Abstract
Nonsteroidal anti-inflammatory drugs are the most widely used medicine to treat pain and inflammation, and to inhibit platelet function. Understanding the expression regulation of enzymes of the prostanoid pathway is of great medical relevance. Histone acetylation crucially controls gene expression. We set out to identify the impact of histone deacetylases (HDACs) on the generation of prostanoids and examine the consequences on vascular function. HDAC inhibition (HDACi) with the pan-HDAC inhibitor, vorinostat, attenuated prostaglandin (PG)E2 generation in the murine vasculature and in human vascular smooth muscle cells. In line with this, the expression of the key enzyme for PGE2 synthesis, microsomal PGE synthase-1 (PTGES1), was reduced by HDACi. Accordingly, the relaxation to arachidonic acid was decreased after ex vivo incubation of murine vessels with HDACi. To identify the underlying mechanism, chromatin immunoprecipitation (ChIP) and ChIP-sequencing analysis were performed. These results suggest that HDACs are involved in the recruitment of the transcriptional activator p300 to the PTGES1 gene and that HDACi prevented this effect. In line with the acetyltransferase activity of p300, H3K27 acetylation was reduced after HDACi and resulted in the formation of heterochromatin in the PTGES1 gene. In conclusion, HDAC activity maintains PTGES1 expression by recruiting p300 to its gene.
Keywords: epigenetics, prostaglandins, prostaglandin E2, vascular biology, smooth muscle cells, histone deacetylase
Prostanoids are important signaling molecules that are not only generated in the vascular system, but also impact on several aspects of vascular biology. While some, such as prostacyclin [prostaglandin (PG)I2], are predominantly endothelium-derived, others, like PGE2, are also produced by vascular smooth muscle cells (VSMCs). Basically all physiologically generated prostanoids alter vascular tone, gene expression, and proliferative state. Vascular PG production is limited by the expression of the key enzymes of the PG pathway and by the availability of arachidonic acid. Altering enzyme expression, therefore, importantly impacts on vascular PG production. Several of these genes are subject to conventional transcription control, for example, by inflammatory transcription factors like NFκB. It is, however, also becoming obvious that large parts of gene expression are controlled by epigenetic mechanisms, which are also important in vascular gene expression control (1, 2)
Of these, histone modifications have a central role in controlling the chromatin structure. Particularly, histone acetylations at lysine residues have a strong impact on gene transcription. Acetylation transforms the condensed chromatin (heterochromatin) into a more relaxed structure (euchromatin) and thereby facilitates the access of the transcriptional machinery that results in increased gene transcription. The degree of acetylation on a lysine is dependent on the activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Both enzyme classes have fundamental roles in biological processes, cell fate decisions, maintenance, survival, and cancer development and, thus, are potential drug candidates (3, 4). Especially, HDAC inhibitors have been introduced in the clinic as novel anti-cancer drugs. Vorinostat (SAHA) was the first HDAC inhibitor approved by the United States Food and Drug Administration and is used for the treatment of cutaneous T-cell lymphoma.
HDAC inhibition (HDACi) also affects the vascular system. In experimental models, HDACi reduces angiogenesis, restenosis, and vascular inflammatory activity (4). HDACi also reduces endothelium-dependent relaxation as it reduces the expression of the endothelium NO synthase (5). Besides NO, arachidonic acid metabolites are the second most important vasoactive autacoids. However, little is known about the epigenetic regulation of the enzymes controlling vascular prostanoid production. For tumor cells, an unusual DNA methylation and histone acetylation has been reported by COX-1 and COX-2 in cancer (6–8). Whether HDACs impact on these enzymes in the vascular system is unknown and was studied here in the murine vasculature and human VSMCs.
MATERIALS AND METHODS
Materials
SAHA, apicidin, PGE2, and diclofenac were purchased from Sigma-Aldrich. Arachchidonic acid and U46619 were from Cayman Chemical. DETA NONOate was acquired from Enzo Life Sciences. Antibodies against H3K9ac (#C15410004), H3K27ac (#pAb-174-050), and RNA polymerase 2 (RNAP2) (C15100055) were from Diagenode. Antibody against p300 was from Bethyl (#A300-358)
siRNA transfection
For siRNA treatment, smooth muscle cells (80–90% confluent) were transfected with Lipofectamine 3000 according to the instructions provided by Thermo Fisher Scientific. siRNAs for HDACs, p300, and control siRNAs (siScrambled/siSCRs) were purchased from Thermo Fisher Scientific (Stealth RNAi), siHDAC1 #HSS104725, siHDAC2 #HSS104728, siHDAC3 #HSS113050, siHDAC4 #HSS114673, siHDAC7 #HSS147499, siHDAC8 #HSS125194, siEP300-1 #HSS103259, siEP300-2 #HSS103258, siSCR1 #12935-300, and siSCR2 #12935112.
Cell culture
Human VSMCs [human aortic smooth muscle cells #354-05a, human coronary artery smooth muscle cells #350-05a, and human carotid smooth muscle cells #3514-05a] were purchased from PELOBiotech (Planegg, Germany). Cells were cultured on collagen type I-coated (#354236; Corning Incorporated, Tewksbury, MA) dishes in smooth muscle cell medium (#PB-MH-200-2190) supplemented with 8% FCS, penicillin (50 U/ml), streptomycin (50 μg/ml), EGF, FGF, glutamine, and insulin from PELOBiotech. A humidified atmosphere of 5% CO2 at 37°C was used. Cells were treated with SAHA (2 μmol/l) or apicidin (200 nmol/l) for 14 h in smooth muscle cell medium with 1% FCS.
Organ chamber experiments
Male C57/BL6 mice (10 weeks of age) were obtained from Charles River Laboratories, Sulzfeld, Germany. Isometric tension recordings were performed in an organ chamber setup with murine carotid rings (1–2 mm) in Krebs Henseleit buffer containing 2.2 g/l glucose at 37°C and 5% CO2. The concentration of the thromboxane (TX) receptor agonist, U46619, used for preconstriction, was adjusted to obtain an identical level of preconstriction (80%) relative to the contraction elicited by KCl (80 mmol/l). Arachidonic acid-induced relaxation was registered in the presence or absence of diclofenac (10 μmol/l).
Organ culture
Murine carotid artery segments were dissected under sterile conditions, cleaned of adherent tissue, and incubated for 14 h under sterile conditions at 37°C in EBM culture medium containing 0.1% BSA, in the presence or absence of SAHA (2 μmol/l) in a conventional incubator at 5% CO2. Subsequently, the tissue was used for organ chamber experiments and molecular biology.
Quantitative RT-PCR
Total RNA was extracted with the RNA Mini Kit (Bio and Sell). cDNA was prepared with Super-Script III reverse transcriptase (Invitrogen) and random hexamer together with oligo(dT) primers (Sigma #O4387). Quantitative real-time PCR was performed with Eva Green Master Mix and ROX as reference dye (Bio and Sell #76.580.5000) in a Mx3005 cycler (Stratagene). Relative expression of target genes was normalized to β-actin and analyzed by the delta-delta Ct method with the MxPro software (Agilent Technologies, Santa Clara, CA). The following two primer pairs were used for quantification of human microsomal PGE synthase-1 (PTGES1): forward (F1) TTG TCG CCT GGA TGC ACT TCC TGG, reverse (R1) AGG TGG CGG GCC GCT TCC CAG AGG; (F2) CGC TGC TGG TCA TCA AGA TGT ACG, (R2) TTT CCT GGG CTT CGT CTA CTC CTT. For murine Ptges1: (F) CAC TGC TGG TCA TCA AGA TGT ACG; (R) AAT GAG TAC ACG AAG CCG AGG AAG. For p300, (F) TAG GAG TTC AAA CGC CGA GTC and (R) GTT GAG CTG CTG TTG GCA TAG
Chromatin immunoprecipitation
Preparation of cell extracts, cross-linking, and isolation of nuclei were performed with the truCHIP™ chromatin shearing kit (Covaris, USA) according to the manufacturer’s protocol. The procedure was similar to that described previously (9). After sonification of the lysates with the Bioruptur Plus (10 cycles, 30 s on, 90 s off, 4°C; Diagenode, Seraing, Belgium), cell debris was removed by centrifugation and the lysates were diluted 1:3 in dilution buffer [20 mmol/l Tris/HCl (pH 7.4), 100 mmol/l NaCl, 2 mmol/l EDTA, 0.5% Triton X-100, and protease inhibitors]. Preclearing was done with 20 μl DiaMag protein A and protein G-coated magnetic beads slurry (Diagenode) for 45 min at 4°C. The samples were incubated as indicated overnight at 4°C with the antibodies indicated. Five percent of the samples served as input. The complexes were collected with 30 μl DiaMag protein A-coated magnetic beads (Diagenode) for 3 h at 4°C, subsequently washed twice for 5 min with each of the three wash buffers [wash buffer 1: 20 mmol/l Tris/HCl (pH 7.4), 150 mmol/l NaCl, 0.1% SDS, 2 mmol/l EDTA, 1% Triton X-100; wash buffer 2: 20 mmol/l Tris/HCl (pH 7.4), 500 mmol/l NaCl, 2 mmol/l EDTA, 1% Triton X-100; wash buffer 3: 10 mmol/l Tris/HCl (pH 7.4), 250 mmol/l lithium chloride, 1% Nonidet p-40, 1% sodium deoxycholate, 1 mmol/l EDTA] and finally washed with TE-buffer (pH 8.0). Elution of the beads was done with elution buffer (0.1 M NaHCO3, 1% SDS) containing 1× proteinase K (Diagenode) and shaking at 600 rpm for 1 h at 55°C, 1 h at 62°C, and 10 min at 95°C. After removal of the beads, the eluate was purified with the QiaQuick PCR purification kit (Qiagen, Hilden, Germany) and subjected to quantitative (q)PCR analysis. The following primers for quantification were used: −500 bp upstream of PTGES1 TSS: (F) CAC AGT CAC GGG TTC TAG GGA TTG, (R) ACT CAG CCT GGA CAA TGG AGC TGC; −1,000 bp upstream of PTGES1 TSS: (F) GCA TTT GAC TGG GGA AAG AGT TC, (R) GCT GTG TTG TTT TAA GCC ACT AAG; exon 1 of PTGES1: (F) TGA TCA CAC CCA CAG TTG AGC TGC, (R) ACA TAC CTT CTT CCG CAG CCT CAC; exon 2 of PTGES1: (F) ACT GGT ATA TTT CAG GCC TTT GC, (R) TGT GCA GAA GAA GTT CTG AAA GG.
Formaldehyde-assisted isolation of regulatory elements
Formaldehyde-assisted isolation of regulatory elements (FAIRE) was performed similarly to the description in (10, 11). VSMCs (2 × 106) were cross-linked with 1% formaldehyde for 5 min and quenched with 125 mM glycine for 5 min. Afterwards, cells were scraped, washed with PBS, and lysed in 2 ml FAIRE buffer 1 [50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100] at 4°C for 10 min followed by centrifugation (5 min, 16,000 g, 4°C). Cell pellets were resuspended and incubated in 2 ml FAIRE buffer 2 [10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA] for 10 min at 22°C followed by centrifugation (5 min, 16,000 g, 4°C). Cell pellets were resuspended in 400 μl FAIRE buffer 3 [10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine]. Samples were sheared (30 s on, 90 s off, 10 cycles, 4°C) with a Bioruptor Plus (Diagenode) to an average length of 200–400 bp. Subsequently, samples were cleared by centrifugation (5 min, 16,000 g, 4°C). Ten percent of each sample was used as input. Samples were further extracted with phenol/chloroform/isoamylalcohol (Roti Phenol/C/I) and the aqueous phase was collected. Input and extracted DNA were de-cross-linked at 65°C for 6 h, followed by DNA purification with the QIAquick PCR purification kit (Qiagen).
Chromatin immunoprecipitation-sequencing
Chromatin immunoprecipitation (ChIP)-sequencing (ChIP-Seq) data of p300 was obtained from ENCODE, track name HeLa-S3 EP300, and visualized by the Integrative Genomics Viewer version 2.3.81 (127).
LC-MS/MS
Sample analysis was performed using LC-ESI-MS/MS. The LC-MS/MS system consisted of a hybrid triple quadrupole-ion trap QTrap 5500 mass spectrometer (Sciex, Darmstadt, Germany) equipped with a Turbo-V-source operating in negative ESI mode, an Agilent 1200 binary HPLC pump and degasser (Agilent, Waldbron, Germany), and an HTC Pal autosampler (Chromtech, Idstein, Germany). A cooling stack was used to store the samples at 4°C in the autosampler. High purity nitrogen for the mass spectrometer was produced by a NGM 22-LC/MS nitrogen generator (CMC Instruments, Eschborn, Germany).
For the chromatographic separation, a Synergi Hydro-RP column and precolumn were used (150 × 2 mm I.D., 4 μm particle size, and 80 Å pore size; Phenomenex, Aschaffenburg, Germany). A linear gradient was employed at a flow rate of 300 μl/min. Mobile phase A was water/formic acid (100:0.0025, v/v, pH 4.0) and mobile phase B was acetonitrile/formic acid (100:0.0025, v/v). Sample solvent was acetonitrile/water/formic acid (20:80:0.0025, v/v, pH 4.0). Total run time was 16 min and injection volume of samples was 20 μl.
The mass spectrometer was operated in the negative ion mode with an electrospray voltage of −4,500 V at 450°C. Multiple reaction monitoring was used for quantification. The mass transitions used were m/z 351.1 → 315.0 for PGE2 and PGD2, m/z 353.1 → 309.2 for PGF2α, m/z 369.1 → 163.0 for 6-keto-PGF1α, m/z 369.1 → 169.1 for TXB2, m/z 355.1 → 275.1 for [2H4]PGE2 and [2H4]PGD2, m/z 357.1 → 313.2 for [2H4]PGF2α, m/z 373.2 → 167.1 for [2H4]6-keto-PGF1α, and m/z 373.1 → 173.1 for [2H4]TXB2 all with a dwell time of 50 ms (all the standards were purchased from Cayman Chemical (Ann Arbor, MI).
All quadrupoles were working at unit resolution. Quantitation was performed with Analyst software V1.5 (Sciex) using the internal standard method (isotope-dilution M). Ratios of analyte peak area and internal standard peak area (y axis) were plotted against concentration (x axis) and calibration curves for each PG were calculated by least square regression with 1/concentration2 weighting.
Statistics
Unless otherwise indicated, data are given as mean ± SEM. Calculations were performed with GraphPad PRISM 5.0. For multiple group comparisons, ANOVA followed by post hoc with Fisher LSD test was performed. In case of multiple testing, Bonferroni correction was applied. Individual statistics of samples were performed by t-test. P < 0.05 was considered as statistically significant. N indicates the number of individual experiments.
RESULTS
Inhibition of HDACs reduces PGE2 in the vasculature
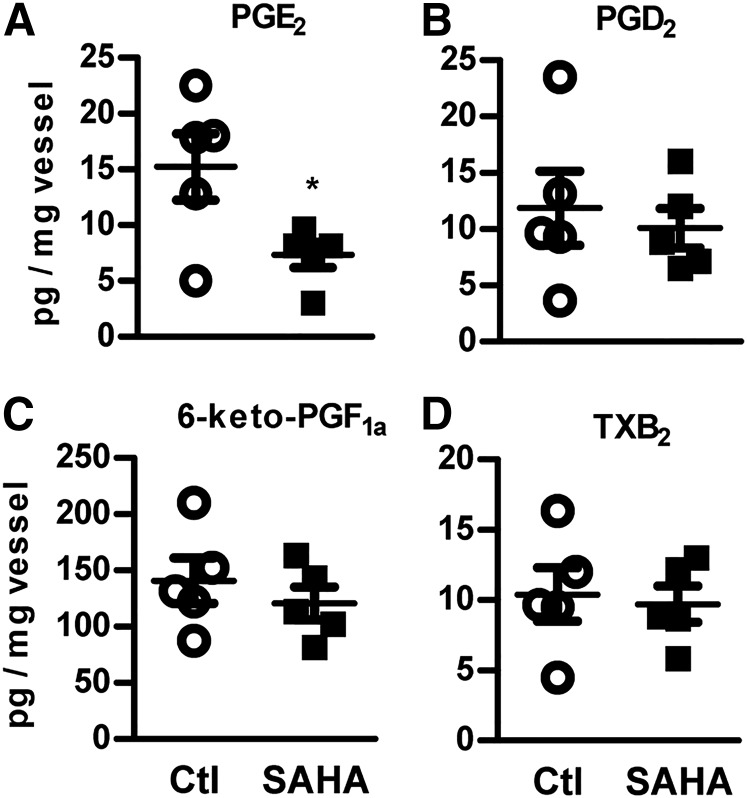
To uncover the role of HDACs in arachidonic acid metabolism, we determined the level of prostanoids in murine vessels incubated ex vivo with the HDAC inhibitor, SAHA (2 μmol/l, 14 h). MS/MS analysis revealed that the abundance of PGE2 was markedly decreased, whereas PGD2, TXB2, and 6-keto-PGF1a, the marker for prostacyclin (PGI2), were not affected by HDACi (Fig. 1). Collectively, these findings suggest that HDAC activities are required for the PGE2 levels in the vascular system.
Fig. 1.
Inhibition of HDACs reduces PGE2 in the vasculature. LC-MS/MS quantification of PGE2 (A), PGD2 (B), 6-keto-PGF1a (C), and TXB2 (D) in mice carotid rings, incubated with SAHA (2 μmol/l, 14 h) or DMSO as control (Ctl). *P < 0.05, n = 5.
PTGES1 expression is decreased by HDAC inhibition
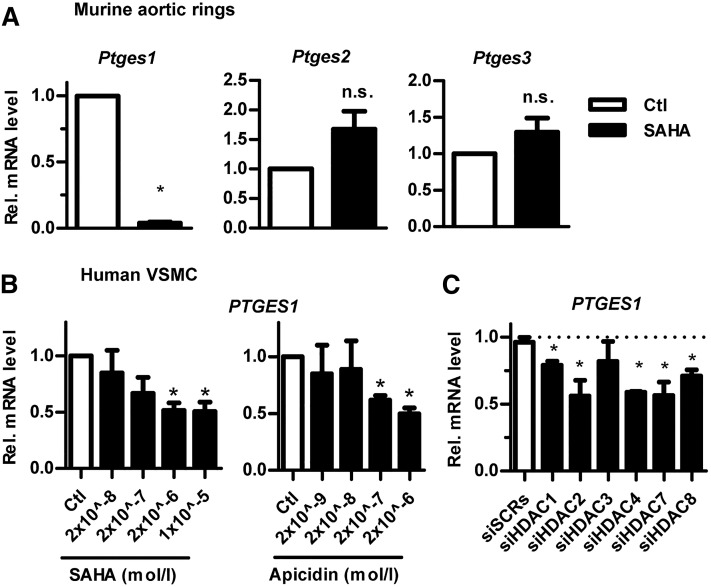
The microsomal PTGES1 is the dominant enzyme responsible for the production of vascular PGE2 from COX-2-derived PGH2 (12). Because PGE2 levels were reduced by HDACi, we suspected that Ptges1 expression was also attenuated by SAHA, which was indeed the case (Fig. 2A). There was, in contrast, a trend toward a slight induction of Ptges2 und Ptges3, which, however, did not reach the significance level. Moreover, these enzymes are not relevant for vascular PG formation. Smooth muscle cells are the predominant cell type in the vasculature. Therefore, we studied PTGES1 expression in combination with HDACi in human VSMCs. Consistent with the data obtained in the mouse vessels, human VSMCs exposed to 2 μmol/l SAHA expressed a significantly lower level of PTGES1, as compared with control-treated cells (Fig. 2B). Thus, our initial observation in murine carotid artery rings is also valid in human VSMCs. To determine whether reduction of PTGES1 was a result of reduced HDAC activity and not an unspecific effect of SAHA, a second inhibitor, apidicin, was tested, which had a similar effect on PTGES1 expression (Fig. 2B). SAHA and apicidin are both canonical inhibitors and, thus, reduce the activity of class 1 and 2 HDACs (13). To identify which HDAC is responsible for maintaining vascular PTGES1 expression, RNAi experiments were performed with two different control scrambled siRNAs. However, knockdown of several HDACs reduced PTGES1 (Fig. 2C), suggesting that the effect of HDACi is a consequence of the concomitant inhibition of several HDACs like HDAC1, -2, -4, -7, and -8.
Fig. 2.
PTGES1 expression is reduced by HDACi (A, B) or depletion (C). A–C: qRT-PCR of the genes indicated (relative to β-actin) from murine carotid rings (A) and from human VSMCs (B, C). A: Effect of SAHA (2 μmol/l, 14 h). B: Effect of SAHA or apicidin (14 h), both relative to the control DMSO (Ctl). C: Effect of two different control scrambled RNAi (siSCRs) or siRNA against HDAC as indicated. *P < 0.05, n ≥ 3.
Inhibition of HDACs attenuates arachidonic acid-induced vessel relaxation
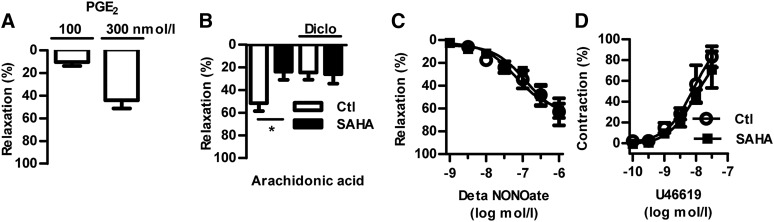
To determine the relevance of HDAC-mediated PTGES1 induction, vascular reactivity assays were performed. Mice lacking Ptges1 exhibit an impaired conversion of arachidonic acid to the vasodilator, PGE2, without changes in the level of other prostanoids (12, 14, 15). Therefore, we exposed preconstricted murine carotid rings to arachidonic acid as well as PGE2 as positive control and analyzed their subsequent relaxation. The carotid vessels responded to PGE2 with relaxation (Fig. 3A). In line with the diminished Ptges1 expression after HDACi, the arachidonic acid-induced relaxation was attenuated by SAHA preincubation (Fig. 3B). Importantly, neither the relaxation to the NO donor, DETA-NONOate, nor the constriction to the TXA2 receptor agonist, U46619, was affected by HDACi (Fig. 3C, D). In order to demonstrate that an attenuated formation of vasodilator PGs, but not unrelated effects of arachidonate, are responsible for the reduced relaxation after HDACi, experiments were performed in the presence of the cyclooxygenase inhibitor, diclofenac. Importantly, diclofenac attenuated the relaxation to arachidonic acid in vessels of the control group, but did not attenuate responses in vessels of the SAHA group. Thus, attenuated metabolism of arachidonic acid to a vasodilator compound is responsible for the HDACi effect (Fig. 3B). These data indicate that reduced HDAC activity in the vascular system attenuates arachidonic acid-induced relaxation, which could be attributed in part to decreased expression of PTGES1.
Fig. 3.
Inhibition of HDACs reduces arachidonic acid-induced vessel relaxation. Vascular response assays of carotid rings. Relaxations to PGE2 (A) and arachidonic acid (100 nmol/l) (B) in combination with and without diclofenac (Diclo; 10 μmol/l) and the NO donor DETA NONOate (C) in U46619 preconstricted vessels. D: Contractions to the TX receptor agonist, U46619. Carotid rings were incubated with SAHA (2 μmol/l, 14 h) or with control solvent DMSO (Ctl). *P < 0.05, n = 7–10.
Inhibition of HDAC abolishes p300 binding to the PTGES1 gene
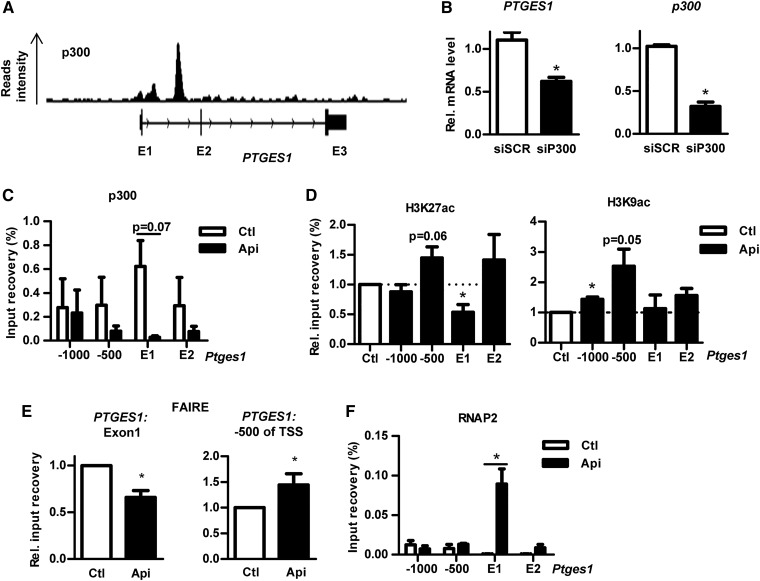
Histone acetylation is regulated by the competing activities of HDACs and HATs. The fact that PTGES1 expression was reduced by HDACi was unexpected, because a direct action of HDACi should rather increase the acetylation of the PTGES1 gene and thus induce its expression. As this was obviously not the case for PTGES1, alternative modes of expression control were sought. For some genes, HDACi decreases rather than increases target acetylation (16, 17), but the mechanism underlying this effect is unclear. Potentially, HDACi attenuates HAT expression and thereby recruitment to its target genes (17). We therefore determined whether HDACi affects the recruitment of the transcriptional activators, HATs. ChIP-Seq data from ENCODE for a putative binding of HATs (GCN5, PCAF, and p300) suggest that the PTGES1 gene is particularly occupied by the p300 HAT (Fig. 4A). Depletion of p300 by RNAi reduced PTGES1 expression in VSMCs, demonstrating that this HAT is indeed important for PTGES1 expression control (Fig. 4B). Moreover, also in our hands, p300 binding to the PTGES1 gene was observed by ChIP (Fig. 4C). In accordance with our hypothesis of an attenuated HAT recruitment of HDACi, p300 abundance at the PTGES1 gene was largely attenuated after HDACi (Fig. 4C). For these experiments, the HDAC inhibitor, apicidin, was used due to its higher potency. In line with a previous report (18), the reduced recruitment of p300 to the PTGES1 gene resulted in a reduced acetylation of the histone mark, H3K27, but not H3K9 (Fig. 4D). Importantly, these effects were specific for the exon 1 of PTGES1 and not observed for the PTGES1 promotor: A strong enriched binding of p300 was not observed 500 and 1,000 bp upstream of PTGES1 TSS, and H3K27 at this region was induced instead (Fig. 4D).
Fig. 4.
Inhibition of HDAC abolishes p300 binding to the PTGES1 gene. A: The p300 ChIP-Seq of HELA cells from ENCODE. B: qRT-PCR of PTGES1 (relative to β-actin) from VSMCs with two different control scrambled siRNAs (siSCRs) or two different siRNAs against p300. ChIP (C, D, F) and FAIRE (E) of VSMCs treated with control (Ctl) solvent (DMSO) or apicidin (200 nmol/l, 14 h) with the antibodies against p300, H3K27ac, H3K9ac, and RNAP2 followed by qPCR for PTGES1 using primers binding at exon 1 (E1), exon 2 (E2), or promoter region (500 or 1,000 bp upstream of TSS). *P < 0.05, n = 3–6.
HDACi not only attenuated acetylation of the PTGES1 exon 1 at H3K27, it also dramatically increased acetylation of H3K9, particularly in the promoter region (Fig. 4D). H3K9ac is an active mark and, thus, should result in PTGES1 expression. Thus, only direct determination of the chromatin state (i.e., hetero vs. euchromatin) can unravel the chromatin situation around the PTGES1 gene. For this purpose, a FAIRE assay was carried out. In accordance with the reduced p300 binding and H3K27ac, more closed chromatin in the PTGES1 exon 1 and more open chromatin in the promoter region was observed (Fig. 4E). Finally, we addressed whether this closed chromatin environment could affect RNAP2 occupancy; as recently, it was reported that HDACi can cause a block in the elongation step of transcription by pausing of RNAP2 (19). In line with this hypothesis, we observed a dramatic accumulation of RNAP2 within the first exon of Ptges1 (Fig. 4F).
Collectively, these findings suggest that the enzymatic activities of HDACs are required for proper binding of the transcription activator, p300, to the PTGES1 gene to facilitate H3K27ac and, thereby, euchromatin to enable proper RNAP2 elongation.
DISCUSSION
In this study, we determined the role of HDACs in some aspect of vascular arachidonic acid metabolism. Inhibition of HDACs reduced vascular PGE2 levels in the murine vasculature and in human VSMCs. This effect was a consequence of reduced p300 recruitment to the Ptges1 gene.
PGE2 is a key product of arachidonic acid in the COX pathway. The lipid mediator has diverse functions, including cytoprotection in the gastrointestinal tract, modulation of the immune system, and induction of the febrile responses. Furthermore, in the vascular system, PGE2 reduced smooth muscle tone and changed proliferation and inflammatory activity (20–23). Here, we observed that HDACi reduced the vascular PGE2 level in the carotid, which resulted in attenuated arachidonic acid-induced relaxation suggesting an attenuated conversion of arachidonic acid to the vasodilator, PGE2 (12, 14, 15). The role of PGE2 in the control of systemic blood pressure is under debate. Infusion of PGE2 into the kidney commonly causes renal vasodilation (20) and mice lacking PTGES1 exhibit an exaggerated hypertensive response to angiotensin II under normal conditions, but not in a hyperlipidemic situation (20, 21, 23). In murine aortic rings, PGE2 directly induces relaxation through the E-prostanoid 4 receptor and, thus, cAMP and also data from our group suggest that the mouse carotid artery, a typical conduit vessel, relaxes to PGE2.
Acetylation of histone marks, H3K27 and H3K9, results in euchromatin and facilitates access of transcription factors to genes. By removing these permissive marks, HDACs are mediators of gene repression. HDACi should therefore induce an open chromatin state. Thus, our observation that HDACi reduced PTGES1 expression in the murine vasculature and in VSMCs was unexpected. A simple explanation would be that HDACi not only induces, but also reduces, acetylation at certain sites. Indeed, recently two reports of mediating HDACi with trichostatin A and SAHA suggested that this can be the case in myocytes and endothelial cells (16, 17). Apparently, a similar mechanism is operative in the present study for PTGES1 in VSMCs.
Gene set enrichment analysis of RNA-Seq suggested that HATs could be influenced by HDACi. Particularly, reduced gene expression of p300-dependent genes was observed as a result of decreased p300 recruitment in trichostatin A-treated cells (17). Consistent with this, we observed that p300 occupancy to PTGES1 was also HDAC dependent. In fact, it was quite remarkable that acetylation of H3K27, a well-established target of p300, was strongly reduced by HDACi (24). In line with this, Jin et al. (18) reported that deletion of GCN5/PCAF, a different HAT, specifically reduces acetylation on histone H3K9, while deletion of p300 specifically reduces acetylation on H3K27. This suggests that due to the selective reduction in the recruitment of p300, the H3K27, but not the H3K9ac, was decreased in response to HDACi. It is certainly unusual that such an epigenetic modification selectively occurs only in the PTGES1 gene, especially in the first exon, but not in the promoter. However, global run-on sequencing revealed that SAHA inhibits transcription of several genes by blocking RNAP2 elongation in the gene body (19). The analysis indicated that repressed genes by HDACi have an impediment within the gene locus. We hypothesize that such an impediment within the gene body could be achieved by changing the H3K27ac level and thus the chromatin environment into a tight state, which results in accumulation of RNAP2. Indeed, such mechanism, e.g., is operating by cotranscriptional RNA splicing, histone acetylation is frequently used to accumulate RNAP2 and the splicing machinery over alternative exons (19, 25, 26)
In conclusion, HDACi results in decreased PTGES1 expression and vascular PGE2 formation as a consequence of attenuated recruitment of p300 to the PTGES1 gene.
Acknowledgments
The authors are grateful for the excellent technical assistance of Tanja Lüneburg, Cindy Höper, Katalin Pálfi, and Susanne Schütz.
Footnotes
Abbreviations:
- ChIP
- chromatin immunoprecipitation
- ChIP-Seq
- chromatin immunoprecipitation-sequencing
- FAIRE
- formaldehyde-assisted isolation of regulatory elements
- HAT
- histone acetyltransferases
- HDAC
- histone deacetylase
- HDACi
- histone deacetylase inhibition
- PG
- prostaglandin
- PTGES1
- microsomal prostaglandin E synthase-1
- RNAP2
- RNA polymerase
- SAHA
- vorinostat
- TX
- thromboxane
- VSMC
- vascular smooth muscle cell
This work was supported by Deutsche Forschungsgemeinschaft SFB1039 (TPA01, TPA02, and Z01) to R.P.B., D.S., and G.G., the Excellence Cluster 147 ECCPS, and the Faculty of Medicine of Goethe University Frankfurt (Nachwuchsforschungsstipendium to C.F.). The authors declare that they have no relevant financial, personal, or professional relationships to disclose that could be perceived as a conflict of interest or as potentially influencing or biasing the authors’ work.
REFERENCES
- 1.Gomez D., Swiatlowska P., and Owens G. K.. 2015. Epigenetic control of smooth muscle cell identity and lineage memory. Arterioscler. Thromb. Vasc. Biol. 35: 2508–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu R., Leslie K. L., and Martin K. A.. 2015. Epigenetic regulation of smooth muscle cell plasticity. Biochim. Biophys. Acta. 1849: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown J. A. L., Bourke E., Eriksson L. A., and Kerin M. J.. 2016. Targeting cancer using KAT inhibitors to mimic lethal knockouts. Biochem. Soc. Trans. 44: 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon S., and Eom G. H.. 2016. HDAC and HDAC inhibitor: from cancer to cardiovascular diseases. Chonnam Med. J. 52: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robb G. B., Carson A. R., Tai S. C., Fish J. E., Singh S., Yamada T., Scherer S. W., Nakabayashi K., and Marsden P. A.. 2004. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J. Biol. Chem. 279: 37982–37996. [DOI] [PubMed] [Google Scholar]
- 6.Cebola I., and Peinado M. A.. 2012. Epigenetic deregulation of the COX pathway in cancer. Prog. Lipid Res. 51: 301–313. [DOI] [PubMed] [Google Scholar]
- 7.Cebola I., Custodio J., Munoz M., Diez-Villanueva A., Pare L., Prieto P., Ausso S., Coll-Mulet L., Bosca L., Moreno V., et al. . 2015. Epigenetics override pro-inflammatory PTGS transcriptomic signature towards selective hyperactivation of PGE2 in colorectal cancer. Clin. Epigenetics. 7: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi T., Itoh F., Toyota M., Suzuki H., Yamamoto H., Fujita M., Hosokawa M., and Imai K.. 2002. Aberrant methylation and histone deacetylation of cyclooxygenase 2 in gastric cancer. Int. J. Cancer. 97: 272–277. [DOI] [PubMed] [Google Scholar]
- 9.Wong M. S., Leisegang M. S., Kruse C., Vogel J., Schurmann C., Dehne N., Weigert A., Herrmann E., Brune B., Shah A. M., et al. . 2014. Vitamin D promotes vascular regeneration. Circulation. 130: 976–986. [DOI] [PubMed] [Google Scholar]
- 10.Simon J. M., Giresi P. G., Davis I. J., and Lieb J. D.. 2012. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 7: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., and Grummt I.. 2015. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 60: 626–636. [DOI] [PubMed] [Google Scholar]
- 12.Liu T., Laidlaw T. M., Feng C., Xing W., Shen S., Milne G. L., and Boyce J. A.. 2012. Prostaglandin E2 deficiency uncovers a dominant role for thromboxane A2 in house dust mite-induced allergic pulmonary inflammation. Proc. Natl. Acad. Sci. USA. 109: 12692–12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieliauskas A. V., and Pflum M. K. H.. 2008. Isoform-selective histone deacetylase inhibitors. Chem. Soc. Rev. 37: 1402–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundequist A., Nallamshetty S. N., Xing W., Feng C., Laidlaw T. M., Uematsu S., Akira S., and Boyce J. A.. 2010. Prostaglandin E(2) exerts homeostatic regulation of pulmonary vascular remodeling in allergic airway inflammation. J. Immunol. 184: 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hristovska A. M., Rasmussen L. E., Hansen P. B., Nielsen S. S., Nusing R. M., Narumiya S., Vanhoutte P., Scott O., and Jensen B. L.. 2007. Prostaglandin E2 induces vascular relaxation by E-prostanoid 4 receptor-mediated activation of endothelial nitric oxide synthase. Hypertension. 50: 525–530. [DOI] [PubMed] [Google Scholar]
- 16.Ooi J. Y. Y., Tuano N. K., Rafehi H., Gao X. M., Ziemann M., Du X. J., and El-Osta A.. 2015. HDAC inhibition attenuates cardiac hypertrophy by acetylation and deacetylation of target genes. Epigenetics. 10: 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rafehi H., Balcerczyk A., Lunke S., Kaspi A., Ziemann M., Kn H., Okabe J., Khurana I., Ooi J., Khan A. W., et al. . 2014. Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res. 24: 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y. and Ge K.. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greer C. B., Tanaka Y., Kim Y. J., Xie P., Zhang M. Q., Park I. H., and Kim T. H.. 2015. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 13: 1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Facemire C. S., Griffiths R., Audoly L. P., Koller B. H., and Coffman T. M.. 2010. The impact of microsomal prostaglandin e synthase 1 on blood pressure is determined by genetic background. Hypertension. 55: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia Z., Guo X., Zhang H., Wang M. H., Dong Z., and Yang T.. 2008. Microsomal prostaglandin synthase-1-derived prostaglandin E2 protects against angiotensin II-induced hypertension via inhibition of oxidative stress. Hypertension. 52: 952–959. [DOI] [PubMed] [Google Scholar]
- 22.Swan C. E., and Breyer R. M.. 2011. Prostaglandin E2 modulation of blood pressure homeostasis: studies in rodent models. Prostaglandins Other Lipid Mediat. 96: 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G., and Chen L.. 2016. An update of microsomal prostaglandin E synthase-1 and PGE2 receptors in cardiovascular health and diseases. Oxid. Med. Cell. Longev. 2016: 5249086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedford D. C., and Brindle P. K.. 2012. Is histone acetylation the most important physiological function for CBP and p300? Aging. 4: 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornblihtt A. R., Schor I. E., Allo M., Dujardin G., Petrillo E., and Munoz M. J.. 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14: 153–165. [DOI] [PubMed] [Google Scholar]
- 26.Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X. D., and Bentley D. L.. 2014. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 28: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]