Abstract
Background
Coagulase-negative staphylococci have become increasingly recognized as the etiological agent of some infections. A significant characteristic of coagulase-negative staphylococci especially strains isolated from animals and clinical samples is their resistance to routinely used antibiotics although, resistant strains isolated from fermented foods have not been fully reported.
Methods
A total of two hundred and fifty-five CoNS isolates were subjected to antimicrobial susceptibility test using the disc diffusion technique. The minimum inhibitory concentration of the isolates to the tested antibiotics was determined using the microbroth dilution method. Methicillin resistant strains were confirmed by detection of methicillin resistant genes (mecA) and also employing cefoxitin screening test.
Results
The isolates were confirmed to be methicillin resistant by the detection of mecA genes and the cefoxitin screening test. The isolates demonstrated appreciable resistance to ampicillin (86.7%), sulfomethoxazole–trimethoprim (74.9%), amoxicillin–clavulanic acid (52.5%) and oxacillin (35.7%). Methicillin resistance was exhibited by 13 out of the 255 isolates although no mecA gene was detected. It was also observed that the methicillin resistant isolates were prevalent in these traditional foods; iru, kindirmo, nono and wara.
Conclusion
This study has ameliorated the incidence of multiple antibiotic resistant coagulase-negative staphylococci in Nigerian fermented foods and if not tackled adequately might lead to horizontal transfer of antibiotic resistance from food to man.
Keywords: Antibiotic resistance, Coagulase-negative staphylococci, Fermented foods, Methicillin resistance, mecA gene
Background
Coagulase-negative staphylococci were previously dismissed as contaminants and were found to occur mostly in hospitalized patients, individuals suffering from nosocomial infections and infections arising from the use of catheter or other intra-uterine devices however, it has been shown that CoNS from fermented foods also exhibit virulent traits [1]. The major challenge of CoNS-related infections has been the difficulty in therapy due to antimicrobial resistance. Antimicrobial agents used in therapy and as feed supplements to promote growth in food animals may increase the spread of drug-resistant bacteria. Such bacteria may contaminate milk or meat and are subsequently found in fermented food made of such raw material [2]. The levels of antibiotic resistant infections in the developing world have increased steadily in the last few decades as a result of combination of microbial characteristics and the selective pressure of antimicrobial use [3]. Microbial mechanisms of overcoming the activities of antimicrobial agents include the production of structure-altering or inactivating enzymes (e.g. beta-lactamase or amino glycoside-modifying enzymes), alteration of penicillin-binding proteins or other cell-wall target sites, altered DNA gyrase targets, permeability mutations, active efflux and ribosomal modification [4–6]. Multidrug-resistant bacteria in both the hospital and community environment are important concern to the clinician, as it is the major cause of failure in the treatment of infectious diseases, increased morbidity, and mortality and the evolution of new pathogens [7, 8].
Penicillin was initially the drug of choice for treatment of infections caused by Staphylococcus however, penicillin resistance in CoNS became very high since 1968 [9]. Nowadays, resistance is around 91% in clinical strains [10]. Two mechanisms confer penicillin resistance in staphylococci; the first and the most important is the production of β-lactamase which inactivates penicillin by the hydrolysis of its β-lactam ring. The second is primarily associated with human isolates and confers resistance due to a penicillin-binding protein, PBP2a, encoded by mecA [2]. The blaZ has also been identified as the cause of penicillin resistance among coagulase-negative staphylococci (CoNS) suggesting that blaZ is one of the main mechanism of penicillin resistance in staphylococci [2]. Methicillin resistance in Staphylococcus is caused by the expression of PBP2a encoded by the mecA gene [11]. Resistance of staphylococci to methicillin and all β-lactam antibiotics is associated with the low affinity of a penicillin-binding protein, PBP2a, which is not present in susceptible staphylococci [12–17]. This protein is encoded by the mecA gene, which is located in the mec region in which the DNA is of foreign origin [18]. There is evidence of horizontal transfer of SCC cassette between staphylococcal species [19] which implies that CoNS could serve as a reservoir for the spread of resistance genes. Transfer of resistance genes between CoNS and S. aureus has been reported thus indicating that CoNS may act as a resistance gene reservoir for S. aureus. It is thus possible that the different species of staphylococci that are present in the same microenvironment, for example on the skin of dairy cows can exchange mecA and blaZ, if the appropriate bacterial factors are met [2].
In this study, the incidence of antibiotic resistance against 9 antibiotics among 255 strains of coagulase-negative staphylococci of fermented food associated CoNS were investigated using disc diffusion technique according to the CLSI guidelines. The antibiotic resistant phenotypes were confirmed molecularly by the detection of mecA genes and cefoxitin screening test.
Methods
CoNS strains used in this study
In total, 255 CoNS strains were used in this study. The strains were isolated from six different Nigerian fermented foods, including kindirmo (66), nono (44) iru (58), wara (32), ogi (28) and kunu (27). The isolates were identified using both conventional and molecular methods employing 16S rRNA sequencing [20].
Antibiotic susceptibility testing
In vitro susceptibility of the test isolates to the antibiotics was determined using Kirby-Bauer disc diffusion [21]. A sterile wire loop was used to pick a discrete colony of the 18 h old culture of each of the test isolate cultured on mannitol salt agar (MSA) and used to inoculate sterile brain heart infusion broth inside a test tube and incubated at 37 °C for 4 h. The inoculum was standardized using the 0.5 McFarland turbidity standard which corresponds to 1.5 × 108 cfu/ml of cells. A sterile cotton swab was dipped into the adjusted suspension and excess inoculum was removed by pressing the swab firmly on the inside wall of the tube. The dried surface of a Mueller–Hinton agar plate was inoculated by streaking the swab over the entire surface. This procedure was repeated by streaking two more times, rotating the plate approximately 60º each time to ensure an even distribution of inoculum. The antimicrobial discs were placed firmly on the surface of the inoculated agar plate using sterile forceps. The plates were left for 1 h after which they were incubated at 35 °C for 18 h. After 16–18 h of incubation, the plates were examined and the diameters of the zones of inhibition were measured. The discs used were ampicillin (30 µg), amoxicillin-clavulanic acid (30 µg), cefotaxime (30 µg), oxacillin (1 µg), ciprofloxacin (5 µg), trimethoprim–sulphomethaxazole (5 µg), erythromycin (25 µg), gentamycin (10 µg) and ofloxacin (5 µg). All the antibiotic discs were procured from Oxoid, Germany. The results were classified as susceptible, intermediate, or resistant according to the approved guidelines of the Clinical and Laboratory Standards Institute [22].
Determination of minimum inhibitory concentration (MIC) using the broth micro-dilution method
The method by [21] was employed. A 96-well microtitre plate was used. Twofold serial dilutions of the different antibiotics were prepared and dispensed into the microtitre plates. The antimicrobial solutions were prepared at twice the desired final concentration and the wells filled with 0.05 ml of the antibiotic instead of 0.1 ml. Each tray labeled had a growth control well and a sterility (uninoculated) well.
The inoculum used for the broth micro-dilution was prepared using the direct colony suspension method. An 18 h old culture of CoNS was grown on blood agar. Distinct colonies were picked and each inoculated into 5 ml of Mueller–Hinton broth in a test tube. The broth culture was incubated at 35 °C for 4 h. The turbidity of the actively growing broth culture was adjusted with sterile broth using 0.5 McFarland standard. The resulting suspension contained approximately 1–2 × 108 cfu/ml. 2 ml of the suspension was dispensed into 38 ml of water (1:20 dilution). The prong of the inoculator was used to transfer 0.01 ml (1:10 dilution) into each well. The MIC panel was inoculated carefully to avoid splashing from one well to another. The microdilution trays were incubated inside a plastic bag at 35 ± 2 °C for 16–20 h in an ambient air incubator within 15 min of adding the inoculum. The amount of growth in the wells containing the antimicrobial agent was compared with the amount of growth in the growth control wells (no antimicrobial agent) used in each set of tests when determining the growth end points. A test was considered valid when acceptable growth was ≥2 mm turbidity at the bottom of the well or when a definite turbidity was observed.
Detection of methicillin resistance genes (mecA)
The methicillin resistant genes present in the coagulase-negative staphylococci strains were detected by polymerase chain reaction according to the method of [23]. DNA was extracted using the QIA Amp mini kit (QIAgen). Polymerase chain reaction for detection of the gene mecA (513 bp) was carried out using the following primers: A22f (5′ AAA ATC GAT GGT AAA GGT TGG C 3′) and A22r (AGT TCT GCA GTA CCG GAT TTG C) as described by [11]. Amplification cycles for mecA was carried out according to the method of [23] Considering 40 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min with a final extension of 72 °C for 5 min. Staphylococcus aureus ATCC43300 mecA + was used as positive control [21]. The amplicons were evaluated by agarose gel electrophoresis followed by staining in ethidium bromide (0.5 mg/ml), visualized on UV transilluminator (UVP, Inc USA) and documented by the program QuantiOne (BioRad) using molecular weight markers of 100 bp (Fermentas ®).
Results
A total of 221 (86.7%) of the isolates were resistant to ampicillin, however majority of the resistant CoNS occurred in wara (93.8%), nono (88.6%), kindirmo (92.4%) and iru (86.2%). 74.9% of the CoNS isolates were resistant to trimethoprim-sulfamethoxazole with high incidence in iru (84.5%), wara (84.4%) and kindirmo (72.7%). The highest resistance of amoxicillin-clavulanic acid was noted in CoNS isolated from ogi (60.7%), iru (60.3%), nono and wara (59.4%). The highest oxacillin resistance isolates were from ogi (42.9%), nono (40.9%) and wara (43.8%). The resistance to the other antibiotics cefotaxime, ciprofloxacin, erythromycin, gentamycin and ofloxacin were not as high as the other antibiotics as shown in Table 1.
Table 1.
Distribution and Percentage Antimicrobial Resistance of Coagulase-Negative Staphylococci from Fermented Food Samples
| Antibiotics | % Resistance of CoNS from foods | ||||||
|---|---|---|---|---|---|---|---|
| Total | Iru | Ogi | Nono | Kindirmo | Kunu zaki | Wara | |
| (255) | n = 58 | n = 28 | n = 44 | n = 66 | n = 27 | n = 32 | |
| Ampicillin | 221 (86.7%) | 50 (86.2%) | 21 (75%) | 39 (88.6%) | 61 (92.4%) | 20 (74.1%) | 30 (93.8%) |
| Trimethoprim–sulfamethoxazole | 191 (74.9%) | 49 (84.5%) | 18 (64.3%) | 31 (70.5%) | 48 (72.7%) | 18 (66.7%) | 27 (84.4%) |
| Amoxicillin–clavulanic acid | 134 (52.5%) | 35 (60.3%) | 17 (60.7%) | 26 (59.1%) | 29 (43.9%) | 8 (29.6%) | 19 (59.4%) |
| Cefotaxime | 9 (3.5%) | 2 (3.4%) | 0 (0%) | 3 (6.8%) | 2 (3.0%) | 1 (3.7%) | 1 (3.1%) |
| Oxacillin | 91 (35.7%) | 17 (29.3%) | 12 (42.9%) | 18 (40.9%) | 21 (31.8%) | 9 (33.3%) | 14 (43.8%) |
| Ciprofloxacin | 61 (23.9%) | 12 (20.7%) | 3 (10.7%) | 10 (22.7%) | 18 (27.3%) | 5 (18.5%) | 13 (40.6%) |
| Erythromycin | 40 (15.7%) | 8 (13.8%) | 2 (7.1%) | 10 (22.7%) | 11 (16.7%) | 0 (0%) | 9 (28.1%) |
| Gentamicin | 29 (11.4%) | 4 (6.9%) | 2 (7.1%) | 9 (20.5%) | 7 (16.7%) | 0 (0%) | 7 (21.9%) |
| Ofloxacin | 18 (7.1%) | 2 (3.4%) | 1 (3.6%) | 4 (9.1%) | 4 (6.1%) | 1 (3.7%) | 6 (18.8%) |
Table 2 shows the resistance phenotype of the CoNS species. Thirty-four (13%) of the isolates were not resistant to any of the antibiotic tested. CoNS species having resistance phenotype to only ampicillin and trimethoprim-sulfamethoxazole were only 57 in number. Staphylococcus epidermidis (92%) demonstrated the highest resistance to ampicillin while S. caprae (69%) had the least percentage of resistance to ampicillin. For trimethoprim-sulfamethoxazole, the highest resistance of 81% was recorded in S. xylosus with S. caprae having the least resistance (53%) to the antibiotic. Staphylococcus simulans (68%) recorded the highest resistance to amoxicillin-clavulanic acid while the least resistance was shown in S. epidermidis (41%). Oxacillin resistance in S. xylosus was 32% which was the highest and the least was in S. kloosii (14%). Staphylococcus caprae exhibited the highest resistance to ciprofloxacin, ofloxacin, gentamycin, erythromycin and cefotaxime as shown in Fig. 1.
Table 2.
Phenotypic antimicrobial resistance patterns of CoNS species from fermented food samples
| Profile | Resistance phenotypes | Number of strains | Number of antibiotic classes |
|---|---|---|---|
| 1 | None | 34 | 0 |
| 2 | AMP | 30 | 1 |
| 3 | AMP, SXT | 57 | 2 |
| 4 | AMP, SXT, AMC | 43 | 2 |
| 5 | AMP, SXT, AMC, OX | 30 | 2 |
| 6 | AMP, SXT, AMC, OX, CIP | 21 | 3 |
| 7 | AMP, SXT, AMC, OX, CIP, E | 11 | 4 |
| 8 | AMP, SXT, AMC, OX, CIP, E, CN | 10 | 5 |
| 9 | AMP, SXT, AMC, OX, CIP, E, CN, OFX | 10 | 5 |
| 10 | AMP, SXT, AMC, OX, CIP, E, CN, OFX, CTX | 9 | 5 |
AMP ampicillin, SXT sulphomethoxazole–trimethoprim, AMC Amoxicillin–clavulanic acid, OX oxacillin, CIP ciprofloxacin, E erythromycin, CN gentamicin, OFX ofloxacin, CTX cefotaxime
Fig. 1.
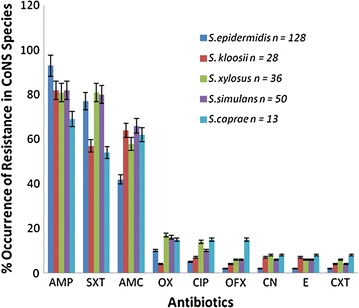
Percentage occurrence of antibiotic resistance in CoNS Species. Error bars represent standard deviation of mean of replicate determinations
Table 3 shows the minimum inhibitory concentration (MIC) distribution of the CoNS species. Based on the Clinical and Laboratory Standards Institute (CLSI) guidelines for MIC reading, the percentage of CoNS species resistant to the ampicillin, trimethoprim-sulfomethoxazole, amoxicillin-clavulanic acid and oxacillin were 85.5, 67.8, 49.8 and 25.9% respectively.
Table 3.
Minimum inhibitory concentration (MIC) of antibiotics against CoNS isolated from fermented food samples
| Antimicrobial agents | Numbers of CoNS with MICa | Resistant isolates (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | ||
| Ampicillin | 24 | 13 | 11 | 62 | 55 | 27 | 0 | 0 | 43 | 18 | 2 | 218 (85.5%) |
| Trimethoprim–sulfomethoxazole | 11 | 37 | 19 | 15 | 0 | 0 | 0 | 0 | 0 | 70 | 103 | 173 (67.8%) |
| Amoxicillin–clavulanic acid | 101 | 0 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 87 | 127 (49.8%) |
| Oxacillin | 53 | 136 | 0 | 0 | 0 | 47 | 0 | 0 | 19 | 0 | 0 | 66 (25.9%) |
| Ciprofloxacin | 84 | 43 | 14 | 0 | 52 | 0 | 7 | 24 | 9 | 9 | 13 | 62 (24.3%) |
| Ofloxacin | 21 | 12 | 22 | 69 | 121 | 0 | 0 | 0 | 0 | 0 | 10 | 10 (3.9%) |
| Gentamicin | 15 | 151 | 16 | 26 | 10 | 0 | 21 | 7 | 0 | 0 | 0 | 37 (14.5%) |
| Erythromycin | 98 | 91 | 15 | 0 | 0 | 0 | 0 | 6 | 0 | 40 | 0 | 46 (18.0%) |
| Cefotaximeb | 0 | 181 | 20 | 0 | 0 | 49 | 0 | 0 | 0 | 3 | 2 | – |
aBased on the CLSI breakpoints. MICs indicative for susceptible isolates are displayed on a white background, those for intermediate on a bold and those for resistant on a italics
bSusceptility of staphylococci to cefotaxime may be detected from testing only penicillin and either cefoxitin or oxacillin
Cefoxitin screening test was carried out on the isolates so as to establish their status as methicillin resistant strains. Thirteen (5.1%) of the tested isolates were positive to cefoxitin screening test and most of them were multidrug resistant with the highest occurrence of the methicillin resistant species in nono. The species were S. kloosii (KIL 4), S. xylosus (WAIL 3, KIL 2 and WAJ 5), S. epidermidis (OGIL 3, IRIL 7, NOMA 10, NOL 3 and IRIL 5), S. simulans (NOJ 6 and KIM 5) and S. caprae (NOMA 5 and NOMA 6) as indicated in Table 4. The molecular expression for mecA genes revealed that none of the isolates possessed the resistant genes (Fig. 2).
Table 4.
Coagulase negative Staphylococci isolates showing methicillin resistance using cefoxitin screening
| ID | Source | C–S | Resistance phenotype |
|---|---|---|---|
| S. xylosus KIL 2 | Kindirmo | + | AMP, OX, SXT, AMC, CIP, E, CXT, CN. OFX |
| S. xylosus WAIL 3 | Wara | + | AMP, OX, SXT, CIP, E, CXT, CN. OFX |
| S. kloosii KIL 4 | Kindirmo | + | AMP, OX, SXT, AMC, CIP, E, CN OFX |
| S. epidermidis OGIL 3 | Ogi | + | AMP, OX, AMC, CIP, E, OFX |
| S. epidermidis IRIL 7 | Iru | + | AMP, OX, SXT, AMC, CIP, CN OFX |
| S. epidermidis NOMA 10 | Nono | + | AMP, OX, SXT, AMC, CIP, E, CXT, CN OFX |
| S. simulans NOJ 6 | Nono | + | AMP, OX, CIP, E, CXT, OFX |
| S. simulans KIM 5 | Kindirmo | + | AMP, OX, SXT, AMC, CIP, E, |
| S. xylosus WAJ 5 | Wara | + | AMP, OX, SXT, AMC, E, CXT |
| S. caprae NOMA 5 | Nono | + | AMP, OX, SXT, AMC, CIP, E, CXT, OFX |
| S. caprae NOMA 6 | Nono | + | AMP, OX, SXT, CIP, E, CN OFX |
| S. epidermidis NOL 3 | Nono | + | AMP, OX, SXT, AMC, E, CXT CN OFX |
| S. epidermidis IRIL 5 | Iru | + | AMP, OX, SXT, CIP, E, CN OFX |
Fig. 2.
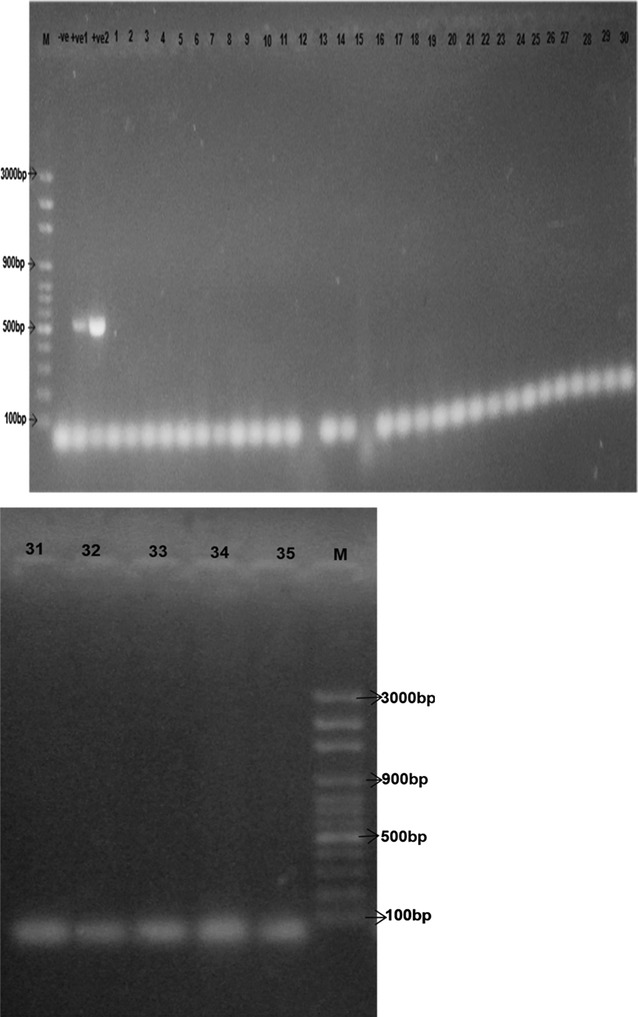
Gel electrophoresis micrograph of PCR product screen for mecA from extracted DNA (1–35) Left to right: M = O’GeneRuler 100 bp plus molecular weight marker (Thermo Scientific Fermentas™), Well 2 = Negative control for mecA gene, well 3 = Positive control 2, well 4 = Positive control 2 for mecA gene, wells 5–30 = Isolates 1–30. Weight of mecA gene = 513 bp. Positive isolates- none
Discussion
Majority of the CoNS strains exhibited resistance to ampicillin, amoxicillin-clavulanic acid, sulphomethoxazole–trimethoprim, and oxacillin. A large percentage of the strains were susceptible to the antibiotics ciprofloxacin, erythromycin, gentamicin, cefotaxime and ofloxacin. Staphylococcus epidermidis demonstrated the highest resistance to ampicillin while S. xylosus exhibited the highest resistance to sulfomethoxazole–trimethoprim. The highest percentage resistance to amoxicillin-clavulanic acid was demonstrated by S. simulans. Oxacillin resistance was highly demonstrated by S. xylosus. The resistance exhibited by a large percentage of CoNS to these routinely used antibiotics in treatment of staphylococcal infections necessitates the search for newer and more effective antibiotics against this group of organisms.
There was discrepancy between the detection of methicillin resistance phenotypically using cefoxitin screening and the absence of mecA gene in the CoNS strains. This may be attributed to methicillin resistance being caused by other mechanisms other than expression of mecA gene [24]. Also the sensitivity of PCR in the detection of mecA may have been compromised by the presence of PCR inhibitors or other physical factors [25, 26]. The work by [27] showed that out of 15 isolates showing oxacillin resistant phenotype, only one possessed the mecA gene, it was noted that there were unusual methicillin resistant CoNS that have a resistance mechanism other than the production of PBP2a and they have been reported as borderline methicillin resistant strains [28]. The borderline methicillin resistant strains are resistant to oxacillin due to their plasmid borne determinants including hyperproduced penicillinases, genes conferring resistance to cadmium or other gene products [29, 30]. It is also possible that the mecA negative oxacillin resistant CoNS may possess mecA alleles which could not be detected by the primers used in this study. Many CoNS strains also show diversity in mecA sequences and have different impact on β-lactam resistance.
Conclusions
The high percentage of antimicrobial resistance demonstrated by the strains shows that food may serve as reservoirs for antibiotic resistance and allow for horizontal gene transfer from farm animals or their products to humans. Indigenous fermented food products may represent a critical risk for transfer of antimicrobial resistance to humans. As a consequence, transfer of antimicrobial resistance genes between bacteria after ingestion by humans may occur. Antimicrobial resistant CoNS present in food constitute a direct risk to public health as they increase the gene pool from which pathogenic bacteria can pick up resistance traits. The resistance of the organisms to routinely used antimicrobials also calls for the search for new antimicrobials and more effective management of diseases caused by CoNS in the event of an outbreak.
Authors’ contributions
FP carried out the laboratory analysis, OS was a major contributor in writing the manuscript. Both authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Funding
This research was self funded.
Abbreviations
- CoNS
coagulase-negative staphylococci
- mecA
methicillin resistant gene
- PBP2a
penicillin binding protein
- DNA
deoxyribonucleic acid
- CLSI
Clinical and Laboratory Standards Institute
- MIC
minimum inhibitory concentration
Contributor Information
P. T. Fowoyo, Phone: +234-706-803-7034, Email: seunpt@yahoo.com
S. T. Ogunbanwo, Email: topzybanwo@yahoo.com
References
- 1.Fowoyo PT, Ogunbanwo ST. Occurrence and characterisation of coagulase-negative staphylococci from Nigerian traditional fermented foods. Food Sci Qual Manag. 2016;50:49–55. [Google Scholar]
- 2.Normanno G, Salandra GL, Dambrosio A, Quaglia NC, Corrente M, Parisi A, Olsen JE, Christensen H, Aarestrup F. Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J Antimicrob Chemother. 2006;57(3):450–460. doi: 10.1093/jac/dki492. [DOI] [PubMed] [Google Scholar]
- 3.Blondeau JM, Tillotson GS. Antimicrobial susceptibility patterns of respiratory pathogens-a global perspective. Semin Respir Infect. 2000;15:195–207. doi: 10.1053/srin.2000.18066. [DOI] [PubMed] [Google Scholar]
- 4.Gold HS, Moellering RC. Antimicrobial-drug resistance. N Engl J Med. 1996;335(19):1445–1453. doi: 10.1056/NEJM199611073351907. [DOI] [PubMed] [Google Scholar]
- 5.Aaterson DL. Extended-spectrum betalactamases: the European experience. Curr Opin Infect Dis. 2001;14:697–701. doi: 10.1097/00001432-200112000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Levy SB. Factors impacting on the problem of antibiotic resistance. J Antimicrob Chemother. 2002;49(1):25–30. doi: 10.1093/jac/49.1.25. [DOI] [PubMed] [Google Scholar]
- 7.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 8.Jones RN, Phaller MA. Bacterial resistance; a worldwide problem. Diagn Microbiol Infect Dis. 1998;31:379–388. doi: 10.1016/S0732-8893(98)00037-6. [DOI] [PubMed] [Google Scholar]
- 9.Corse J, Williams REO. Antibiotic resistance of coagulase-negative staphylococci and micrococci. J Clin Pathol. 1968;21:722. doi: 10.1136/jcp.21.6.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koksal F, Yasar H, Samasti M. Antibiotic resistance patterns of coagulase negative Staphylococcus strains isolated from blood cultures of septicemic patients in Turkey. Microbiol Res. 2007;164(4):404–410. doi: 10.1016/j.micres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Kumurya AS. Loss of the meca gene during storage of methicillin resistant Staphylococcus aureus isolates in Northwestern Nigeria. J Public Health Epidemiol. 2013;5(10):410–415. [Google Scholar]
- 12.Hartman BJ, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierre J, Williamson R, Bornet M, Gutmann L. Presence of an additional penicillin-binding protein in methicillin-resistant Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, and Staphylococcus simulans with a low affinity for methicillin, cephalothin, and cefamandole. Antimicrob Agents Chemother. 1990;34:1691–1694. doi: 10.1128/AAC.34.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambers HF. Coagulase-negative staphylococci resistant to beta-lactam antibiotics in vivo produce penicillin-binding protein 2a. Antimicrob Agents Chemother. 1987;31:919–924. doi: 10.1128/aac.31.12.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chambers HF. Penicillin-binding protein-mediated resistance in pneumococci andstaphylococci. J Infect Dis. 1999;179(Suppl. 2):S353–S359. doi: 10.1086/513854. [DOI] [PubMed] [Google Scholar]
- 17.Mohammad R, Mahmood Y, Au F. Comparison of different laboratory methods for detection of MRSA. Pak J Med Sci. 2006;22(4):442–445. [Google Scholar]
- 18.Matsuhashi M, Song MD, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to P-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986;167:975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanssen A, Kjeldsen G, Sojlid JUE. Local variants of Staphylococcal Cassette chromosome mec in Sporadic methicillin-resistant Staphylococcus aureus and Methicillin-resistant coagulase-negative Staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother. 2004;48(1):285–296. doi: 10.1128/AAC.48.1.285-296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowoyo PT, Ogunbanwo ST. Virulence and toxigenicity of coagulase-negative staphylococci in Nigerian traditional fermented foods. Can J Microbiol. 2016;62:1–7. doi: 10.1139/cjm-2015-0752. [DOI] [PubMed] [Google Scholar]
- 21.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by standardized single disc method. Am J Clin Pathol. 1996;44:493–496. [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute (CLSI). M100-S22. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. Clinical and Laboratory Standards Institute, Wayne, 2012.
- 23.Coelho SMO, Reinoso E, Pereira IA, Soares LC, Demo M, Bogni C, Souza MMS. Virulence factors and antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis in Rio de Janeiro. Pesq Vet Bras. 2009;29:369–374. doi: 10.1590/S0100-736X2009000500002. [DOI] [Google Scholar]
- 24.Gradelski E, Aleksunes L, Valera L, Bonner D, Fung- Tomc J. Correlation between genotype and phenotypic categorization of Staphylococci based on methicillin susceptibility and resistance. J Clin Microbiol. 2001;39(8):2961–2963. doi: 10.1128/JCM.39.8.2961-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingato SP, Kimangá N, Omuse G, Kariuki S, Gunturu R, Dinda V. Characteristics of archived coagulase negative staphylococci isolates at a University hospital, Nairobi, Kenya. Kenya. Open J Med Microbiol. 2014;4:236–241. doi: 10.4236/ojmm.2014.44026. [DOI] [Google Scholar]
- 26.Procop GW, Shrestha NK, Tuohy MJ, Hall GS, Isada CM. Rapid identification of Staphylococcus aureus and the mecA Gene from BacT/ALERT blood culture bottles by using the light cycler system. J Clin Microbiol. 2002;40(7):2659–2661. doi: 10.1128/JCM.40.7.2659-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han JE, Hwang SY, Kim JH, Shin SP, Jun JW, Chai JY, Park YH, Park SC. CPRMethicillin resistant coagulase-negative staphylococci isolated from South Korean ducks exhibiting tremor. Acta Vet Scand. 2013;55(1):88. doi: 10.1186/1751-0147-55-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki E, Hiramatsu K, Yokota T. Survey of methicillin—resistant clinical strains of coagulase negative for mecA gene distribution. Antimicrob Agent Chemother. 1992;36:429–434. doi: 10.1128/AAC.36.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massida O, Montanari MP, Mingoia M, Viraldo PE. Borderline methicillin susceptible Staphylococcus aureus strains have more in common than reduced susceptibility to penicillase-resistant penicillins. Antimicrob Agents Chemother. 1996;40:2769–2774. doi: 10.1128/aac.40.12.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massida O, Mingoia M, Fadda D, Whalen MB, Montanari MP, Varaldo PE. Analysis of the beta lactamase plasmid of borderline methicillin susceptible Staphylococcus aureus: focus on bla complex genes and cadmium resistance determinants cadD and cadX. Plasmid. 2006;55:114–127. doi: 10.1016/j.plasmid.2005.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].


