Abstract
Hepatitis C virus (HCV) is an enveloped RNA virus responsible for 170 million cases of viral hepatitis worldwide. Over 50% of chronically infected HCV patients develop hepatic steatosis, and steatosis can be induced by expression of HCV core protein (core) alone. Additionally, core must associate with cytoplasmic lipid droplets (LDs) for steatosis development and viral particle assembly. Due to the importance of the LD as a key component of hepatic lipid storage and as a platform for HCV particle assembly, it seems this dynamic subcellular organelle is a gatekeeper in the pathogenesis of viral hepatitis. Here, we hypothesized that core requires the host LD scaffold protein, perilipin (PLIN)3, to induce hepatic steatosis. To test our hypothesis in vivo, we have studied core-induced hepatic steatosis in the absence or presence of antisense oligonucleotide-mediated knockdown of PLIN3. PLIN3 knockdown blunted HCV core-induced steatosis in transgenic mice fed either chow or a moderate fat diet. Collectively, our studies demonstrate that the LD scaffold protein, PLIN3, is essential for HCV core-induced hepatic steatosis and provide new insights into the pathogenesis of HCV.
Keywords: triglyceride, lipid droplets, lipoproteins/metabolism, liver
The hepatitis C virus (HCV) affects 3% of the world’s population and is a leading cause of end stage liver failure, presenting a considerable global healthcare burden (1). Liver tissue is the primary target for HCV infections, where the virus elegantly co-opts the hepatic lipid metabolic processes to promote viral assembly (2, 3). Steatosis is a hallmark of HCV infection, occurring in 50–70% of chronically infected individuals (4). Importantly, there is an inverse correlation between steatosis and response to antiviral treatment and hepatic steatosis increases the risk for steatohepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) (4). Hepatic steatosis is characterized by a pathological accumulation of liver lipids resulting in increases in the size and number of lipid droplets (LDs). Although it is well-appreciated that HCV infection promotes hepatic steatosis, the exact mechanism is incompletely understood. The HCV core protein (core) is a viral structural protein that serves to form a capsid around the viral RNA genome and has been shown to induce steatosis in various models (5, 6). Core is the first viral protein translated and undergoes two proteolytic processing events at the endoplasmic reticulum membrane (7). Once core undergoes the second cleavage by signal peptide peptidase, it is able to associate with host cytoplasmic LDs via a C-terminal binding domain composed of two amphipathic helices separated by a hydrophobic loop (8, 9). The processing events that allow core to associate with LDs have been shown to be essential for steatosis formation (10). Once core is associated with the LD, newly synthesized viral RNA is trafficked, by HCV nonstructural protein NS5A, to core where core forms a protein capsule (nucleocapsid) around the HCV genome (11). Importantly, nucleocapsid formation by core occurs at the LD, which serves as the primary site of HCV viral particle assembly. The importance of the core-LD interaction is highlighted by independent studies demonstrating that LDs are required for HCV production and that disruption of core from the LD inhibits virus production (12, 13). There is now unequivocal evidence that the ability of core to interact with host LDs is critical for both viral particle assembly as well as associated hepatic steatosis (10, 12, 13).
Host liver LDs, which are essential for HCV particle production, are dynamic organelles storing neutral lipids within a hydrophobic core. The neutral lipid core, composed mainly of TGs and cholesterol esters, is surrounded by a phospholipid monolayer that is decorated by a unique proteome (14). LDs expand and contract based on cellular metabolic demand. When fatty acids are in excess, they can be incorporated into TGs by enzymes, such as diacylglycerol acyl transferase (DGAT), which catalyze the terminal step in TG synthesis (15). When the cellular energy state is diminished, lipolytic enzymes can hydrolyze TGs from within the LD to release free fatty acids for β-oxidation. One of the primary lipolytic enzymes for TG hydrolysis is adipose TG lipase (ATGL), along with its cofactor, comparative gene identification 58 (CGI-58), which cooperate to release fatty acids from the hydrophobic TG-rich LD core (16–18). Importantly, enzymes such as the ATGL-CGI-58 complex must gain access to the neutral lipid core of the LD in order to release fatty acids. Notably, certain LD proteins, specifically the perilipin (PLIN), adipocyte differentiation-related protein (ADRP), and tail-interacting protein at 47 kDa (TIP47) (PAT) family proteins, can act as scaffolding proteins and help regulate LD size by serving as gatekeepers that regulate access to the neutral lipid core and allow for the assembly of lipogenic/lipolytic enzymes (19).
In liver, the two main LD-associated scaffolding proteins are PLIN3, also known as TIP47, and PLIN2, also known as ADRP (19, 20). PLIN3 is a highly exchangeable protein involved in multiple processes, including lipid storage, lipid mobilization, and LD biogenesis (21, 22). PLIN2 is a nonexchangeable protein that is degraded if displaced from the LD (23). Expression levels of PLIN2 and/or PLIN3 on the LD surface can vary in response to changing cellular metabolic needs to alter TG storage levels in the cell (24–27) by regulating accessibility of enzymes involved in lipid synthesis or lipolysis. Interestingly, previous evidence has shown that expression of core alters the normal composition of LD scaffolding proteins. Core expression in hepatoma cells resulted in increased PLIN3 expression and decreased PLIN2 expression (28). Furthermore, core expression has been shown to cause a redistribution of LDs from the cell periphery to the perinuclear region, which is associated with the displacement of PLIN2 from redistributed LDs (29). The observation that core expression alters important LD scaffolding protein expression and changes the LD proteome may highlight a mechanism involved in core-induced steatosis, which is the focus of the current study.
Previous studies have shown that core’s association with the LD is an indispensable part of HCV replication and persistence, making it an attractive target for discovery of new targets to inhibit HCV infections (12, 13, 30). Although the core-LD association is a critical part of the HCV life cycle, it is still unclear as to how the viral protein interacts with the LD resulting in a pathological accumulation of lipids in the liver. In order to understand the mechanisms involved in HCV-mediated pathogenesis, we have focused our studies on core-induced hepatic steatosis in vivo, given that hepatoma cell models have dramatically altered lipid metabolic processes when compared with primary hepatocytes. Core serves as a critical component in the viral life cycle of HCV and by defining how core interacts with the LD, we should be able to identify innovative targets to disrupt core’s association with the LD and impair HCV infections. Given that core expression has been shown to increase expression of the LD scaffolding protein, PLIN3 (28), we hypothesized that PLIN3 is an important component for core to induce steatosis. To test our hypothesis, we have generated a hepatocyte-specific HCV core transgenic mouse model and studied core-induced steatosis with normal or diminished levels of PLIN3.
MATERIALS AND METHODS
Generation of HCV core transgenic mice
Although HCV genotype 3a is most closely related to hepatic steatosis risk in people, we chose to use genotype 1b to allow comparisons to the vast majority of cell biology work with HCV core. The full-length coding sequence of HCV core (genotype 1b) was subcloned from the pCAGC191 vector (31) into the MluI- and ClaI-digested pLiv11 vector (32). The pLiv11 vector contains the human apoE promoter along with a 3′-hepatic control region, which selectively drives transgene expression in hepatocytes (32, 33). The pLiv11-HCVcore transgenic cassette was separated from the vector backbone by digestion with restriction enzymes NotI (5′) and SpeI (3′). This linearized vector was then microinjected into fertilized embryos of B6D2 F1J mice. PCR was used to confirm the presence of the HCV core gene, apoE promoter, and the hepatic control region (data not shown) in founder mice. Genotyping was performed by PCR analysis on genomic DNA isolated from ear snips, as previously described (34), using primers specific for the core construct as follows: primer 1, 5′-GAG CAC AAA TCC TAA ACC CCA AAG-3′, primer 2, 5′-GAT GGT CAA ACA GGA CAG CAG AG-3′. From this effort, we established three independent founder strains with low (HCVcoreTg29), medium (HCVcoreTg15), or high (HCVcoreTg3) core protein expression.
Animal studies
At the age of 6–8 weeks, subgroups of mice were fed ad libitum with either chow or a moderate fat diet (MFD) containing 20% of energy as lard and added cholesterol, 0.1% (w/w), for a total of 6 or 8 weeks. Mice treated with antisense oligonucleotide (ASO) received 50 mg/kg of either PLIN3 ASO or control (nontargeting) ASO via intraperitoneal injection once a week (total dose of 50 mg/kg per week) for 6–8 weeks. Second-generation ASOs were synthesized by Ionis Pharmaceuticals (Carlsbad, CA) and formulated in PBS (35). The PLIN3 ASO, Ionis 409003 (5′-CACAGTGTTGTCTAGGGCCT-3′), is a second-generation oligonucleotide that incorporates several chemical modifications to improve potency, duration of action, and tolerability. All of the internucleotide phosphates are chemically modified with a phosphorothioate substitution, in which one of the nonbridging oxygen atoms is substituted with sulfur. Additionally the compound incorporates five 2′-O-(2-methoxyethyl)-modified ribonucleosides at the 3′ and 5′ ends with ten 2′-O-deoxyribonucleosides in between to support RNaseH-1-mediated target mRNA degradation. These modifications improve the binding affinity for target mRNA as well as stability against exonuclease-mediated degradation. A control oligonucleotide, Ionis 141923 (5′-CCTTCCCTGAAGGTTCCTCC-3′), contains the same chemical modifications, with no complementarity to known genes, including LD proteins (26). For liver X receptor (LXR) agonist and fasting studies, 6-week-old C57BL/6 mice were fed ad libitum with chow for a period of 6 weeks while receiving treatment with either control or PLIN3 ASOs. For studies with LXR agonist, T0901317 was suspended in a vehicle containing 1.0% carboxymethylcellulose and 0.1% Tween 80. For a period of seven days mice were gavaged once daily with either vehicle or 25 mg/kg T0901317, as previously described (36, 37). For fasting studies, subgroups of mice were fasted for 18 h prior to necropsy.
All mice used in the studies were housed in a pathogen-free barrier facility at Wake Forest University School of Medicine or the Cleveland Clinic with the approval of the American Association for Accreditation of Laboratory Animal Care. The Institutional Animal Care and Use Committee from Wake Forest University or the Cleveland Clinic approved all protocols before execution of the studies.
Immunoblotting and real-time PCR
Proteins were resolved by SDS-PAGE, transferred to PVDF membrane (Millipore), and detected after incubation with the indicated antibodies using LiCor Odyssey Infrared Imaging system. The antibodies used include: anti-protein disulfide isomerase rabbit polyclonal (Cell Signaling #2446), anti- GAPDH rabbit monoclonal (Cell Signaling #5174), anti-α tubulin mouse monoclonal (Cell Signaling #3873), and anti-PLIN3 rabbit polyclonal (Proteintech #10694-1-AP). The monoclonal HCV core antigen antibody (C7-50) was obtained from Thermo Scientific. Densitometry was determined using Image Studio version 4.0.21. Tissue RNA extraction and quantitative (q)PCR were conducted as previously described (38, 39). Cyclophilin A was used for an invariant control and expression levels were calculated based on the ΔΔCT method. qPCR was conducted using the Applied Biosystems 7500 real-time PCR system. All qPCR primers are available upon request.
Liver histology
Portions of livers were fixed in 10% buffered formalin and processed for hematoxylin and eosin (H&E) staining by the Clinical Pathology Laboratory in the Department of Pathology at Wake Forest University School of Medicine or the Imaging Core in the Lerner Research Institute at the Cleveland Clinic Foundation.
Hepatic lipid quantification
Liver lipid extracts were made and TG, total cholesterol, free cholesterol, and cholesterol ester were measured using detergent-solubilized enzymatic assays as previously described (34, 38, 40).
Plasma lipid and lipoprotein analyses
Plasma TG levels were quantified enzymatically (L-Type TG M; Wako Diagnostics, Richmond, VA). Plasma lipoproteins were fractionated by size from 2.5 μl of individual plasma samples using a Superose 6 PC 3.2/30 column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) followed by on-line determination of TGs and cholesterol, as previously described (41). The lipid concentrations of the different lipoprotein fractions were calculated after integration of individual chromatograms.
Liver LD isolation
Hepatic LDs were isolated by sucrose gradient centrifugation essentially as described in (42). Approximately 100 mg of tissue was minced with a razor blade on a cold surface. Minced tissue was transferred to a Potter-Elvehjem homogenizer, and then 200 μl of 60% sucrose was added to the tissue sample and incubated on ice for 10 min. Next, 800 μl of lysis buffer was added and mixed, and then incubated on ice for 10 min. Samples were homogenized with five strokes of a Teflon® pestle and transferred to a 2 ml centrifuge tube. Lysis buffer (600 μl) was carefully layered on top of homogenate and centrifuged for 2 h at 20,000 g at 4°C. The tube was then frozen at −80°C and cut at the 1,000 μl mark. The bottom piece of the centrifuge tube contained the non-LD fraction, which was allowed to thaw before being transferred to a new tube. The LD fraction was collected by cutting an ∼4–6 mm piece from the top of the ice cylinder and placing it in a new 2 ml tube. To increase the purity of the LD fraction, this process was repeated once more. Briefly, 200 μl of 60% sucrose was added to the LD fraction. Next, 800 μl of lysis buffer was added and mixed followed by careful layering with 600 μl of lysis buffer and then centrifugation for 2 h at 20,000 g at 4°C. After freezing at −80°C, the tube was cut and the LD fraction was collected by cutting an ∼4–6 mm piece from the top of the ice cylinder and placing it in a new tube. Protein analysis was performed using the modified Lowry assay, as previously described (43). Lipids in the LD fraction were extracted according to the Folch method (44) and then determined using detergent-solubilized enzymatic assays, as previously described (34, 38, 40).
Statistical analyses
All graphs were plotted by GraphPad Prism 6.0e (45). Unless indicated, data are expressed as the mean ± SEM, and were analyzed using either a one- or two-way ANOVA followed by Tukey’s post hoc analysis using JMP version 10.0.2d1 software (SAS Institute, Cary, NC) as previously described (36, 46).
RESULTS
Expression of core in liver induces a dose-dependent increase in liver lipid accumulation
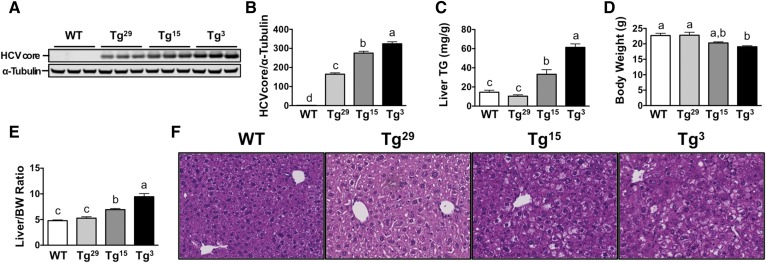
In order to study hepatic steatosis induced by core in vivo, we generated multiple lines of mice with hepatocyte-specific transgenic expression of core. To ensure hepatocyte-specific expression of core, we inserted the full-length sequence of genotype 1b into the pLiv11 vector (21). We used the sequence of genotype 1b because it is the most common genotype in chronic HCV infections (47). After backcrossing founder mice to a C57BL/6 background we analyzed liver lysates for expression of core and identified three distinct HCV core transgenic lines with increased core expression (Fig. 1A, B): HCVcoreTg29 (low core expression); HCVcoreTg15 (moderate core expression); and HCVcoreTg3 (high core expression). To examine the effect of core expression on liver lipid accumulation, we performed biochemical analysis of liver TGs (Fig. 1C) and found that HCVcoreTg15 and HCVcoreTg3 had significantly increased liver TGs relative to WT mice, with a 2.29- and 4.23-fold increase, respectively. While there was only a slight decrease in body weight (Fig. 1D), only reaching significance in HCVcoreTg3 mice relative to control, there was increased liver size in both HCVcoreTg15 and HCVcoreTg3 mice (Fig. 1E). H&E staining on fixed liver sections also showed a dose-dependent increase in liver lipid accumulation with increasing levels of core (Fig. 1F).
Fig. 1.
Expression of core in liver induces a dose-dependent increase in liver lipid accumulation. Male mice from each line were maintained on a chow diet until 6–8 weeks of age. A: Western blot analysis of liver homogenate of each transgenic line from (B). B: Quantitative analysis of liver immunoblot. C: Enzymatic determination of TGs from liver lipid extract (n = 3–7 per group). All hepatic lipid values were normalized to tissue weight. D, E: Total body weight and liver size (expressed as a ratio to body weight) at necropsy. F: Liver sections of each line were stained with H&E for morphological analysis (20× magnification). Data shown represent mean ± SEM. Levels not connected by the same letter are significantly different.
HCVcoreTg15 mice exhibit hepatic steatosis, which is exacerbated after feeding a MFD
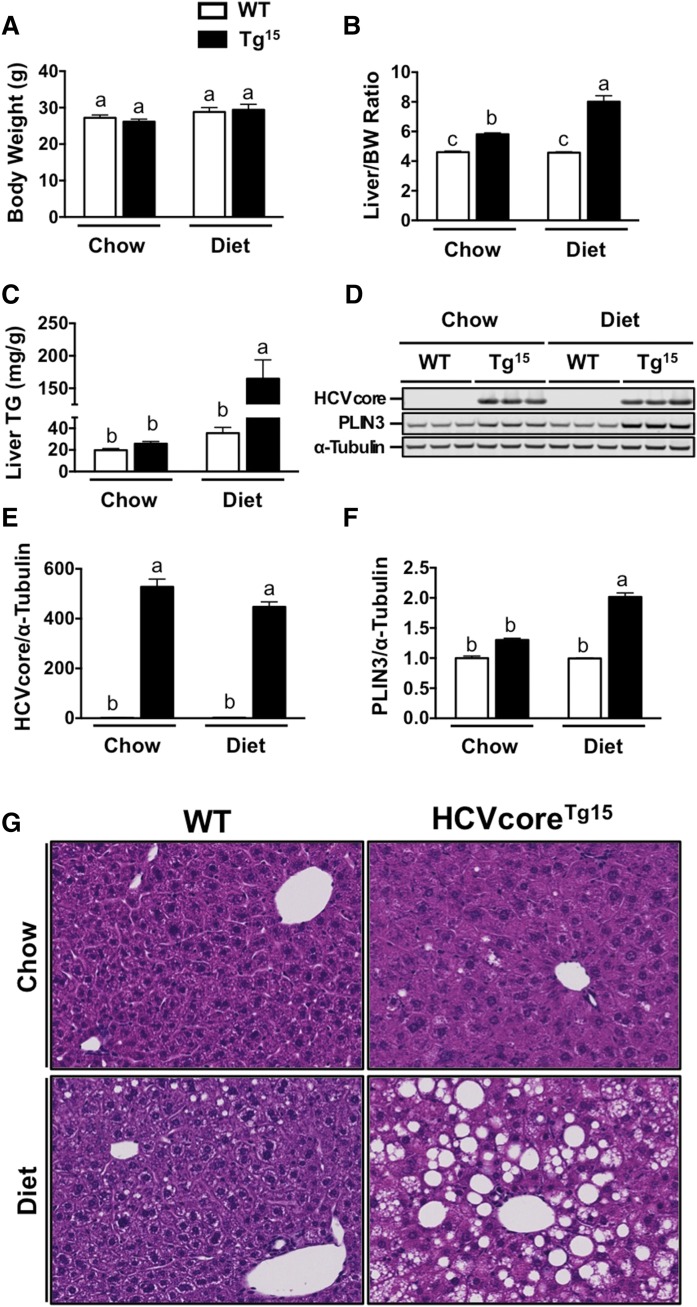
Initial studies were performed to determine the optimal dietary background to study HCV core-induced hepatic steatosis. In order to differentiate between core-induced steatosis and diet-induced steatosis, we elected to feed mice a MFD containing 20% kcal from lard and 0.1% added cholesterol (w/w) or a chow diet for six weeks (Fig. 2). There was no significant difference in body weight between WT littermates and HCVcoreTg15 mice on chow or MFD, although both groups had 5–12% increased body weight after 6 weeks of MFD (Fig. 2A). Additionally, MFD feeding did not alter liver size in WT mice, measured by liver to body weight ratio; however, HCVcoreTg15 mice consistently displayed a larger liver by 26% on chow and 74% on MFD (Fig. 2B). In addition, we found that HCVcoreTg15 mice had a significant increase in liver TGs after addition of MFD with a 4.64-fold increase relative to WT mice on MFD (Fig. 2C). Western blot analysis showed HCVcoreTg15 mice had 30% increased PLIN3 expression on a chow diet compared with WT; however, HCVcoreTg15 mice had a 2-fold increase in PLIN3 expression relative to WT mice on MFD (Fig. 2D–F). H&E staining of liver sections confirmed the increase in liver TG storage in HCVcoreTg15 mice compared with WT, which was greatly exacerbated by MFD feeding, as seen by an increase in LD size and number (Fig. 2G).
Fig. 2.
HCVcoreTg15 mice have a considerable increase in liver TGs after feeding with MFD (Diet). At 6 weeks of age, male WT or HCVcoreTg15 mice were placed on chow or MFD (20% kcal lard, 0.1% cholesterol) for a period of 6 weeks. A, B: Total body weight and liver size (expressed as a ratio to body weight) at necropsy. C: Enzymatic determination of TGs from liver lipid extract (n = 4–5 per group). D: Western blot analysis of liver homogenates from mice fed chow or MFD. E, F: Quantitative analysis of liver immunoblot for core (E) and PLIN3 (F). G: Liver sections of chow- and MFD-fed mice were stained with H&E for morphological analysis (20× magnification). Data shown represent mean ± SEM. Levels not connected by the same letter are significantly different.
ASO-mediated knockdown of PLIN3 expression decreases steatosis in HCVcoreTg15 mice on chow and MFD
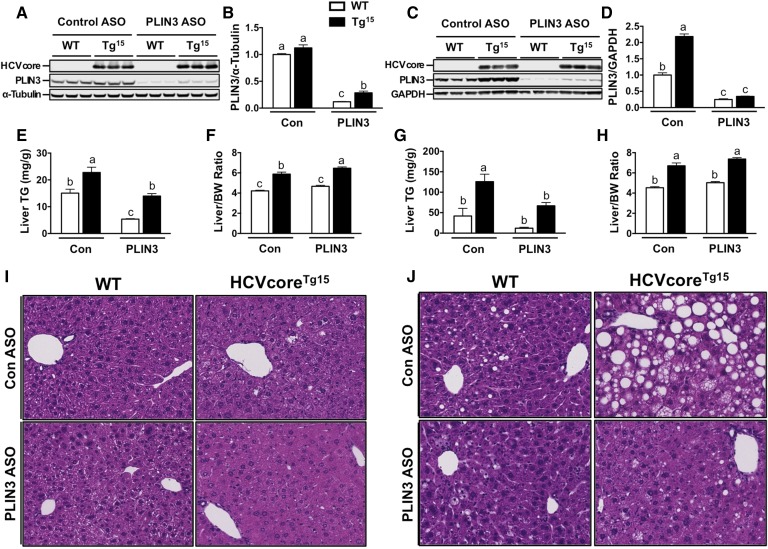
Next, to test the importance of PLIN3 expression in HCV core-induced hepatic steatosis, we used ASOs to knockdown expression of PLIN3. We observed PLIN3 ASO reduced expression of liver PLIN3 protein to ∼12–25% of the respective control ASO groups (Fig. 3A, B); however, HCVcoreTg15 mice given PLIN3 ASO did have 2.4-fold increased expression relative to WT mice on PLIN3 ASO (Fig. 3B). On control ASO, HCVcoreTg15 mice had 51% more liver TGs than WT mice; however, knockdown of PLIN3 reduced liver TGs to similar levels of WT mice given control ASO (Fig. 3C). Additionally, PLIN3 ASO reduced liver TGs in WT mice by approximately 66% (Fig. 3C). Interestingly, despite the significant decreases in liver TGs in both WT and HCVcoreTg15 mice, there was no decrease in liver size (Fig. 3D). In fact, HCVcoreTg15 mice had a small, but significant, 10% increase in liver/body weight ratio when given PLIN3 ASO (Fig. 3D). Liver sections with H&E staining did show some decreased lipid storage; however, sections were mostly unremarkable from mice on chow (Fig. 3E).
Fig. 3.
ASO-mediated knockdown of PLIN3 expression decreases steatosis in HCVcoreTg15 mice on chow and MFD. At 6 weeks of age, male WT and HCVcoreTg15 mice were treated with either control (Con) or PLIN3 ASO for 6 weeks while being maintained on either chow or MFD. A, B: Western blot (A) and quantitative analysis (B) of PLIN3 in liver homogenates from mice fed chow. C, D: Enzymatic determination of TGs from liver lipid extract (C) and liver size [expressed as a ratio to body weight (BW)] at necropsy from mice fed chow (D). E: Representative pictures of H&E staining performed on fixed liver sections (20× magnification) from mice fed chow. F, G: Western blot (F) and quantitative analysis (G) of PLIN3 in liver homogenates from mice fed MFD. H, I: Enzymatic determination of TGs from liver lipid extract (H) and liver size (expressed as a ratio to body weight) at necropsy from mice fed MFD (I). J: Representative pictures of H&E staining performed on fixed liver sections (20× magnification) from mice fed MFD. All hepatic lipid values were normalized to tissue weight. Data shown represent mean ± SEM. Levels not connected by the same letter are significantly different.
Our previous studies showed that the MFD exacerbated the steatosis phenotype in HCVcoreTg15 mice (Fig. 2), so we wanted to determine the effect of PLIN3 ASO on this exacerbated phenotype. Analysis of protein expression showed that HCVcoreTg15 mice had a 2.19-fold increase in PLIN3 expression compared with WT mice; however, both genotypes had expression levels reduced to 25–34% of those of WT mice when given PLIN3 ASO (Fig. 3F, G). On control ASO, HCVcoreTg15 mice had an approximately 3-fold increase in liver TGs compared with WT mice (Fig. 3H). Importantly, knockdown of PLIN3 significantly reduced liver TGs by almost 50% in HCVcoreTg15 mice. WT mice receiving PLIN3 ASO also had reduced liver TGs; however, this did not reach significance. Interestingly, while both groups did have reduced liver TGs with PLIN3 ASO treatment, there was no change in liver size, with transgenic mice maintaining an approximately 50% increase in liver/body weight ratio (Fig. 3I). Liver sections with H&E staining showed that HCVcoreTg15 mice on control ASO had much greater lipid accumulation compared with WT mice, as evidenced by increased LD size and number; however, treatment with PLIN3 ASO completely blunted this effect (Fig. 3J), which was confirmed by a pathologist’s histological examination (data not shown).
ASO-mediated knockdown of PLIN3 expression decreases steatosis in HCVcoreTg3 mice on chow diet
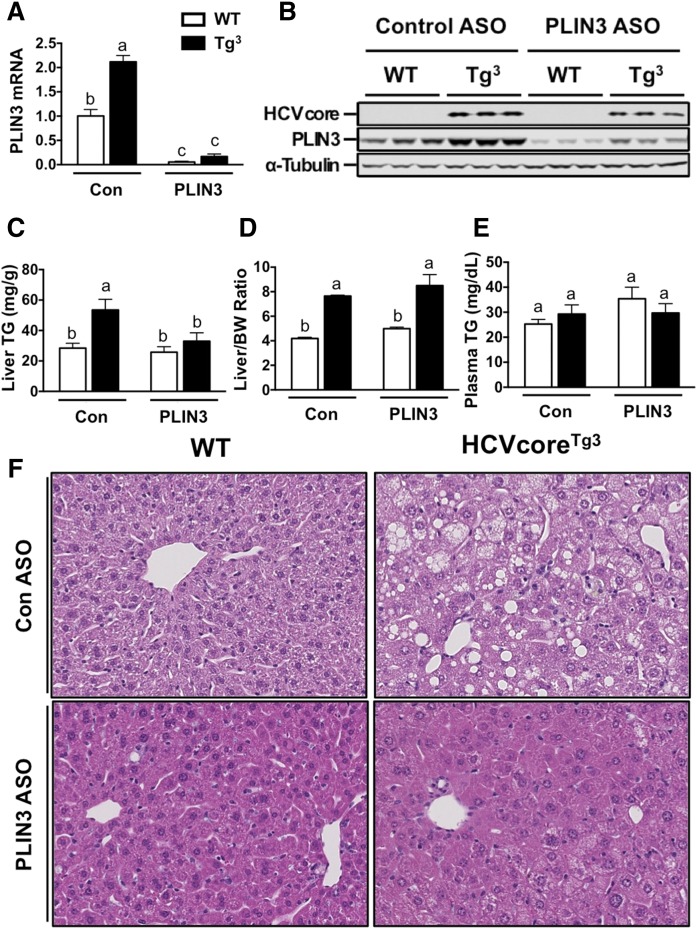
In order to confirm our results, we performed a similar study in a separate line of HCV core transgenic mice using HCVcoreTg3 mice (high core expression) fed chow. Female HCVcoreTg3 mice were maintained on a chow diet for 8 weeks while receiving biweekly injections with either a nontargeting control ASO or an ASO directed against PLIN3. As expected, HCVcoreTg3 mice had higher levels of PLIN3 mRNA in the liver (2.1-fold increased) compared with WT mice (Fig. 4A). However, both WT and HCVcoreTg3 mice receiving PLIN3 ASO had significantly reduced PLIN3 mRNA, which was ∼5–10% of that in respective control ASO treatment groups (Fig. 4A). Western blotting confirmed knockdown of PLIN3 protein expression in both WT and HCVcoreTg3 mice receiving the PLIN3 ASO (Fig. 4B). Interestingly, HCVcoreTg3 mice with PLIN3 knockdown had a 38% reduction in liver TGs, and TG mass was statistically similar to that of WT mice (Fig. 4C). There was no significant difference between body weight of WT and HCVcoreTg3 mice on either ASO treatment regimen (data not shown); however, HCVcoreTg3 mice exhibited an ∼2-fold increase in liver/body weight ratio compared with WT mice, which was not altered by PLIN3 knockdown (Fig. 4D). Next, to assess the effect of PLIN3 knockdown on liver morphology, we performed H&E staining on fixed liver sections and observed increased steatosis in HCVcoreTg3 mice compared with WT mice in the control ASO group (Fig. 4F). Remarkably, HCVcoreTg3 mice receiving PLIN3 ASO had diminished steatosis and lipid accumulation that was comparable to WT mice (Fig. 4F). Collectively, knockdown of PLIN3 effectively blunted HCV core-induced hepatic steatosis in our transgenic mice that had the highest amount of core expression.
Fig. 4.
ASO-mediated knockdown of PLIN3 expression decreases steatosis in HCVcoreTg3 mice on chow diet. At 8 weeks of age, female WT and HCVcoreTg3 mice were treated with either control (Con) or PLIN3 ASO for 8 weeks while being maintained on a chow diet. A: Relative levels of liver PLIN3 mRNA were quantified by real-time PCR, normalized to levels of cyclophilin A, and expressed relative to levels in WT mice given control ASO (n = 4 per group). B: PLIN3 protein expression determined by Western blot analysis of liver homogenates. C: Enzymatic determination of TGs from liver lipid extract (n = 4–6 per group). D: Liver size (expressed as a ratio to body weight) of mice at necropsy. E: Measurement of plasma TGs determined enzymatically. F: Representative pictures of H&E staining performed on fixed liver sections (20× magnification). All hepatic lipid values were normalized to tissue weight. Data shown represent mean ± SEM from four to six mice per group. Levels not connected by the same letter are significantly different.
Knockdown of PLIN3 reduces hepatic steatosis due to LXR agonist, but not steatosis due to fasting
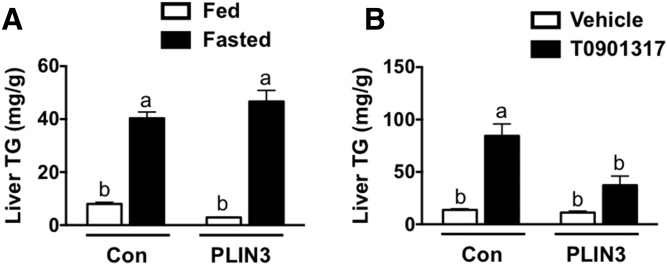
Given that treatment with PLIN3 ASO effectively diminished hepatic steatosis due to core protein expression and MFD, we wished to address whether knockdown of PLIN3 could prevent hepatic steatosis that occurred by alternative mechanisms. To do this, we first used the synthetic LXR agonist, T0901317, to induce steatosis after treating male C57BL/6 mice with either control or PLIN3 ASO for 6 weeks. Upon examination of liver TGs, control ASO-treated mice receiving the LXR agonist had a greater than 6-fold increase in liver TGs (Fig. 5A). However, when mice given the LXR agonist were treated with PLIN3 ASO, there was a significant reduction of liver TGs by 56%. Next, we tested whether PLIN3 knockdown would have an effect on hepatic steatosis-induced fasting. After treating male C57BL/6 mice with either control or PLIN3 ASO for 6 weeks, we fasted subgroups of mice for 18 h. As expected, in the control ASO-treated group, fasted mice had a 5-fold increase in liver TGs compared with fed mice (Fig. 5B). Amazingly, treatment with PLIN3 ASO had no effect on liver TGs in fasted mice, with similar values to the control ASO group. Overall, these results indicate that ASO-mediated knockdown of PLIN3 selectively reduces hepatic steatosis due to certain conditions.
Fig. 5.
Knockdown of PLIN3 reduces LXR agonist-induced hepatic steatosis, but not steatosis due to fasting. A: Liver TGs were measured enzymatically from extracts made from male C57BL/6 mice fed a chow diet and treated with either a control (Con) or PLIN3 ASO for 6 weeks. During the last week of treatment, mice were also gavaged orally with either a vehicle or exogenous LXR agonist (T0901317) (n = 5 per group). B: Enzymatic determination of liver TGs from lipid extracts made from male C57BL/6 mice fed a chow diet and treated with either a control or PLIN3 ASO for 6 weeks and necropsied following an 18 h fast (n = 5 per group).
Knockdown of PLIN3 alters plasma VLDL and HDL lipids
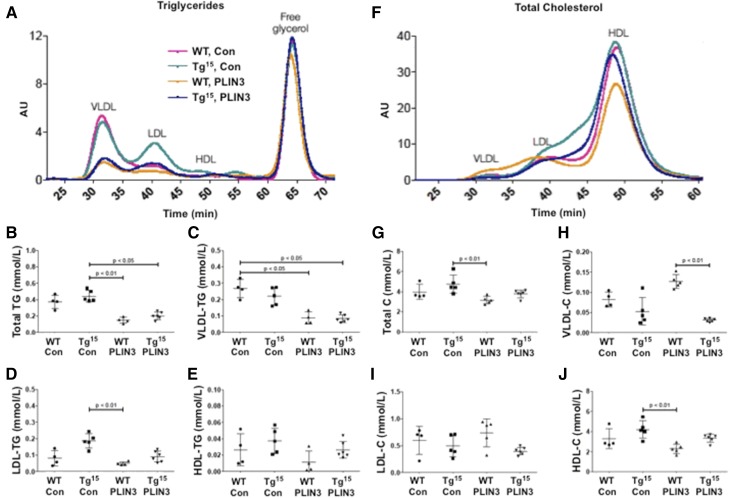
Expression of core in the liver has previously been shown to impair the secretion of VLDL (48); therefore, we wanted to examine whether knockdown of PLIN3 had an effect on plasma lipoprotein profiles in our mice. To do this, we examined the mass of lipids in different fractions of lipoproteins isolated by HPLC in male WT and HCVcoreTg15 mice fed a MFD (Fig. 6). Overall, both WT and HCVcoreTg15 mice treated with PLIN3 ASO had significantly reduced plasma TGs, primarily due to a decrease in the VLDL fraction (Fig. 6A–C). Additionally, HCVcoreTg15 mice did show a trend for slightly increased LDL TGs in the control ASO group, however there were no significant changes in LDL or HDL TGs within groups (Fig. 6D, E). Interestingly, when looking at lipoprotein fractions of total cholesterol, HCVcoreTg15 mice seemed to have increased amounts of intermediate particles, seen as small LDL/large HDL (Fig. 6F). While WT mice treated with PLIN3 ASO had an overall trend for decreased plasma total cholesterol, VLDL particles from these mice had increased total cholesterol, while HDL cholesterol trended to decrease (Fig. 6E–J). Overall, these data indicate that knockdown of PLIN3 reduces overall plasma TGs while having an inverse effect on plasma total cholesterol between WT and HCVcoreTg15 mice on a MFD.
Fig. 6.
Knockdown of PLIN3 alters plasma VLDL and HDL levels. Plasma from male WT and HCVcoreTg15 mice, 6 weeks of age, treated with either control (Con) or PLIN3 ASO for 6 weeks while being maintained on a MFD (n = 4–6 per group). Levels not connected by the same letter are significantly different. A: Plasma lipoprotein profile of TGs. B: Total plasma TGs. C: Total plasma VLDL TGs. D: Total plasma LDL TGs. E: Total plasma HDL TGs F: Lipoprotein profile of plasma total cholesterol. G: Total plasma cholesterol (Total C). H: Total plasma VLDL cholesterol (VLDL-C). I: Total plasma LDL cholesterol (LDL-C). J: Total plasma HDL cholesterol (HDL-C).
PLIN3 knockdown reduces core expression on the LD
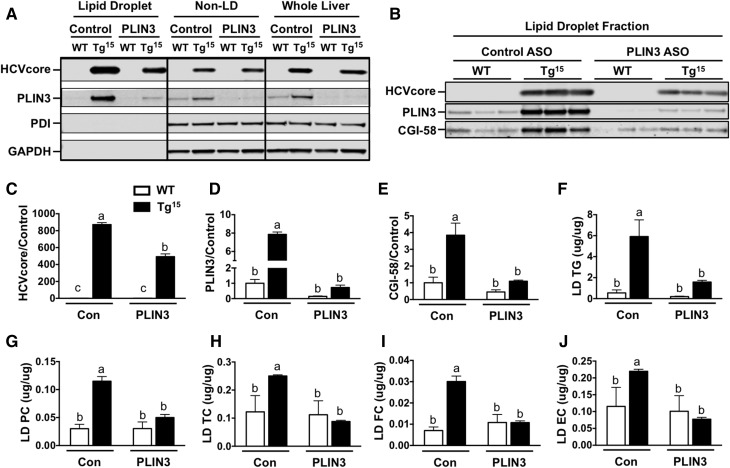
Given that both PLIN3 and HCV core interface with LDs, we examined LD proteins in core transgenic mice with normal or reduced PLIN3 levels. To do this, we performed LD isolation from livers of HCVcoreTg15 mice fed a MFD. As expected, in the LD fraction, we saw an enrichment of LD proteins, PLIN3, and core compared with the non-LD fraction and whole liver (Fig. 7A). Furthermore, the LD fraction contained little to no expression of the endoplasmic reticulum protein, protein disulfide isomerase, or the cytosolic protein, GAPDH. Additionally, administration of PLIN3 ASO resulted in substantial knockdown of PLIN3 in all three fractions. To examine the changes in LD proteins more closely, we performed Western blot on the LD fraction of individual animals (Fig. 7B). Consistently, PLIN3 protein expression was dramatically decreased in the LDs of WT and HCVcoreTg15 animals with PLIN3 ASO treatment. Interestingly, HCVcoreTg15 mice had a significant reduction of 44% in expression of core on the LD when treated with PLIN3 ASO (Fig. 7C). Both WT and HCVcoreTg15 mice had approximately 85–91% reduced expression of PLIN3 on the LD (Fig. 7D). Intriguingly, HCVcoreTg15 mice had a 3.85-fold increase in CGI-58 expression compared with WT animals in the control ASO group; however, knockdown of PLIN3 reduced the expression of CGI-58 on the LD to similar levels between HCVcoreTg15 and WT animals (Fig. 7E). Analysis of lipids extracted from the liver LD fraction showed that HCVcoreTg15 mice given control ASO had an overall significant increase in TGs, phosphatidylcholine, total cholesterol, free cholesterol, and esterified cholesterol (Fig. 7F–J).
Fig. 7.
Decreased core accumulation on hepatic LDs of HCVcoreTg15 mice with ASO-mediated knockdown of PLIN3. LD isolation from livers of male WT and HCVcoreTg15 mice, 6 weeks of age, treated with either control (Con) or PLIN3 ASO for 6 weeks while being maintained on a MFD (n = 3 per group). A: Western blot analysis was performed on fractions. B–E: Western blot analysis was performed on the liver LD fraction of individual animals and quantitative analysis was performed relative to the WT control group on expression of core (C), PLIN3 (D), and CGI-58 (E). F–J: Lipids were extracted from LD fractions and lipid content was determined enzymatically for TGs (F), phosphatidylcholine (PC) (G), total cholesterol (TC) (H), free cholesterol (FC) (I), and esterified cholesterol (EC) (J). Data shown represent mean ± SEM. Levels not connected by the same letter are significantly different.
HCV core-induced hepatomegaly does not involve PLIN3
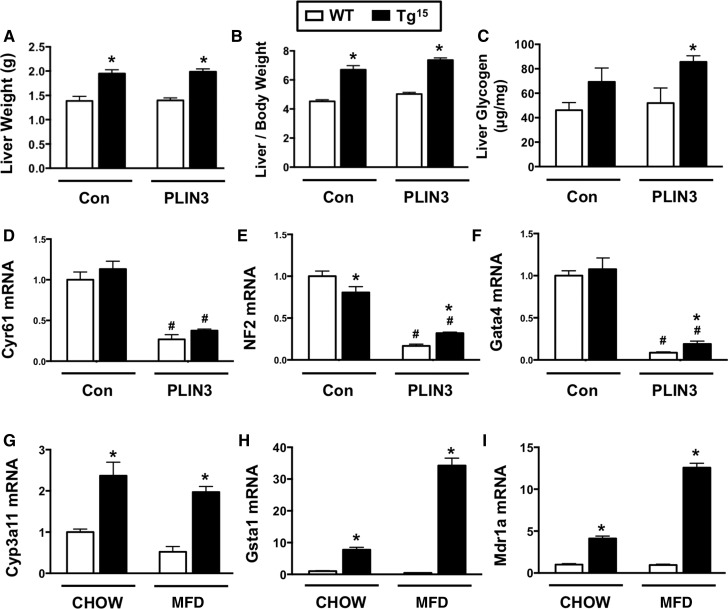
Although PLIN3 knockdown protects against HCV core-induced hepatic steatosis (Figs. 3, 4), hepatomegaly driven by HCV does not depend on PLIN3 (Fig. 8). HCVcoreTg15 mice exhibited significant increases in liver weight (Fig. 8A), even when normalized to body weight (Fig. 8B). However, PLIN3 knockdown did not alter HCV core-induced hepatomegaly (Fig. 8A, B). To examine PLIN3-independent mechanisms underlying HCV core-induced hepatomegaly, we examined hepatic glycogen storage and activation of transcriptional programs that are known to regulate liver size (49, 50). HCVcoreTg15 mice had modestly increased levels of hepatic glycogen, which were significantly elevated with PLIN3 ASO treatment (Fig. 8C). Given the key role that the Hippo/Yap/Taz pathway plays in liver size (49), we examined the hepatic expression of key genes driven by this pathway. HCVcoreTg15 mice did not have significant increases in Hippo/Yap/Taz target genes (Cyr61, NF2, or Gata4), yet PLIN3 knockdown was unexpectedly associated with decreased expression of Hippo/Yap/Taz target genes (Fig. 8D–F). Another key transcriptional pathway known to regulate liver size is activation of the nuclear hormone receptor, pregnane X receptor (PXR) (50), and HCVcoreTg15 mice exhibited marked increases in several PXR target genes (Fig. 8G–I). Collectively, these data suggest that HCV core-induced hepatomegaly is independent of PLIN3, and may involve increased glycogen storage and aberrant activation of PXR signaling.
Fig. 8.
HCV core-induced hepatomegaly does not involve PLIN3. At 8 weeks of age, male WT and HCVcoreTg15 mice were treated with either control (Con) or PLIN3 ASO for 8 weeks while being maintained on a MFD. A, B: Total liver weight (A) and liver size (expressed as a ratio to body weight) of mice at necropsy (B). C: Enzymatic determination of glycogen from liver extracts and normalized to tissue weight (n = 5 per group). D–F: Relative levels of liver mRNA were quantified by real-time PCR, normalized to levels of cyclophilin A, and expressed relative to levels in WT mice given control ASO (n = 5 per group). G–I: At 6 weeks of age, male WT or HCVcoreTg15 mice were placed on chow or MFD for a period of 6 weeks and relative levels of liver mRNA were quantified by real-time PCR, normalized to levels of cyclophilin A, and expressed relative to levels in WT mice given control ASO (n = 4–5 per group). Data shown represent mean ± SEM. Student’s t-test analysis was performed. *Significantly different from WT within each group (P < 0.05). #Significantly different from WT in control ASO group (P < 0.05).
DISCUSSION
Here, we demonstrate that transgenic expression of HCV core results in increased expression of the LD-associated protein, PLIN3, in vivo. Furthermore, HCV core-induced hepatic steatosis is diminished by ASO-mediated knockdown of PLIN3. This result was confirmed in two distinct HCV core transgenic lines and under different dietary conditions. Additionally, we provide evidence that core expression is associated with increased expression of PLIN3 on the LD and that knockdown of PLIN3 expression results in a significant decrease in core expression on the LD. Lastly, knockdown of PLIN3 expression selectively reduces steatosis due to core expression and LXR agonist treatment, but has no effect on fasting-induced fatty liver. Overall, these studies reinforce the significant role the LD plays in HCV-mediated pathogenesis, and identifies the host LD protein, PLIN3, as a factor driving HCV core-induced pathology.
It is well-known that core expression alone is sufficient to induce hepatic steatosis (5), a common feature of chronically infected HCV patients. Uncovering the exact mechanism of core-induced steatosis has broad clinical implications, but is still not completely understood. Multiple mechanisms by which core induces steatosis have been proposed such as: increased lipid synthesis (51); decreased lipolysis (10); decreased fatty acid oxidation (52); and decreased secretion of lipids via VLDL (53). Although core expression can alter many lipid metabolic pathways, one important aspect to consider is that core must associate with LDs in order to stimulate LD accumulation and steatosis (9, 10). Previous work by the Ott group showed that core’s association with cytosolic LDs blocked the turnover of TG resulting in steatosis (10). In a later study examining lipid storage and release from LDs in murine liver and cell culture, the group conclusively showed that core expression primarily resulted in decreased TG hydrolysis and that this required the lipolytic enzyme, ATGL (54). Paradoxically, in vitro studies revealed that although core inhibits lipolysis at LDs, core expression was associated with increased abundance of the TG hydrolase complex, ATGL-CGI-58, on the LD (34). Based on this evidence, the authors concluded that core altered properties of the LD, resulting in enhanced and prolonged interaction between ATGL and CGI-58, thereby preventing access of the complex to TG within the LD (54). Similar to these cell-based findings, we have found that transgenic expression of HCV core in vivo results in increased LD-associated CGI-58 (Fig. 7E). Furthermore, HCVcoreTg15 mice treated with PLIN3 ASO had reduced CGI-58 expression on the LD; however, this may be part of an overall trend for decreases in LD protein expression on isolated liver LDs with PLIN3 knockdown.
Although CGI-58 expression is not increased on the LD of HCVcoreTg15 mice treated with PLIN3 ASO, it is possible that core expression causes an imbalance in localization of scaffolding proteins on the LD, which prevents access of lipolytic enzymes to TG within the LD. Previous evidence in cell models shows that core expression alters the expression of LD scaffolding proteins (28, 29) and our results confirm, in vivo, that liver PLIN3 expression is increased in addition to increased PLIN3 on the LD (Fig. 7). Knocking down expression of PLIN3 in HCVcoreTg mice possibly restores the balance of LD scaffolding proteins, allowing for lipolytic enzymes to access the neutral lipid core. The Farese and Walther group have recently shown the importance of protein crowding on determining LD protein composition. Their seminal study showed that LD proteins compete for limited binding sites on the LD surface, and during lipolysis, the LD binding affinity plays a large role in determining localization (55). Additionally, this work suggests that there is a competition between LD proteins, when the LD surface is at a steady state, increasing levels of proteins with high affinities can change the composition of LD proteins and PAT proteins, possibly serving a regulatory function related to crowding (55). Accordingly, it seems reasonable to postulate that core outcompetes other LD proteins for the limited space on the LD due to the high affinity of core for the LD. As core expression on the LD increases, it causes a change in the LD protein composition, which we observed as expression of the PAT family member PLIN3 increased. These changes likely alter the normal LD protein composition and prevent lipolytic enzymes from accessing lipids within the LD, resulting in lipid accumulation and steatosis formation.
Importantly, our studies show that only certain types of hepatic steatosis are resolved by PLIN3 knockdown. In addition to HCV core-induced hepatic steatosis, we have shown that PLIN3 ASO effectively blunts LXR agonist-stimulated fatty liver (Fig. 5A). Additionally, previous work has shown that PLIN3 ASO effectively reduces diet-induced fatty liver in addition to improving insulin sensitivity and glucose tolerance (26). Interestingly, knockdown of PLIN3 expression had no significant effect on hepatic steatosis due to fasting (Fig. 5B). It is likely that the source of lipids that accumulate in the liver play a major role in the type of LD scaffolding proteins that accumulate on the LD. Activation of LXR in the liver leads to an increase in genes involved in lipid synthesis, such as SREBP-1, FAS, and ACC (56). Additionally, HCV core expression has also been shown to increase lipogenesis to some degree (51, 57). Previous studies using PLIN3 ASO have demonstrated a decrease in liver Gpat and Dgat2 expression (26), which are two major enzymes required for the incorporation of de novo synthesized fatty acids into monoacylglycerol and triacylglycerol, respectively (58, 59). Interestingly, PLIN3 ASO treatment was only shown to have a significant impact on Dgat2. This is unique because, while both DGAT1 and DGAT2 are able to synthesize TG, only DGAT2 can be found localized to the surface of LDs (15) and DGAT2 primarily synthesizes TG from endogenous fatty acids, while DGAT1 plays a greater role in incorporating exogenous fatty acids into TG (58). Additionally, previous evidence shows that lipogenic enzymes can relocalize to the LD in order to facilitate LD growth (60). It is possible that PLIN3 is an important factor for localization of lipid synthetic enzymes on the LD, and knockdown via PLIN3 ASO reduces the incorporation of lipids into the LD and blunts steatosis.
While knockdown of PLIN3 does significantly reduce HCV core- and LXR agonist-induced hepatic steatosis, there seems to be no impact on fatty liver that results from prolonged fasting. Previous work has shown that fasting seems to have little impact on PLIN3 expression in the liver, whereas PLIN2 is substantially upregulated in mice fasted for 24 h (61). This possibly indicates that PLIN3 is primarily essential for the incorporation of de novo synthesized lipids, whereas PLIN2 may be important in the storage of lipids taken up as a result of adipose lipolysis. Regardless of the mechanism, it is important to recognize that PLIN3 ASO effectively reduces many types of hepatic steatosis, in addition to improving insulin sensitivity, and may provide an effective means in treating patients affected by nonalcoholic fatty liver disease. HCV patients who develop steatosis respond poorly to treatment (4), and reduction of steatosis may be able to augment certain treatment regimens. Furthermore, due to the evidence that hepatic steatosis may be considered a prerequisite for enhanced liver disease (62), PLIN3 ASO therapy might provide treatment against cirrhosis and HCC, although further studies are required.
While therapy with PLIN3 ASO may provide an effective means against many types of fatty liver disease, it is also critical to consider this as an alternative treatment for HCV-infected patients. Importantly, previous studies demonstrate that the association of core on the LD is essential for the assembly of infectious HCV particles (12, 13), and our data show that knockdown of PLIN3 expression significantly reduces core expression on the LD (Fig. 7B, C). Furthermore, studies using HCV infectious cell culture systems show that PLIN3 is associated with released HCV particles (63), and silencing PLIN3 effectively abrogates the assembly and release of infectious viral particles (64, 65). Although recently developed direct acting antiviral treatments are very effective, Scheel and Rice (66) point out that even if current therapies were to achieve a success rate of 95% in all infected patients, there would still be more than 10 million individuals requiring treatment. The statement points to the need for innovative cures incorporated to include HCV infections of all genotypes, in various disease states, and circumvent the appearance of drug-resistant strains. Furthermore, even though new direct acting antiviral treatments are successful, there is evidence that treatment with direct acting antivirals may enhance the prevalence of HCC (67), although studies are conflicting (68, 69).
Overall, by taking advantage of ASO technology, we demonstrate the importance of the LD scaffolding protein, PLIN3, in a mouse model of HCV core-induced hepatic steatosis. Our studies further clarify mechanisms underlying the association of core with host LDs, and show that core induces a change in LD proteins in vivo. The change in LD proteins may be a major factor in the importance of the core-LD association for HCV particle assembly by forming a stable assembly platform. Due to the importance of LD binding affinity for determination of protein localization on LDs, it seems reasonable why the core sequence is so highly conserved across multiple HCV genotypes (70, 71). Additionally, while core has not been shown to directly interact with PLIN3, previous in vitro studies have shown that PLIN3 is critical for trafficking of NS5A to LDs (65). Furthermore, it has been determined that the functionally similar capsid of dengue virus binds to LDs via PLIN3 (72). Because PLIN3 knockdown diminished HCV core-driven hepatic steatosis, coupled with the importance of the LD for assembly of HCV viral particles, it is tempting to speculate that knocking down PLIN3 may hold therapeutic benefit for other viruses that co-opt similar LD-associated viral particle assembly. Collectively, this work demonstrates that targeting host LD-associated proteins may be an effective means to prevent HCV-induced pathology. Although effective anti-HCV therapies are currently available (66), this work provides additional broader implications for other pathogens that use the LD for enhanced persistence (73).
Note added in proof
Daniela S. Allende was inadvertently omitted as an author from the version of this manuscript that was published as a Paper in Press on December 10, 2016. This error has now been corrected.
Acknowledgments
The authors would like to thank Dr. Ryosuke Suzuki (National Institute of Infectious Diseases, Tokyo, Japan) for providing the pCAGC191 vector containing the full-length sequence of core (genotype 1b), as described in (31). They would also like to thank Dr. Greg Shelness (Wake Forest University School of Medicine) for providing the pLiv11 expression vector, as described in (32).
Footnotes
Abbreviations:
- ADRP
- adipocyte differentiation-related protein
- ASO
- antisense oligonucleotide
- ATGL
- adipose TG lipase
- CGI-58
- comparative gene identification 58
- core
- hepatitis C virus core protein
- DGAT
- diacylglycerol acyl transferase
- HCC
- hepatocellular carcinoma
- HCV
- hepatitis C virus
- H&E
- hematoxylin and eosin
- LD
- lipid droplet
- LXR
- liver X receptor
- MFD
- moderate fat diet
- PAT
- perilipin, adipocyte differentiation-related protein, and tail-interacting protein at 47 kDa
- PLIN
- perilipin
- PXR
- pregnane X receptor
- TIP47
- tail-interacting protein at 47 kDa
This work was supported in part by National Institutes of Health Grants R00 HL096166 (J.M.B.), R01 HL122283 (J.M.B.), P50 AA024333 (J.M.B.), and 2T32AI007401-21 (D.F.) and institutional startup funds from the Cleveland Clinic. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Mohd Hanafiah K., Groeger J., Flaxman A. D., and Wiersma S. T.. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 57: 1333–1342. [DOI] [PubMed] [Google Scholar]
- 2.Syed G. H., Amako Y., and Siddiqui A.. 2010. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol. Metab. 21: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herker E., and Ott M.. 2011. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol. Metab. 22: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lonardo A., Adinolfi L. E., Restivo L., Ballestri S., Romagnoli D., Baldelli E., Nascimbeni F., and Loria P.. 2014. Pathogenesis and significance of hepatitis C virus steatosis: an update on survival strategy of a successful pathogen. World J. Gastroenterol. 20: 7089–7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moriya K., Yotsuyanagi H., Shintani Y., Fujie H., Ishibashi K., Matsuura Y., Miyamura T., and Koike K.. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78: 1527–1531. [DOI] [PubMed] [Google Scholar]
- 6.Roingeard P., and Hourioux C.. 2008. Hepatitis C virus core protein, lipid droplets and steatosis. J. Viral Hepat. 15: 157–164. [DOI] [PubMed] [Google Scholar]
- 7.Pène V., Hernandez C., Vauloup-Fellous C., Garaud-Aunis J., and Rosenberg A. R.. 2009. Sequential processing of hepatitis C virus core protein by host cell signal peptidase and signal peptide peptidase: a reassessment. J. Viral Hepat. 16: 705–715. [DOI] [PubMed] [Google Scholar]
- 8.Boulant S. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281: 22236–22247. [DOI] [PubMed] [Google Scholar]
- 9.McLauchlan J., Lemberg M. K., Hope G., and Martoglio B.. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21: 3980–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris C., Herker E., Farese R. V., and Ott M.. 2011. Hepatitis C virus core protein decreases lipid droplet turnover: a mechanism for core-induced steatosis. J. Biol. Chem. 286: 42615–42625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartenschlager R., Penin F., Lohmann V., and André P.. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19: 95–103. [DOI] [PubMed] [Google Scholar]
- 12.Boulant S., Targett-Adams P., and McLauchlan J.. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88: 2204–2213. [DOI] [PubMed] [Google Scholar]
- 13.Miyanari Y. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9: 1089–1097. [DOI] [PubMed] [Google Scholar]
- 14.Yang L., Ding Y., Chen Y., Zhang S., Huo C., Wang Y., Yu J., Zhang P., Na H., Zhang H., et al. 2012. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 53: 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yen C-L. E., Stone S. J., Koliwad S., Harris C., and Farese R. V.. 2008. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 49: 2283–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann R. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 17.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., and Zechner R.. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metab. 3: 309–319. [DOI] [PubMed] [Google Scholar]
- 18.Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., and Zimmermann R.. 2006. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 281: 40236–40241. [DOI] [PubMed] [Google Scholar]
- 19.Bickel P. E., Tansey J. T., and Welte M. A.. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791: 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell M., Wang H., Chen H., McLenithan J. C., Gong D-W., Yang R-Z., Yu D., Fried S. K., Quon M. J., Londos C., et al. 2008. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 57: 2037–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skinner J. R., Shew T. M., Schwartz D. M., Tzekov A., Lepus C. M., Abumrad N. A., and Wolins N. E.. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284: 30941–30948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulankina A. V., Deggerich A., Wenzel D., Mutenda K., Wittmann J. G., Rudolph M. G., Burger K. N. J., and Höning S.. 2009. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185: 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda Y., Itabe H., Odaki M., Hama K., Fujimoto Y., Mori M., Sasabe N., Aoki J., Arai H., and Takano T.. 2006. ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J. Lipid Res. 47: 87–98. [DOI] [PubMed] [Google Scholar]
- 24.Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., and Brown D. A.. 2007. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J. Lipid Res. 48: 2751–2761. [DOI] [PubMed] [Google Scholar]
- 25.Goodman J. M. 2009. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50: 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carr R. M., Patel R. T., Rao V., Dhir R., Graham M. J., Crooke R. M., and Ahima R. S.. 2012. Reduction of TIP47 improves hepatic steatosis and glucose homeostasis in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 302: R996–R1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang B. H. J., Li L., Paul A., Taniguchi S., Nannegari V., Heird W. C., and Chan L.. 2006. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 26: 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato S. 2006. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J. Biochem. 139: 921–930. [DOI] [PubMed] [Google Scholar]
- 29.Boulant S., Douglas M. W., Moody L., Budkowska A., Targett-Adams P., and McLauchlan J.. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 9: 1268–1282. [DOI] [PubMed] [Google Scholar]
- 30.Filipe A., and McLauchlan J.. 2015. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol. Med. 21: 34–42. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki R., Tamura K., Li J., Ishii K., Matsuura Y., Miyamura T., and Suzuki T.. 2001. Ubiquitin-mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology. 280: 301–309. [DOI] [PubMed] [Google Scholar]
- 32.Cheng D., MacArthur P. S., Rong S., Parks J. S., and Shelness G. S.. 2010. Alternative splicing attenuates transgenic expression directed by the apolipoprotein E promoter-enhancer based expression vector pLIV11. J. Lipid Res. 51: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonet W. S., Bucay N., Lauer S. J., and Taylor J. M.. 1993. A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-I genes in transgenic mice. J. Biol. Chem. 268: 8221–8229. [PubMed] [Google Scholar]
- 34.Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A, Davies J. P., Nilsson L. M., and Yu L.. 2007. Hepatic Niemann-Pick C1-like 1 regulates biliary cholesterol concentration and is a target of ezetimibe. J. Clin. Invest. 117: 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooke R. M., Graham M. J., Lemonidis K. M., Whipple C. P., Koo S., and Perera R. J.. 2005. An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J. Lipid Res. 46: 872–884. [DOI] [PubMed] [Google Scholar]
- 36.Warrier M., Shih D. M., Burrows A. C., Ferguson D., Gromovsky A. D., Brown A. L., Marshall S., McDaniel A., Schugar R. C., Wang Z., et al. The TMAO-generating enzyme flavin monooxygenase 3 is a central regulator of cholesterol balance. Cell Rep. January 14, 2015; doi:10.1016/j.celrep.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Temel R. E., Sawyer J. K., Yu L., Lord C., Degirolamo C., McDaniel A., Marshall S., Wang N., Shah R., Rudel L. L., et al. 2010. Biliary sterol secretion is not required for macrophage reverse. Cell Metab. 12: 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown J. M., Bell T. A., Alger H. M., Sawyer J. K., Smith T. L., Kelley K., Shah R., Wilson M. D., Davis M. A., Lee R. G., et al. 2008. Targeted depletion of hepatic ACAT2-driven cholesterol esterification reveals a non-biliary route for fecal neutral sterol loss. J. Biol. Chem. 283: 10522–10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Temel R. E., Lee R. G., Kelley K. L., Davis M. A., Shah R., Sawyer J. K., Wilson M. D., and Rudel L. L.. 2005. Intestinal cholesterol absorption is substantially reduced in mice deficient in both ABCA1 and ACAT2. J. Lipid Res. 46: 2423–2431. [DOI] [PubMed] [Google Scholar]
- 40.Carr T. P., Andresen C. J., and Rudel L. L.. 1993. Enzymatic determination of triglyceride, free cholesterol, and total cholesterol in tissue lipid extracts. Clin. Biochem. 26: 39–42. [DOI] [PubMed] [Google Scholar]
- 41.Parini P., Johansson L., Broijersen A., Angelin B., and Rudling M.. 2006. Lipoprotein profiles in plasma and interstitial fluid analyzed with an automated gel-filtration system. Eur. J. Clin. Invest. 36: 98–104. [DOI] [PubMed] [Google Scholar]
- 42.Harris L-A. L. S., Shew T. M., Skinner J. R., and Wolins N. E.. 2012. A single centrifugation method for isolating fat droplets from cells and tissues. J. Lipid Res. 53: 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Temel R. E., Parks J. S., and Williams D. L.. 2003. Enhancement of scavenger receptor class B type I-mediated selective cholesteryl ester uptake from apoA-I-/- high density lipoprotein (HDL) by apolipoprotein A-I requires HDL reorganization by lecithin cholesterol acyltransferase. J. Biol. Chem. 278: 4792–4799. [DOI] [PubMed] [Google Scholar]
- 44.Folch J., Lees M., and Stanley G. H. S.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 45.Brown J. M., Chung S., Sawyer J. K., Degirolamo C., Alger H. M., Nguyen T., Zhu X., Duong M-N., Wibley A. L., Shah R., et al. 2008. Inhibition of stearoyl-coenzyme A desaturase 1 dissociates insulin resistance and obesity from atherosclerosis. Circulation. 118: 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas G., Betters J. L., Lord C. C., Brown A. L., Marshall S., Ferguson D., Sawyer J., Davis M. A., Melchior J. T., Blume L. C., et al. 2013. The serine hydrolase ABHD6 is a critical regulator of the metabolic syndrome. Cell Reports. 5: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansaldi F., Orsi A., Sticchi L., Bruzzone B., and Icardi G.. 2014. Hepatitis C virus in the new era: Perspectives in epidemiology, prevention, diagnostics and predictors of response to therapy. World J. Gastroenterol. 20: 9633–9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perlemuter G., Sabile A., Letteron P., Vona G., Topilco A., Chretien Y., Koike K., Pessayre D., Chapman J., Barba G., et al. 2002. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 16: 185–194. [DOI] [PubMed] [Google Scholar]
- 49.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., and Pan D.. 2007. Elucidation of a universal size-control mechanism in Drosophila. Cell. 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall A. P., Elcombe C. R., Foster J. R., Harada T., Kaufmann W., Knippel A., Kuttler K., Malarkey D. E., Maronpot R. R., Nishikawa A., et al. 2012. Liver hypertrophy: a review of adaptive (adverse and non-adverse) changes–conclusions from the 3rd International ESTP Expert Workshop. Toxicol. Pathol. 40: 971–994. [DOI] [PubMed] [Google Scholar]
- 51.Kim K. H., Hong S. P., Kim K., Park M. J., Kim K. J., and Cheong J.. 2007. HCV core protein induces hepatic lipid accumulation by activating SREBP1 and PPARγ. Biochem. Biophys. Res. Commun. 355: 883–888. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka N., Moriya K., Kiyosawa K., Koike K., Gonzalez F. J., and Aoyama T.. 2008. PPAR alpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J. Clin. Invest. 118: 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirandola S., Bowman D., Hussain M. M., and Alberti A.. 2010. Hepatic steatosis in hepatitis C is a storage disease due to HCV interaction with microsomal triglyceride transfer protein (MTP). Nutr. Metab. (Lond). 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camus G., Schweiger M., Herker E., Harris C., Kondratowicz A. S., Tsou C. L., Farese R. V. Jr., Herath K., Previs S. F., Roddy T. P., et al. 2014. The hepatitis C virus core protein inhibits adipose triglyceride lipase (ATGL)-mediated lipid mobilization and enhances the ATGL interaction with comparative gene identification 58 (CGI-58) and lipid droplets. J. Biol. Chem. 289: 35770–35780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kory N., Thiam A. R., Farese R. V. Jr., and Walther T. C.. 2015. Protein crowding is a determinant of lipid droplet protein composition. Dev. Cell. 34: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukasawa M., Tanaka Y., Sato S., Ono Y., Nitahara-Kasahara Y., Suzuki T., Miyamura T., Hanada K., and Nishijima M.. 2006. Enhancement of de novo fatty acid biosynthesis in hepatic cell line Huh7 expressing hepatitis C virus core protein. Biol. Pharm. Bull. 29: 1958–1961. [DOI] [PubMed] [Google Scholar]
- 58.Qi J., Lang W., Geisler J. G., Wang P., Petrounia I., Mai S., Smith C., Askari H., Struble G. T., Williams R., et al. 2012. The use of stable isotope-labeled glycerol and oleic acid to differentiate the hepatic functions of DGAT1 and -2. J. Lipid Res. 53: 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wendel A. A., Cooper D. E., Ilkayeva O. R., Muoio D. M., and Coleman R. A.. 2013. Glycerol-3-phosphate acyltransferase (GPAT)-1, but not GPAT4, incorporates newly synthesized fatty acids into triacylglycerol and diminishes fatty acid oxidation. J. Biol. Chem. 288: 27299–27306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., Uchida A., Cheng J. X., Graham M., Christiano R., Frohlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalen K. T., Ulven S. M., Arntsen B. M., Solaas K., and Nebb H. I.. 2006. PPARα activators and fasting induce the expression of adipose differentiation-related protein in liver. J. Lipid Res. 47: 931–943. [DOI] [PubMed] [Google Scholar]
- 62.Brunt E. M. 2007. Pathology of fatty liver disease. Mod, Pathol. 20(Suppl 1): S40–S48. [DOI] [PubMed] [Google Scholar]
- 63.Ploen D., Hafirassou M. L., Himmelsbach K., Schille S. A., Biniossek M. L., Baumert T. F., Schuster C., and Hildt E.. 2013. TIP47 is associated with the hepatitis C virus and its interaction with Rab9 is required for release of viral particles. Eur. J. Cell Biol. 92: 374–382. [DOI] [PubMed] [Google Scholar]
- 64.Ploen D., Hafirassou M. L., Himmelsbach K., Sauter D., Biniossek M. L., Weiss T. S., Baumert T. F., Schuster C., and Hildt E.. 2013. TIP47 plays a crucial role in the life cycle of hepatitis C virus. J. Hepatol. 58: 1081–1088. [DOI] [PubMed] [Google Scholar]
- 65.Vogt D. A., Camus G., Herker E., Webster B. R., Tsou C. L., Greene W. C., Yen T. S., and Ott M.. 2013. Lipid droplet-binding protein TIP47 regulates hepatitis C virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog. 9: e1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheel T. K., and Rice C. M.. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 19: 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conti F., Buonfiglioli F., Scuteri A., Crespi C., Bolondi L., Caraceni P., Foschi F. G., Lenzi M., Mazzella G., Verucchi G., et al. 2016. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct acting antivirals. J. Hepatol. 65: 727–733. [DOI] [PubMed] [Google Scholar]
- 68.Llovet J. M., and Villanueva A.. 2016. Liver cancer: effect of HCV clearance with direct-acting antiviral agents on HCC. Nat. Rev. Gastroenterol. Hepatol. 13: 561–562. [DOI] [PubMed] [Google Scholar]
- 69.Kobayashi M., Suzuki F., Fujiyama S., Kawamura Y., Sezaki H., Hosaka T., Akuta N., Suzuki Y., Saitoh S., Arase Y., et al. Sustained virologic response by direct antiviral agents reduces the incidence of hepatocellular carcinoma in patients with HCV infection. J. Med. Virol. Epub ahead of print. August 17, 2016; doi:10.1002/jmv.24663. [DOI] [PubMed] [Google Scholar]
- 70.Khaliq S., Jahan S., and Pervaiz A.. 2011. Sequence variability of HCV core region: Important predictors of HCV induced pathogenesis and viral production. Infect. Genet. Evol. 11: 543–556. [DOI] [PubMed] [Google Scholar]
- 71.Bukh J., Purcell R. H., and Miller R. H.. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA. 91: 8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carvalho F. A., Carneiro F. A., Martins I. C., Assunção-Miranda I., Faustino A. F., Pereira R. M., Bozza P. T., Castanho M. A. R. B., Mohana-Borges R., Da Poian A. T., et al. 2012. Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. J. Virol. 86: 2096–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saka H. A., and Valdivia R.. 2012. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu. Rev. Cell Dev. Biol. 28: 411–437. [DOI] [PubMed] [Google Scholar]