Abstract
The gut microbiota influences the development and progression of metabolic diseases partly by metabolism of bile acids (BAs) and modified signaling through the farnesoid X receptor (FXR). In this study, we aimed to determine how the human gut microbiota metabolizes murine BAs and affects FXR signaling in colonized mice. We colonized germ-free mice with cecal content from a mouse donor or feces from a human donor and euthanized the mice after short-term (2 weeks) or long-term (15 weeks) colonization. We analyzed the gut microbiota and BA composition and expression of FXR target genes in ileum and liver. We found that cecal microbiota composition differed between mice colonized with mouse and human microbiota and was stable over time. Human and mouse microbiota reduced total BA levels similarly, but the humanized mice produced less secondary BAs. The human microbiota was able to reduce the levels of tauro-β-muricholic acid and induce expression of FXR target genes Fgf15 and Shp in ileum after long-term colonization. We show that a human microbiota can change BA composition and induce FXR signaling in colonized mice, but the levels of secondary BAs produced are lower than in mice colonized with a mouse microbiota.
Keywords: bile acids and salts, biosynthesis and metabolism, enzyme mechanisms and regulation, farnesoid X receptor agonist, farnesoid X receptor antagonist, gut microbiota, intestine, humanized mouse models
The gut microbiota influences the development and progression of metabolic diseases such as obesity, diabetes, and atherosclerosis (1). The influence of the gut microbiota on metabolic diseases may in part be mediated by modification of bile acids (BAs), which are now recognized to have a central role as signaling molecules through activation of receptors involved in metabolic pathways such as the nuclear farnesoid X receptor (FXR) and G-protein-coupled receptor TGR5 (2–4). FXR is a powerful regulator of lipid and glucose metabolism and is activated by the primary BAs chenodeoxycholic acid (CDCA) and cholic acid (CA), the secondary BA deoxycholic acid (DCA), and to a lesser extent by the secondary BA lithocholic acid (LCA) (5, 6). The conjugated forms of these BAs, amidated with glycine (G) or taurine (T), can also activate FXR (5, 6). BA composition differs between humans and mice; in addition to the BAs found in humans, mice have the primary BAs α- and β-muricholic acid (α/βMCA), which in their T-conjugated forms function as FXR antagonists (7, 8). The gut microbiota deconjugates and subsequently metabolizes primary BAs to secondary BAs in the gut and will thereby modify the extent of FXR activation (9).
BA synthesis is regulated by FXR mainly via two downstream targets, fibroblast growth factor 19 (FGF19; in mice, FGF15) in ileum and small heterodimer partner (SHP) in the liver, both of which inhibit expression of the rate-limiting enzyme in BA synthesis, cholesterol 7α-hydroxylase (CYP7A1) in liver. We have previously shown that differences in BA metabolism between germ-free (GF) and conventionally raised (CONV-R) mice are dependent on the FXR antagonist tauro-β-muricholic acid (TβMCA) (7). TβMCA is not metabolized in the absence of bacteria. Thus, GF mice show accumulation of TβMCA and reduced FXR signaling (7) and, consequently, increased BA synthesis.
GF mice colonized with a human microbiota (here termed humanized mice) have been used to study the effect of environmental and genetic factors on gut microbiota and host physiology. These studies showed that the human microbiota is able to establish a stable community in the recipient mice with similarities to the community in the donor sample (10–12). However, it is unclear if a human gut microbiota can metabolize murine BAs and induce FXR signaling. This would be important to elucidate in order to improve translation of results from mouse models into a human setting. Here we aimed to determine whether a human gut microbiota can deconjugate and metabolize primary murine BAs and thus increase FXR activation in the recipient mice.
MATERIALS AND METHODS
Mice and colonization
GF female Swiss Webster mice were maintained in flexible plastic gnotobiotic isolators under a strict 12 h light cycle and fed an autoclaved chow diet (Lab diet, St. Louis, MO) ad libitum. GF isolators were routinely tested for sterility by culturing and PCR analysis of feces amplifying the 16S rRNA gene.
Colonization was performed by diluting human fecal or mouse cecal samples (∼0.5 g) in 5 ml reduced PBS; 0.2 ml of this suspension was introduced by gavage into each GF mouse. The human fecal sample from the first donor was obtained from a healthy 38-year-old female volunteer, and the sample from the second donor was obtained from a healthy 40-year-old male volunteer. The samples were obtained shortly before colonization and immediately (within 5 min) diluted and introduced into the GF mice by gavage within 2 h after dilution. The mouse cecal sample was obtained from a 9-week-old CONV-R Swiss Webster female mouse. The GF and humanized (CONV-H) mice were maintained in the isolators, and the conventionalized (CONV-M) mice were transferred from the isolators into conventional cages after colonization.
Mice were colonized at 8–15 weeks of age and maintained for 2 weeks or 15 weeks after colonization. Blood was collected from the portal vein and inferior caval vein under deep isoflurane-induced anesthesia following 4 h of fasting. The mice were then euthanized, and tissues (liver, gallbladder, ileum, and cecum) were harvested. All tissues were immediately frozen in liquid nitrogen and stored at −80°C until further processed.
All experiments were performed in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animal using protocols approved by the University of Gothenburg Animal Studies Committee. The use of human feces from healthy volunteers was approved by the Regional Ethics Committee in Gothenburg.
Gut microbiota analysis
Genomic DNA was isolated, and the V4 region of the bacterial 16S rRNA gene was amplified with PCR (see supplemental data). The amplified DNA was sequenced at Genomic Core Facility at University of Gothenburg, and the sequencing data were analyzed using the software package Quantitative Insights into Microbial Ecology (QIIME), version 1.8.0. Linear discriminant analysis effect size (LEfSe) algorithm (13) was used to identify taxa that discriminated cecal microbiota profiles according to the colonization origin (see supplemental data).
BA analysis
BAs in liver, gallbladder, cecum, and serum from portal and caval veins were analyzed using ultra-performance LC/MS/MS and quantified using a combination of unlabeled standards and deuterium-labeled internal standards [see supplemental data and Ref. (14)]. The whole gallbladder, ∼50 mg of liver and cecum, and 50 µl serum were used for the analyses.
In vitro assay of BA metabolism by a human microbiota
Suspensions with fecal samples from the first human donor were incubated with TCA or CA to test the capacity of a human microbiota to metabolize BAs in vitro (see supplemental data).
Quantitative real-time PCR
Approximately 30 mg of liver or distal ileum was homogenized using TissueLyzer (Qiagen), and total RNA was isolated using RNeasy kit (Qiagen). High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used to synthesize 20 µl cDNA templates from 500 ng purified RNA using random hexamer primers, and the products were diluted 7× before use in subsequent reactions. 1× SYBR Green Master Mix buffer (Thermo Scientific) was used for quantitative RT-PCR at final reaction volumes of 10 µl. Gene-specific primers (900 nM) were used in each reaction, and all results were normalized to the ribosomal protein L32 mRNA [primer sequences were previously described (7)]. Assays were performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems) or CFX96 Real-Time System (Bio-Rad Laboratories). The reactions were analyzed using the ΔΔCT analysis method.
Statistical analyses
Data are presented as mean ± SEM. Resource equation method was used to determine the adequate sample size (15). Significant differences between the groups were analyzed with one-way ANOVA followed by Tukey’s honestly significant difference using R for the BA analysis and one-way ANOVA followed by Dunnett’s multiple comparison tests using GraphPad Prism 6 software for the gene expression analysis.
RESULTS
Cecal microbiota differs between mice colonized with mouse or human microbiota
We analyzed cecal microbiota after short-term (2 weeks) or long-term (15 weeks) colonization of GF mice with cecal content from a mouse donor (recipient mice termed CONV-M) or feces from a human donor (recipient mice termed CONV-H), to investigate if there was a difference in their gut microbiota composition.
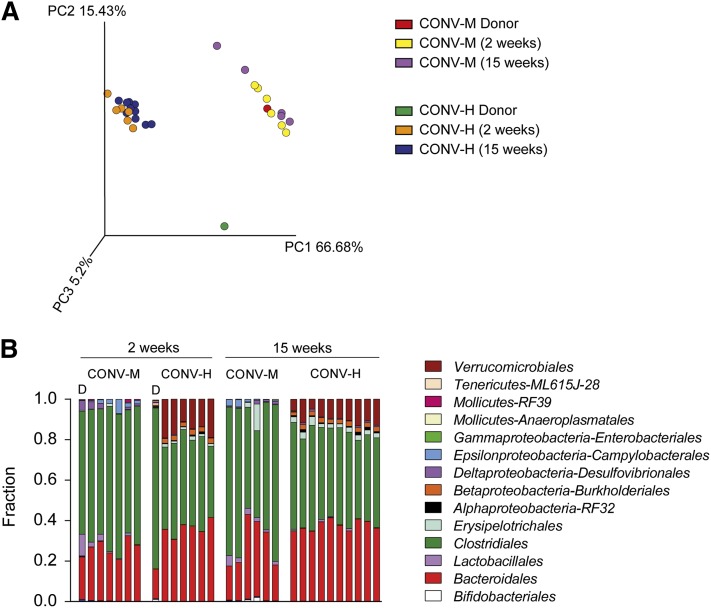
Principal coordinate analysis of weighted UniFrac distances showed a clear separation between the microbial communities in the mice driven by the origin of the donor microbiota observed at the first principal coordinate (x-axis), which explained more than 66% of the variance (Fig. 1A). The second principal coordinate (y-axis) accounted for 15% of the variance and separated the microbial communities by host (human vs. mouse) (Fig. 1A). In particular, the community of the human donor sample separated from the communities of CONV-H mice, CONV-M mice, and the mouse donor sample. In contrast, the community of the mouse donor sample and the CONV-M recipient mice clustered together (Fig. 1A).
Fig. 1.
Gut microbiota composition after colonization with human or mouse microbiota. A: Mouse cecal bacterial communities were clustered using principal coordinates analysis of the UniFrac weighted distance matrix. The percentage of the variation explained by the plotted principal coordinates is indicated in the axis labels. Each dot represents a cecal community. B: Relative abundance of orders in cecal bacteria from CONV-M and CONV-H mice colonized for 2 weeks or 15 weeks. n = 6–10 mice /group.
Next, we analyzed the microbiota composition of the donors and recipient mice and found that at order level the cecal microbiota of the CONV-M mice had a higher representation of Clostridiales, whereas CONV-H mice were characterized by a more pronounced representation of Verrucomicrobiales, Burkoholderiales, and Erysipelotrichales (Fig. 1B). The lower abundance of Clostridiales in the humanized mice was not caused by low representation of this taxon in the human donor samples. In fact, Clostridiales was highly abundant in the human donor sample, in agreement with a previous report (12). In contrast, the human donor sample had relatively low abundance of Verrucomicrobiales, Burkoholderiales, Erysipelotrichales, and Bacteroidales. The microbiota composition of CONV-M mice was similar to the mouse donor sample apart from a lower representation of Lactobacillales and higher representation of Campylobacterales. The mice colonized for 15 weeks had similar microbiota composition at the order level as the mice colonized for 2 weeks, which indicates that the microbiota composition remains stable over time (Fig. 1B). These findings were also confirmed in mice colonized with a second human donor (supplemental Fig. S1A).
To identify taxonomic differences in the microbiota between mice colonized with mouse and human microbiota, we applied an LEfSe algorithm (13). After 2 weeks’ colonization, the cecal microbiota of CONV-M mice was enriched in members of Bacilli such as Lactobacillus and Staphylococcus and the Epsilonproteobacteria-Helicobacteraceae (supplemental Fig. S2). Several members of the Bacteroidetes phylum such as Prevotella, Parabacteroides, and Bacteroides, as well as an unclassified RF39 Mollicutes and a member of the Proteobacteria phylum, Desulfovibrio, were also enriched in the CONV-M mice after 2 weeks of colonization (supplemental Fig. S2). The taxa enriched in the cecal microbiota of CONV-H mice after 2 weeks of colonization were affiliated to Akkermansia from the Verrucomicrobia phylum, several members of class Erysipelotrichi such as Allobaculum, Coprobacillus, Eubacterium dollichum and cc115 and Oxalobacter formigenes, Suturella and Bilophila from the Proteobacteria phylum.
Most of the taxonomic clades that were differently enriched after 2 weeks’ colonization showed a similar pattern after 15 weeks’ colonization demonstrating stability of the bacterial communities in the colonized mice over time. Some of the additional differences we found after 15 weeks’ colonization were enrichment of Bifidobacterium, the genus Anaeroplasma from the Tenerecutes phylum and Allobaculum in the CONV-M mice and enrichment of Enterobacterales in the CONV-H mice (supplemental Fig. S3).
In summary, we show that the microbiota composition differs between mice colonized with mouse and human microbiota, and the major differences between the groups are preserved over long-term colonization.
BA composition differs between mice colonized with mouse or human microbiota
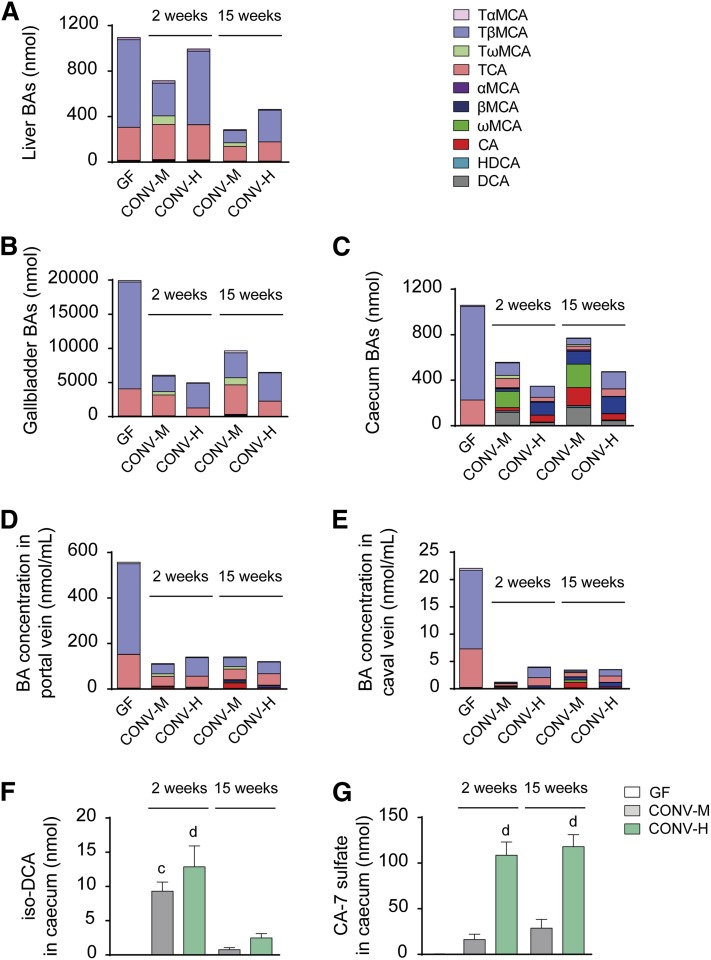
Next, we evaluated how colonization with mouse or human microbiota affected BA levels and composition in liver, gallbladder, and cecum of the recipient mice. Because of the major differences in relative gallbladder and cecum weights between the GF and colonized mice (supplemental Tables S2 and S3), we calculated the amounts of BAs in the whole organ rather than the amount of BAs per milligram of tissue. Total BA levels (referring to the total amounts of BAs analyzed) in the liver did not change significantly after the short-term colonization, but after 15 weeks’ colonization, the total BAs were reduced in both CONV-M and CONV-H mice compared with GF counterparts (Fig. 2A; supplemental Table S1). In gallbladder, total BAs were significantly reduced in all colonized mice at both time points (Fig. 2B; supplemental Table S2), and BAs levels in cecum were reduced in all groups except for CONV-M mice after 15 weeks (Fig. 2C; supplemental Table S3). Analysis of individual BAs showed that TβMCA and TCA were the predominant BAs in all compartments analyzed in GF mice (Fig. 2A–E; supplemental Tables S1–S5). In the liver, short-term colonization with mouse or human microbiota did not change the levels of TβMCA or TCA significantly, but colonization with mouse microbiota resulted in the presence of tauro-ω-muricholic acid (TωMCA), a secondary BA derived from TβMCA (Fig. 2A; supplemental Table S1). After 15 weeks’ colonization, the levels of TβMCA decreased in both CONV-M and CONV-H mice, while the levels of TCA remained unchanged. In gallbladder, the levels of TβMCA were significantly reduced after 2 weeks’ colonization with either mouse or human microbiota, while the levels of TCA were reduced only in the CONV-H mice (Fig. 2B; supplemental Table S2).
Fig. 2.
Changes in BAs composition in different compartments after colonization. Whole organ amounts of BAs in liver (A), gallbladder (B), and cecum (C). Concentrations of BAs in portal vein (D) and caval vein (E). For statistics on specific BA levels, see supplemental Tables 1–5. Whole organ amounts of iso-DCA (F) and CA-7 sulfate (G) in cecum. Mean values ± SEM are plotted; n = 4–9 samples/group; a P < 0.05, b P < 0.01, c P < 0.001, d P < 0.0001 indicate differences versus GF with ANOVA and Dunnett’s multiple comparisons test. HDCA, hyodeoxycholic acid.
In cecum, mouse microbiota had a large effect on BA composition. The major cecal BAs in CONV-M mice were ωMCA and DCA (secondary BAs derived from TβMCA and TCA, respectively) (Fig. 2C; supplemental Table S3). In cecum of the humanized mice, the most prevalent BA was βMCA followed by TβMCA, and there were only small amounts of DCA and almost no ωMCA (Fig. 2C; supplemental Table S3). To rule out the possibility that the low amounts of secondary BAs in the humanized mice were specific to this particular human donor, we colonized GF mice with human feces from a second donor and analyzed the BA composition. Levels of total and secondary BAs in cecum of mice colonized with feces from the second human donor did not differ significantly from the first donor (supplemental Fig. S1B) indicating that low production of secondary BAs in humanized mice might be a general finding. The relative amount of secondary BAs in cecum was low in the humanized mice, while secondary BAs constituted more than 50% of the cecal BAs in mice colonized with a mouse microbiota (supplemental Fig. S4A–F). In addition, the relative amount of unconjugated BAs was higher in the CONV-M mice after 15 weeks’ colonization compared with the humanized mice, suggesting a difference in bile salt hydrolase activity between the mouse and human microbiota (supplemental Fig. S5A–F).
We also analyzed the BA composition in portal vein serum, which contains reabsorbed BAs from the intestine and thus reflects the BA composition in ileum. Total BA levels and, in particular, TβMCA levels were dramatically reduced in the portal vein serum by colonization with mouse or human microbiota (Fig. 2D; supplemental Table S4). We found a similar pattern of BA changes in caval vein serum, with particularly low BA levels in the CONV-M mice 2 weeks after colonization (Fig. 2E; supplemental Table S5). The total concentration of BAs were >30-fold lower in the peripheral circulation compared with the levels in portal vein demonstrating the capacity of the liver to maintain BAs within the enterohepatic circulation. Furthermore, we found high levels of CA in both portal and caval vein serum after 15 weeks’ colonization with mouse microbiota (Fig. 2D, E; supplemental Tables S4 and S5).
In addition to the regular BAs we also analyzed 3β-hydroxylated iso-BAs in cecum and found significant amounts of iso-DCA after 2 weeks’ colonization both in CONV-M and CONV-H mice and after 15 weeks’ colonization the levels were decreased in both groups of colonized mice (Fig. 2F).
We also investigated the levels of sulfated BAs in cecum and found that mice colonized with a human microbiota had four times higher levels of CA-7 sulfate than mice colonized with mouse microbiota (Fig. 2G).
Mice colonized with the second human donor had similar levels of CA-7 sulfate as mice colonized with the first human donor, but the levels of iso-DCA were lower at 2 weeks’ colonization (supplemental Fig. S1C, D). As expected, we did not detect any iso-DCA in the GF mice and only traces of CA-7 sulfate (Fig. 2F, G; supplemental Fig. S1C, D). Thus, CONV-H mice deconjugated primary BAs as CONV-M counterparts but only metabolized CA and not βMCA. In addition, humanized mice generated more CA-7 sulfate than mice colonized with a mouse microbiota.
Metabolism of TCA by a human microbiota in vitro
To further investigate the potential of a human microbiota to deconjugate, dehydroxylate, and sulfate TCA, we performed in vitro experiments with fecal samples from the first human donor in culture medium supplemented with TCA or CA. BA analysis showed that the human microbiota sufficiently deconjugated TCA into CA, but there was only minor conversion into DCA and no production of CA-7 sulfate (supplemental Fig. S6A, B).
Colonization with a human microbiota induces FXR signaling
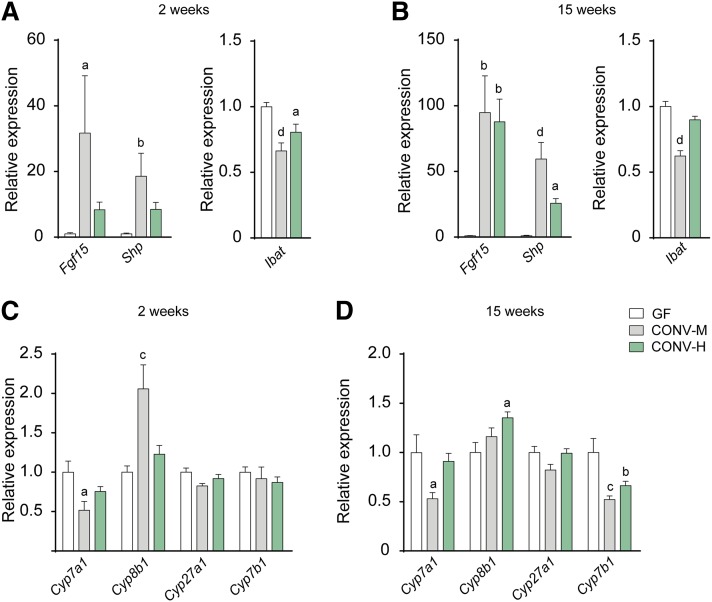
To evaluate if the changes in BAs composition following colonization with a human microbiota would be sufficient to increase FXR signaling, we analyzed gene expression of FXR target genes in ileum and liver. Colonization with mouse microbiota induced expression of both Fgf15 and Shp in ileum already after 2 weeks’ colonization (Fig. 3A), and the expression was further increased after 15 weeks’ colonization (Fig. 3B), which is in agreement with our previous study (7). Colonization with a human microbiota showed a tendency of induction after 2 weeks’ colonization and reached significant levels after 15 weeks’ colonization. We also analyzed gene expression of the ileal BA transporter Ibat and found a significant reduction in the mice colonized with a mouse microbiota at both time points and a minor reduction in the humanized mice (Fig. 3A, B).
Fig. 3.
Microbiota-induced changes in FXR signaling. Gene expression of FXR target genes Fgf15, Shp, and Ibat in ileum after 2 weeks (A) or 15 weeks (B) of colonization. Expression of genes involved in BA synthesis in liver after 2 weeks (C) or 15 weeks (D) of colonization. Gene expression is presented as relative expression compared with the mean expression of the GF control group. Mean values ± SEM are plotted; n = 4–9 samples/group; a P < 0.05, b P < 0.01, c P < 0.001, d P < 0.0001 indicate differences versus GF with ANOVA and Dunnett’s multiple comparisons test.
Furthermore, we analyzed gene expression of liver enzymes involved in BA synthesis and found that colonization with mouse microbiota (both short term and long term) promoted decreased gene expression of the rate-limiting enzyme CYP7A1, and the humanized mice showed a similar expression pattern, although the changes were not significant (Fig. 3C, D). The changes in Cyp7a1 expression corresponded inversely with the changes in Fgf15 expression in ileum; this reciprocal pattern has been observed in previous studies (7, 8, 16–19). The gene expression of sterol 12-α-hydroxylase (CYP8B1), which is required for CA synthesis, was increased after colonization (Fig. 3C, D). The expression of Cyp7a1 and Cyp8b1 is regulated by a negative feedback mechanism involving FXR and SHP in the liver (20–22). However, our finding that Cyp7a1 and Cyp8b1 are differently regulated is supported by several previous studies (7, 23–26). The genes encoding the cytochrome enzymes involved in the alternative BA synthesis, sterol 27-hydroxylase (CYP27A1) and oxysterol 7-α-hydroxylase (CYP7B1), were unchanged after 2 weeks’ colonization with mouse or human microbiota (Fig. 3C), and after 15 weeks’ colonization, the expression of CYP27A1 was still unchanged while CYP7B1 expression was reduced in both CONV-M and CONV-H mice (Fig. 3D).
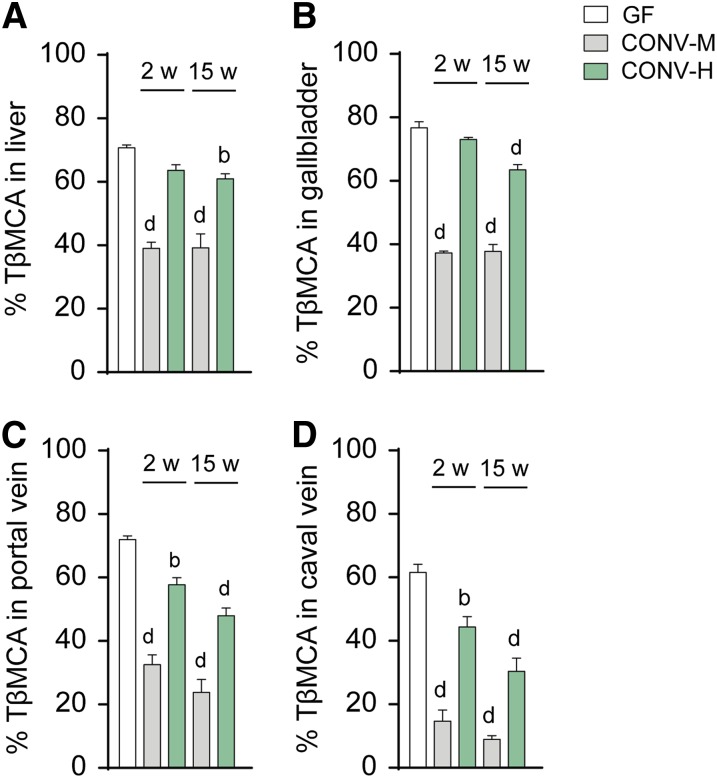
To further pinpoint the relationship between BA composition and FXR signaling in mice colonized with mouse versus human microbiota, we calculated the percentage of the FXR antagonist TβMCA in liver, gallbladder, and portal and caval veins and investigated if the levels of TβMCA would reflect the differences in expression of FXR target genes. Indeed, the reduction in percentage of TβMCA corresponded inversely with the increased expression of Fgf15 and Shp in ileum. In the CONV-M mice, there was a significant reduction in TβMCA levels in liver and gallbladder already after 2 weeks, while in the CONV-H mice a significant reduction of TβMCA was not seen until after 15 weeks’ colonization (Fig. 4A, B). In portal and caval veins, the percentage of TβMCA was significantly reduced in all the colonized mice after 2 weeks’ colonization and further reduced after 15 weeks’ colonization (Fig. 4C, D). Because FXR activation is dependent on the levels of both agonists and antagonists, we also calculated the ratio between FXR agonistic (TCA, TCDCA, TDCA, TLCA, CA, CDCA, DCA, LCA) and antagonistic BAs (TαMCA, TβMCA). We found an increased ratio in the CONV-M mice already after 2 weeks’ colonization and a minor but significant increase of the ratio in gallbladders of the humanized mice after 15 weeks’ colonization (supplemental Fig. S7A–D), which is in agreement with the changes in FXR target gene expression.
Fig. 4.
Changes in TβMCA levels after colonization. Percentage of the FXR antagonist TβMCA in liver (A), gallbladder (B), portal vein (C), and caval vein (D). Mean values ± SEM are plotted; n = 4–9 samples/group; a P < 0.05, b P < 0.01, c P < 0.001, d P < 0.0001 indicate differences versus GF with ANOVA and Dunnett’s multiple comparisons test. 2w, 2 weeks; 15w, 15 weeks.
DISCUSSION
We show that colonization of mice with a human microbiota reduces total BA levels to a similar extent as colonization with a mouse microbiota, but BA composition differs and fewer secondary BAs are present in the humanized mice. Furthermore, we show that a human microbiota can reduce the levels of TβMCA sufficiently to induce FXR signaling and increase expression of FXR target genes in ileum.
Nevertheless, it appears that humanized mice lack specific bacteria important for secondary BA production. In particular, human microbiota seems to lack capacity to generate secondary BAs from the murine primary BAs, which has been indicated in earlier studies (27, 28). This may be explained by the fact that the human microbiome has never been exposed to murine BAs and is therefore not adapted to them. We tested the hypothesis that the human microbiota might require a longer time to adjust to the environment of the mouse gut and colonized some of the mice for 15 weeks. Long-term colonization resulted in lower levels of the FXR antagonist TβMCA, but the levels of secondary BAs did not increase significantly, and we did not observe major changes in the microbiota composition of the humanized mice after long-term colonization.
One notable difference between the microbiota composition of CONV-M and CONV-H mice was the higher representation of Clostridiales in the mice colonized with mouse microbiota even though the Clostridiales was highly abundant in the human donor sample. This finding is in agreement with a previous study on mice colonized with human microbiota, which reported relatively high abundance of Clostridia and low abundance of Erysipelotrichi in the human donor sample and the opposite (low Clostridia and high Erysipelotrichi abundance) in the recipient mice (11). These results indicate that some species-specific divergences in microbiota composition cannot be transferred from one mammalian host to another, likely due to the environment in the recipient gut, and hence the microbiota of the donor sample is more preserved when transferred to a recipient of the same species. Previous studies have shown that bacterial species (e.g., Lactobacillus reuteri) can have subpopulations with strong host specificity and a gene content reflecting the niche characteristics for the specific hosts (29). This phenomenon might explain why Clostridiales from human feces colonize mice in lower abundance than Clostridiales with mouse origin. In addition, the mouse diet differs from the human diet and is less diverse, which can also influence the microbiota composition.
Previous studies have reported low levels of secondary BAs upon colonization of GF mice with human infant microbiota, and this response was attributed to the fact that infants have low bacterial diversity (30, 31). However, our findings suggest that not only infant but also adult microbiota has reduced capacity to metabolize murine BAs.
Both HDCA and ωMCA have been reported as possible secondary BAs generated from βMCA (32–34). Others have reported high levels of HDCA and no ωMCA in CONV-R mice (35), in contrast to our finding of predominantly ωMCA and very low levels of HDCA in mice colonized with mouse microbiota. Variations in the microbiota community between different mouse facilities might explain these discrepancies because specific bacteria possess different abilities to generate these two secondary BAs (generation of HDCA from βMCA requires both 7β-dehydroxylation and 6β-epimerization, whereas generation of ωMCA only requires 6β-epimerization).
DCA is the secondary BA generated from CA, and it is common for both humans and mice. Nevertheless, the humanized mice produced lower levels of DCA than mice with a mouse microbiota, which may be explained by the relatively low levels of Clostridiales. In contrast, the levels of iso-DCA were somewhat higher in the humanized mice indicating an alternative metabolic pathway of CA in these mice. Iso-DCA is a common BA produced by bacteria in the human gut, and the levels can vary largely between different individuals, which could explain why the mice colonized with the second human donor have lower levels of this BA metabolite (36–38).
Furthermore, we found that the human microbiota induced sulfation of CA. The presence of CA-7 sulfate in mice has previously been reported by Eyssen et al. (39, 40). BA sulfation can occur in the liver as an alternative pathway in BA metabolism when the enterohepatic circulation is interrupted (41). However, we did not detect CA-7 sulfate in liver or gallbladder in the colonized mice, and we only detected traces in the cecum of GF mice, which suggests that the CA-7 sulfate might be produced by bacteria in the gut. Another possibility is that sulfation might occur in the intestinal epithelial cells, which have been suggested previously (37, 42). Our in vitro assays showed that a human microbiota sufficiently deconjugated TCA but did not generate any CA-7 sulfate, which supports the hypothesis that CA-7 sulfate might be produced by the host rather than the microbiota. The low levels of DCA in the humanized mice could in part be explained by increased sulfation of CA at the 7-hydroxyl group, instead of common 7α-dehydroxylation. Another possible contribution to the decreased DCA formation could be inhibition of CA transport into 7α-dehydroxylating bacteria.
Our finding that Fgf15 and Shp expression in ileum was increased after colonization shows that a human microbiota can enhance FXR signaling, but not to the same extent as observed with a mouse microbiota. It is noteworthy that even though the human microbiota cannot metabolize TβMCA to secondary murine BAs, it can still induce a change in BA composition that permits FXR signaling. We hypothesize that deconjugation of TβMCA is sufficient to remove the FXR antagonistic effect and initiate the negative-feedback inhibition of BA synthesis. This is in line with other studies and our previous findings showing that only the T-conjugated form of βMCA displays FXR antagonistic properties (7, 8, 43).
In conclusion, we show that a human microbiota can establish a functional community in the mouse gut and that it reduces total BA levels to the same extent as a mouse microbiota. Furthermore, a human microbiota can reduce TβMCA levels and induce FXR signaling. These are important findings that justify the use of humanized mice for studies on FXR-dependent interactions between a human microbiota and host metabolism. However, it is important to keep in mind that when we introduce human bacteria into GF mice with the aim to study the effect of specific bacteria on metabolic functions, the starting condition is inhibition of FXR by TβMCA, which is absent in humans. CYP2C70 has recently been suggested as the enzyme responsible for the formation of murine BAs by catalyzing 6β-hydroxylation of CDCA and ursodeoxycholic acid (UDCA) into α- and βMCA respectively (44). Thus, mice with a deletion in the Cyp2c gene cluster (including Cyp2c70) exhibit a BA pool without murine BAs and solely CA, CDCA, and UDCA as primary BAs, although still with almost exclusively T conjugates. To use GF Cyp2c70-deficient mice and introduce human microbiota into these mice might be an improved model to study the effect of a human microbiota on metabolic functions.
Supplementary Material
Acknowledgments
The authors thank Rosie Perkins for editing the manuscript; Anna Hallén for excellent technical assistance; Rozita Akrami and Valentina Tremaroli for preprocessing of the raw sequencing data; Carina Arvidsson, Sara Nordin-Larsson, Caroline Wennberg, and Ulrica Enqvist for superb mouse husbandry; Genomic Core Facility of Gothenburg University for 16S rRNA sequencing; and Tord Inghardt at AstraZeneca in Mölndal for helping with the synthesis of CA-7 sulfate.
Footnotes
Abbreviations:
- BA
- bile acid
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- CONV-H
- humanized
- CONV-M
- conventionalized
- CONV-R
- conventionally raised
- CYP7A1
- cholesterol 7α-hydroxylase
- CYP8B1
- sterol 12-α-hydroxylase
- DCA
- deoxycholic acid
- FGF19/15
- fibroblast growth factor 19/15
- FXR
- farnesoid X receptor
- G
- glycine
- GF
- germ free
- HDCA
- hyodeoxycholic acid
- LCA
- lithocholic acid
- SHP
- small heterodimer partner
- T
- taurine
- α/βMCA
- α- and β-muricholic acid
- ωMCA
- ω-muricholic acid.
This study was supported by the Swedish Research Council, Hjärt-Lungfonden, Torsten Söderbergs Stiftelse, Ragnar Söderbergs stiftelse, IngaBritt och Arne Lundbergs Forskningsstiftelse, Swedish Foundation for Strategic Research, Knut and Alice Wallenberg Foundation, and the regional agreement on medical training and clinical research (ALF) between Västra Götalandsregionen and Sahlgrenska Universitetssjukhuset. F.B. is a recipient of ERC Consolidator Grant (European Research Council, Consolidator Grant 615362 - METABASE).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Karlsson F., Tremaroli V., Nielsen J., and Backhed F.. 2013. Assessing the human gut microbiota in metabolic diseases. Diabetes. 62: 3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre P., Cariou B., Lien F., Kuipers F., and Staels B.. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191. [DOI] [PubMed] [Google Scholar]
- 3.Ma H., and Patti M. E.. 2014. Bile acids, obesity, and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 28: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato H., Macchiarulo A., Thomas C., Gioiello A., Une M., Hofmann A. F., Saladin R., Schoonjans K., Pellicciari R., and Auwerx J.. 2008. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J. Med. Chem. 51: 1831–1841. [DOI] [PubMed] [Google Scholar]
- 5.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- 6.Wang H., Chen J., Hollister K., Sowers L. C., and Forman B. M.. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553. [DOI] [PubMed] [Google Scholar]
- 7.Sayin S. I., Wahlstrom A., Felin J., Jantti S., Marschall H. U., Bamberg K., Angelin B., Hyotylainen T., Oresic M., and Backhed F.. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 8.Li F., Jiang C. T., Krausz K. W., Li Y. F., Albert I., Hao H. P., Fabre K. M., Mitchell J. B., Patterson A. D., and Gonzalez F. J.. 2013. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4: 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahlström A., Sayin S., Marschall H., and Bäckhed F.. 2016. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24: 41–50. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., and Gordon J. I.. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., and Gordon J. I.. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seedorf H., Griffin N. W., Ridaura V. K., Reyes A., Cheng J., Rey F. E., Smith M. I., Simon G. M., Scheffrahn R. H., Woebken D., et al. 2014. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 159: 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., and Huttenhower C.. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tremaroli V., Karlsson F., Werling M., Stahlman M., Kovatcheva-Datchary P., Olbers T., Fandriks L., le Roux C. W., Nielsen J., and Backhed F.. 2015. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 22: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Festing M. F., and Altman D. G.. 2002. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J. 43: 244–258. [DOI] [PubMed] [Google Scholar]
- 16.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. 2003. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17: 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 18.Lundåsen T., Galman C., Angelin B., and Rudling M.. 2006. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J. Intern. Med. 260: 530–536. [DOI] [PubMed] [Google Scholar]
- 19.Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., and Gonzalez F. J.. 2007. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48: 2664–2672. [DOI] [PubMed] [Google Scholar]
- 20.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., and Mangelsdorf D. J.. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: 507–515. [DOI] [PubMed] [Google Scholar]
- 21.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526. [DOI] [PubMed] [Google Scholar]
- 22.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 23.Out C., Hageman J., Bloks V. W., Gerrits H., Sollewijn Gelpke M. D., Bos T., Havinga R., Smit M. J., Kuipers F., and Groen A. K.. 2011. Liver receptor homolog-1 is critical for adequate up-regulation of Cyp7a1 gene transcription and bile salt synthesis during bile salt sequestration. Hepatology. 53: 2075–2085. [DOI] [PubMed] [Google Scholar]
- 24.Inoue Y., Yu A. M., Yim S. H., Ma X., Krausz K. W., Inoue J., Xiang C. C., Brownstein M. J., Eggertsen G., Bjorkhem I., et al. 2006. Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4alpha. J. Lipid Res. 47: 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abrahamsson A., Gustafsson U., Ellis E., Nilsson L. M., Sahlin S., Bjorkhem I., and Einarsson C.. 2005. Feedback regulation of bile acid synthesis in human liver: importance of HNF-4alpha for regulation of CYP7A1. Biochem. Biophys. Res. Commun. 330: 395–399. [DOI] [PubMed] [Google Scholar]
- 26.Song P., Rockwell C. E., Cui J. Y., and Klaassen C. D.. 2015. Individual bile acids have differential effects on bile acid signaling in mice. Toxicol. Appl. Pharmacol. 283: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacquet E. C., Gadelle D. P., Riottot M. J., and Raibaud P. M.. 1984. Absence of transformation of beta-muricholic acid by human microflora implanted in the digestive tracts of germfree male rats. Appl. Environ. Microbiol. 47: 1167–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacquet E., Parquet M., Riottot M., Raizman A., Nordlinger B., and Infante R.. 1985. Metabolism of beta-muricholic acid in man. Steroids. 45: 411–426. [DOI] [PubMed] [Google Scholar]
- 29.Frese S. A., Benson A. K., Tannock G. W., Loach D. M., Kim J., Zhang M., Oh P. L., Heng N. C., Patil P. B., Juge N., et al. 2011. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 7: e1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin F. P., Dumas M. E., Wang Y., Legido-Quigley C., Yap I. K., Tang H., Zirah S., Murphy G. M., Cloarec O., Lindon J. C., et al. 2007. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin F. P., Wang Y., Sprenger N., Yap I. K., Lundstedt T., Lek P., Rezzi S., Ramadan Z., van Bladeren P., Fay L. B., et al. 2008. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 4: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Limaye P. B., Renaud H. J., and Klaassen C. D.. 2014. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol. Appl. Pharmacol. 277: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sacquet E. C., Raibaud P. M., Mejean C., Riottot M. J., Leprince C., and Leglise P. C.. 1979. Bacterial formation of omega-muricholic acid in rats. Appl. Environ. Microbiol. 37: 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eyssen H., De Pauw G., Stragier J., and Verhulst A.. 1983. Cooperative formation of omega-muricholic acid by intestinal microorganisms. Appl. Environ. Microbiol. 45: 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degirolamo C., Rainaldi S., Bovenga F., Murzilli S., and Moschetta A.. 2014. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Reports. 7: 12–18. [DOI] [PubMed] [Google Scholar]
- 36.Bayerdörffer E., Mannes G. A., Ochsenkühn T., Dirschedl P., Wiebecke B., and Paumgartner G.. 1995. Unconjugated secondary bile acids in the serum of patients with colorectal adenomas. Gut. 36: 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton J. P., Xie G., Raufman J. P., Hogan S., Griffin T. L., Packard C. A., Chatfield D. A., Hagey L. R., Steinbach J. H., and Hofmann A. F.. 2007. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G256–G263. [DOI] [PubMed] [Google Scholar]
- 38.Devlin A. S., and Fischbach M. A.. 2015. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11: 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyssen H. J., Parmentier G. G., and Mertens J. A.. 1976. Sulfate bile acids in germ-free and conventional mice. Eur. J. Biochem. 66: 507–514. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y., and Klaassen C. D.. 2010. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51: 3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alnouti Y. 2009. Bile Acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108: 225–246. [DOI] [PubMed] [Google Scholar]
- 42.Hagey L. R., and Krasowski M. D.. 2013. Microbial biotransformations of bile acids as detected by electrospray mass spectrometry. Adv. Nutr. 4: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang C., Xie C., Li F., Zhang L., Nichols R. G., Krausz K. W., Cai J., Qi Y., Fang Z. Z., Takahashi S., et al. 2015. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest. 125: 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi S., Fukami T., Masuo Y., Brocker C. N., Xie C., Krausz K. W., Wolf C. R., Henderson C. J., and Gonzalez F. J.. 2016. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57: 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.