions of >98% and 80–98% identity, respectively. White regions correspond to strain specific loci. c Venn diagram analysis of S. thermophilus strains KLDS 3.1003, JIM 8232 and ACA-DC 2, as implemented in EDGAR
The 137 unique genes of strain ACA-DC 2 were found to be involved in diverse functions (Fig. 6). At least some of those genes may be the result of horizontal gene transfer (HGT). HGT acquired genes may play a role in the technological properties of S. thermophilus strains [11]. Another analysis that may also reveal regions of HGT in the bacterial chromosome is the identification of GIs [38]. Twelve integrated GIs were predicted in the genome of S. thermophilus ACA-DC 2 (Fig. 6), containing a total of 213 genes also involved in diverse functions (Fig. 6). Detailed analysis of genes either unique or in the GIs could relate some of them to important technological traits. For example, we determined genes coding for cold shock proteins CspA and CspG (STACADC2_0749-0750), acid resistance locus arl7 (STACADC2_0743), putative bacteriocin peptides (STACADC2_1453 and STACADC2_1458) and a type I RM system (STACADC2_0642, STACADC2_0645, STACADC2_0648). A putative agmatinase gene (STACADC2_0818) that may play a role to protocooperation of S. thermophilus and L. bulgaricus during polyamine metabolism, was also detected in ACA-DC 2 strain [10]. Furthermore, genes implicated in zinc and heavy metals transport (STACADC2_0165-0166, STACADC2_0752), in DNA repair and metabolism (STACADC2_1696, STACADC2_1716, STACADC2_1719, STACADC2_1754) as well as several ribosome binding proteins, were also identified (STACADC2_0137, STACADC2_1568-1569, STACADC2_1667, STACADC2_1669-1671, STACADC2_1675-1695, STACADC2_1717, STACADC2_1732-1733, STACADC2_1752, STACADC2_1755).
Fig. 6.
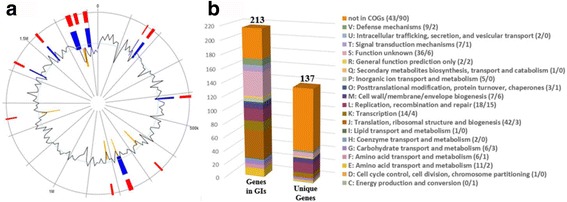
Additional genomic features of S. thermophilus ACA-DC 2. a Circular map of the S. thermophilus ACA-DC 2 genome as generated by IslandViewer 3. Highlighted regions correspond to GIs. GIs are colored within the circular map according to the prediction method used: five GIs in orange and eight GIs in blue were predicted with SIGI-HMM and IslandPath-DIMOB, respectively. Twelve integrated GIs are presented on the periphery in red. The black line plot represents the GC content (%) of the genomic sequence. b Distribution of genes in GIs and unique genes of S. thermophilus ACA-DC 2 into COG categories
Conclusions
The genome of S. thermophilus ACA-DC 2 presents characteristics in accordance with its adaptation to the milk environment including a high percentage of pseudogenes and absence of pathogenic features. Our analysis revealed that the strain carries genes involved in lactose and galactose catabolism and protein degradation that may accommodate its growth during milk fermentation. Stress response related genes that may contribute to survival under technological hurdles were also detected. Whole genome phylogeny suggested that S. thermophilus strains may diversify in three phylogenetic clades. Comparative analysis of genomes representative of each clade, including strain ACA-DC 2, revealed a number of unique genes for the latter. Furthermore, certain unique genes or genes belonging to GIs could be related to technological properties important for starter cultures. Theoretically, such genes could have been acquired through HGT. These findings render S. thermophilus ACA-DC 2 an appropriate candidate for use in the production of fermented dairy products.
Acknowledgements
We would like to thank Dr. Nikos Kyrpides at the Joint Genome Institute (United States Department of Energy) for the analysis of the ACA-DC 2 genome with the GenePrimp server. Furthermore we would like to thank Prof. Konstantinos Fasseas at the Faculty of Crop Science (Agricultural University of Athens) for the transmission electron microscopy experiment.
Funding
The present work was co-financed by the European Social Fund and the National resources EPEAEK and YPEPTH through the Thales project.
Authors’ contributions
VA performed genome analysis and participated in the writing of the manuscript. MK performed genome analysis and participated in the writing of the manuscript. JB performed genome analysis. BP performed genome analysis. ET characterized strain ACA-DC 2, conceived the project and participated in the writing of the manuscript. KP conceived the project, performed genome analysis and participated in the writing of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- ACA-DC
Agricultural College of Athens - Dairy Collection
- CLR
Continuous long read
- COG
Clusters of Orthologous Groups
- CRISPR
Clustered regularly interspaced short palindromic repeats
- ENA
European nucleotide archive
- EPS
Exopolysaccharide
- GenePRIMP
Gene prediction improvement pipeline
- GI
Genomic Island
- GRAS
Generally regarded as safe
- HGT
Horizontal gene transfer
- PTA
Phosphotungstic acid
- RAST
Rapid annotation using subsystem technology
- RM
Restriction-modification
- SMRT
Single molecule real time
References
- 1.Papadimitriou K, Pot B, Tsakalidou E. How microbes adapt to a diversity of food niches. Curr Opin Food Sci. 2015;2:29–35. doi: 10.1016/j.cofs.2015.01.001. [DOI] [Google Scholar]
- 2.Poornachandra Rao K, Chennappa G, Suraj U, Nagaraja H, Charith Raj AP, Sreenivasa MY. Probiotic potential of Lactobacillus strains isolated from sorghum-based traditional fermented food. Probiotics Antimicrob Proteins. 2015;7:146–56. [DOI] [PubMed]
- 3.Topisirovic L, Kojic M, Fira D, Golic N, Strahinic I, Lozo J. Potential of lactic acid bacteria isolated from specific natural niches in food production and preservation. Int J Food Microbiol. 2006;112:230–5. doi: 10.1016/j.ijfoodmicro.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Broadbent JR, McMahon DJ, Welker DL, Oberg CJ, Moineau S. Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review. J Dairy Sci. 2003;86:407–23. [DOI] [PubMed]
- 5.Leroy F, De Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Tech. 2004;15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 6.Krzysciak W, Pluskwa KK, Jurczak A, Koscielniak D. The pathogenicity of the Streptococcus genus. Eur J Clin Microbiol Infect Dis. 2013;32:1361–76. [DOI] [PMC free article] [PubMed]
- 7.Iyer R, Tomar SK, Uma Maheswari T, Singh R. Streptococcus thermophilus strains: multifunctional lactic acid bacteria. Int Dairy J. 2010;20:133–41.
- 8.Rossi F, Marzotto M, Cremonese S, Rizzotti L, Torriani S. Diversity of Streptococcus thermophilus in bacteriocin production; inhibitory spectrum and occurrence of thermophilin genes. Food Microbiol. 2013;35:27–33. [DOI] [PubMed]
- 9.Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol. 2004;22:1554–8. [DOI] [PMC free article] [PubMed]
- 10.Goh YJ, Goin C, O’Flaherty S, Altermann E, Hutkins R. Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb Cell Fact.2011;10(Suppl 1):S22. [DOI] [PMC free article] [PubMed]
- 11.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, et al. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev.2005;29:435–63. [DOI] [PubMed]
- 12.Prajapati JB, Nathani NM, Patel AK, Senan S, Joshi CG. Genomic analysis of dairy starter culture Streptococcus thermophilus MTCC 5461. J Microbiol Biotechnol. 2013;23:459–66. [DOI] [PubMed]
- 13.Tsakalidou E, Manolopoulou E, Kabaraki E, Zoidou E, Pot B, Kersters K, et al. The combined use of whole cell protein extracts for the identification (SDS-PAGE) and enzyme activity screening of lactic acid bacteria isolated from traditional Greek dairy products. System Appl Microbiol. 1994;17:444–58. doi: 10.1016/S0723-2020(11)80062-7. [DOI] [Google Scholar]
- 14.Tsakalidou E, Zoidou E, Kalantzopoulos G. SDS-Polyacrylamide gel electrophoresis of cell proteins from Lactobacillus delbreuckii subsp. bulgaricus and Streptococcus salivarius subsp. thermophilus strains isolated from yoghurt and cheese. Milchwissenschaft. 1992;47:296–8.
- 15.Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–6. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- 16.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. [DOI] [PMC free article] [PubMed]
- 19.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–96. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW, editor. Metagenomics and its applications in agriculture, biomedicine and environmental studies. New York: Nova Science Publishers; 2011. pp. 61–78. [Google Scholar]
- 22.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, et al. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods. 2010;7:455–7. doi: 10.1038/nmeth.1457. [DOI] [PubMed] [Google Scholar]
- 24.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–93. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007;35:W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–85. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta A, Kapil R, Dhakan DB, Sharma VK. MP3: a software tool for the prediction of pathogenic proteins in genomic and metagenomic data. PLoS One. 2014;9:e93907. doi: 10.1371/journal.pone.0093907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–6. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013;41:W448–53. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blom J, Kreis J, Spanig S, Juhre T, Bertelli C, Ernst C, et al. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016;44:W22–8. doi: 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darzentas N. Circoletto: visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–1. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- 34.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, et al. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 2015;43:W104–8. doi: 10.1093/nar/gkv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–4. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Z, Chen X, Wang J, Zhao W, Shao Y, Wu L, et al. Complete genome sequence of Streptococcus thermophilus strain ND03. J Bacteriol.2011;193:793–4. [DOI] [PMC free article] [PubMed]
- 37.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–12. [DOI] [PMC free article] [PubMed]
- 38.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–93. [DOI] [PMC free article] [PubMed]
- 39.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibbons NE, Murray RGE. Proposals concerning the higher taxa of Bacteria. Int J Syst Bacteriol. 1978;28:1–6. doi: 10.1099/00207713-28-1-1. [DOI] [Google Scholar]
- 42.Garrity G, Holt J. The road map to the manual. In: Boone D, Castenholz R, Garrity G, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2001. pp. 119–66. [Google Scholar]
- 43.Ludwig W, Schleifer KH, Whitman WB. Class I. Bacilli class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, et al., editors. Bergey’s manual of systematic bacteriology. vol. 3. 2nd ed. New York: Springer; 2009;19–20.
- 44.Euzéby J. List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol. 2010;60:469–72. doi: 10.1099/ijs.0.022855-0. [DOI] [PubMed] [Google Scholar]
- 45.Ludwig W, Schleifer KH, Whitman WB. Order II. Lactobacillales ord. nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, et al., editors. Bergey’s manual of systematic bacteriology. vol. 3. 2nd ed. New York: Springer; 2009;464.
- 46.Goodfellow M, Kämpfer P, De Vos P, Rainey F, Schleifer KH, Whitman WB. Family VI. Streptococcaceae fam. nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, et al., editors. Bergey’s manual of systematic bacteriology. vol. 3. 2nd ed. New York: Springer; 2009;655.
- 47.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 48.Deibel RH, Seeley HW. Family II. Streptococcaceae. In: Buchanan RE, Gibbons NE, editors. Bergey’s manual of determinative bacteriology. 8. Baltimore: The Williams and Wilkins Co; 1974. pp. 490–515. [Google Scholar]
- 49.Whiley RA, Hardie JM. Genus I. Streptococcus gen. nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, et al., editors. Bergey’s manual of systematic bacteriology. vol. 3. 2nd ed. New York: Springer; 2009. p. 655–710.
- 50.Rosenbach FJ. In: Bergmann JF, editor. Microorganismen bei den Wund-Infections-Krankheiten des Menschen. Wiesbaden: JF Bergmann; 1884;1–122.
- 51.Schleifer KH, Ehrmann M, Krusch U, Neve H. Revival of the species Streptococcus thermophilus (ex OrIa-Jensen, 1919) nom. rev. System App Microbiol. 1991;14:386–8.
- 52.Orla-Jensen S. The lactic acid bacteria. 1919. pp. 1–196. [Google Scholar]
- 53.Beal C, Louvet P, Corrieu G. Influence of controlled pH and temperature on the growth and acidification of pure cultures of Streptococcus thermophilus 404 and Lactobacillus bulgaricus 398. Appl Microbiol Biotechnol.1989;32:148–54.
- 54.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]


