Abstract
Efficient delivery of biomacromolecules (e.g., proteins, nucleic acids) into cell cytosol remains a critical challenge for the development of macromolecular therapeutics or diagnostics. To date, most common approaches to assess cytosolic delivery rely on fluorescent labeling of macromolecules with an “always on” reporter and subcellular imaging of endolysosomal escape by confocal microscopy. This strategy is limited by poor signal-to-noise ratio and only offers low throughput, qualitative information. Herein we describe a quantitative redox-activatable sensor (qRAS) for the real-time monitoring of cytosolic delivery of macromolecules. qRAS-labeled macromolecules are silent (off) inside the intact endocytic organelles, but can be turned on by redox activation after endolysosomal disruption and delivery into in the cytosol, thereby greatly improving the detection accuracy. In addition to confocal microscopy, this quantitative sensing technology allowed for a high-throughput screening of a panel of polymer carriers toward efficient cytosolic delivery of model proteins on a plate reader. The simple and versatile qRAS design offers a useful tool for the investigation of new strategies for endolysosomal escape of biomacromolecules to facilitate the development of macromolecular therapeutics for a variety of disease indications.
Keywords: high-throughput screening, activatable sensor, FRET, endolysosomal disruption, cytosolic delivery
Graphical Abstract
A quantitative redox-activatable sensor (qRAS) for the real-time monitoring of cytosolic delivery of macromolecules was developed. The qRAS probe allowed for a high-throughput screening of polymers toward efficient cytosolic delivery of macromolecules on a plate reader. This new sensing platform will help accelerate the development of macromolecular therapeutics for various diseases.
Cytosolic delivery of biomacromolecules (e.g., proteins, peptides, nucleic acids) is critically important to achieve biological efficacy in immunotherapy, gene therapy and RNA interference.[1] Macromolecular agents are typically taken up by the target cells through endocytosis or macropinocytosis, where escape from endolysosomes is essential to prevent proteolytic degradation inside the lysosomes.[2] To achieve this goal, extensive efforts have been devoted to the development of cytosolic delivery strategies that allow endolysosomal escape of biomacromolecules to reach their targets in the cytoplasm.[3]
Despite great advances, the lack of a broad and quantitative detection assay presents a major challenge in the rapid discovery of new strategies for the cytosolic delivery of macromolecules with different size, charge and physical properties.[4] Conventional method utilizes confocal microscopy to investigate the spatial and temporal distribution of fluorescently labeled macromolecules.[5] Typical fluorescent labels employ “always on” reporter molecules where detection intensity is solely dependent on the probe concentration. Such imaging strategies have low signal-to-noise ratio and lack detection accuracy due to the extensive dilution of probe in the cytosol, strong signal in the endocytic vesicles, and a confounding effect of cytosolic autofluorescence background. Furthermore, they are not compatible with high-throughput assays such as plate readers when quantification of subcellular distribution is not feasible. Therefore, a simple, quantifiable cytosolic sensing assay is urgently needed to achieve high-throughput screening and microscopic examination of endolysosomal escape and cytosolic delivery of macromolecules in living cells.
Previous studies have reported that cell cytosol is a reducing environment with high concentrations (1–10 mM) of glutathione (GSH) and neutral pH (7.4) that are optimal to reduce disulfides or reactive oxygen species.[6] In contrast, the endocytic vesicles are much more oxidative with 100-fold lower concentration of GSH and acidic pH as low as 4.5. As a result, disulfide bonds from exogenic molecules are stable in the endosomes or lysosomes, but can be efficiently cleaved in the cell cytosol.[7] Several drug delivery systems have employed disulfide bonds for cytosolic delivery of therapeutics to improve biological efficacy.[8]
In this study, to achieve an accurate measurement of cytosolic delivery efficiency of biomacromolecules, we established a disulfide-based, redox-activatable fluorescent sensor that stays off in the extracellular space and along the endocytic pathway, but can be turned on after endolysosomal disruption and reaching the cell cytosol (Scheme 1). The quantitative redox-activatable sensor (qRAS) is synthesized by conjugating a fluorescent donor and acceptor pair onto the same cysteine (Cys) residue, one of which is through a disulfide bond (Figure 1). At the off state with an intact disulfide bond, hetero-Förster resonance energy transfer (hetero-FRET) abolishes the fluorescence signals of the donor dye. Upon disulfide cleavage by GSH in the cell cytosol, the fluorescence signal is turned on as a result of donor/acceptor separation. The donor-Cys-acceptor design maximizes atom efficiency while allowing short distance of donor and acceptor molecules to achieve superior FRET quenching efficiency.[9] Moreover, the available carboxylic acid group on the Cys residue can be activated by the formation of N-hydroxysuccinimide (NHS) esters and offers a convenient strategy to conjugate qRAS to various amine-containing macromolecules.
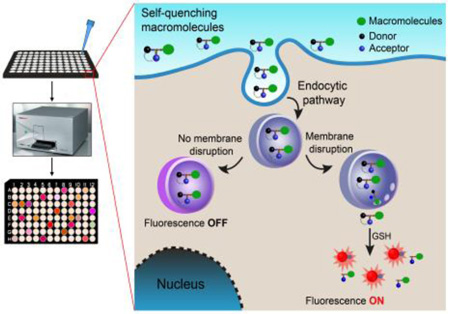
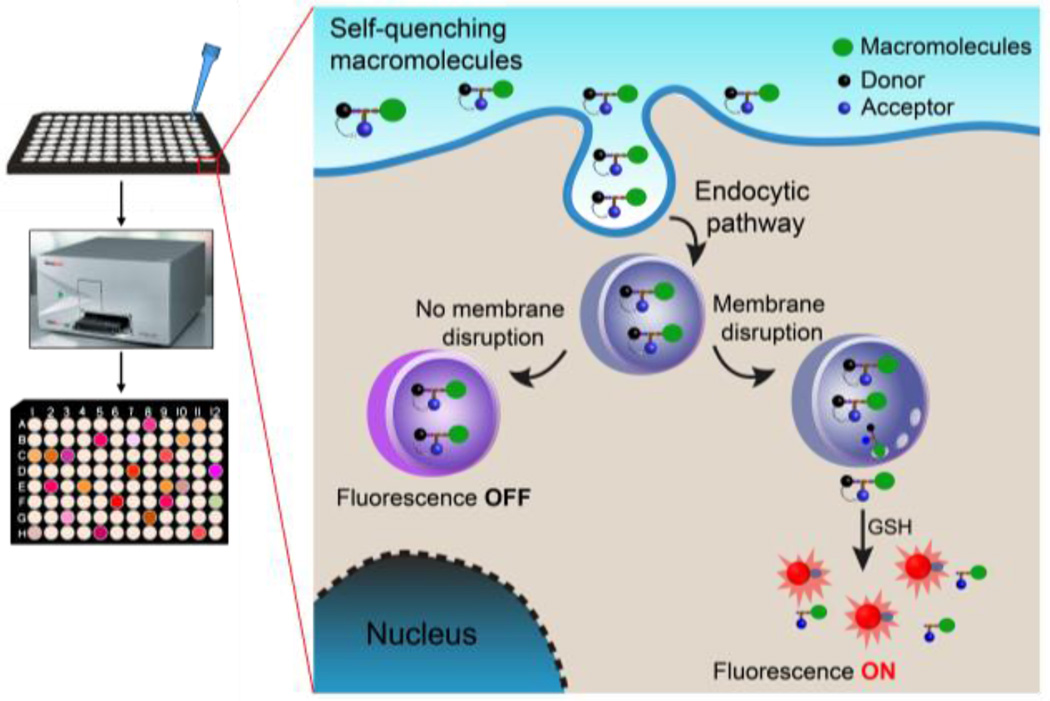
Scheme 1.
High-throughput assay of cytosolic delivery of biomacromolecules labeled with a redox-activatable sensor using a plate reader. The fluorescence signal is off in the extracellular environment and through the endocytic pathway. After endolysosomal disruption and reaching to the cell cytosol, the fluorescence signal is turned on by cytosolic GSH activation.
Figure 1.
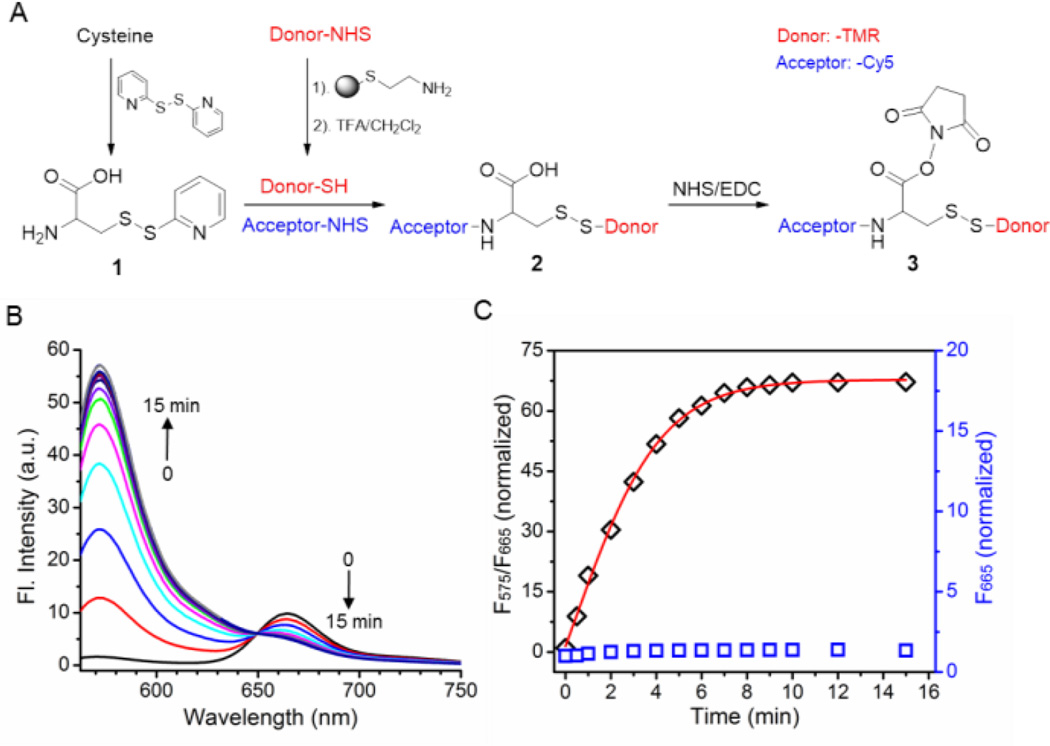
Synthesis and characterization of a representative qRAS molecule. A) Scheme of qRAS synthesis using TMR and Cy5 as model fluorescent donor and acceptor, respectively. B) The fluorescence spectra of qRAS after the addition of TCEP to cleave the disulfide bond. The sample was excited at 550 nm. C) Ratio of F575/F665 over time in response to redox cleavage by TCEP (black diamonds). Data was obtained from 1B. The emission intensity of Cy5 (F665, excited at 640 nm; blue squares) was used for comparison. Both F575/F665 and F665 values were normalized to those at t = 0.
Figure 1A illustrates the synthesis of an exemplary qRAS molecule, where tetramethyl rhodamine (TMR) and Cyanine 5 (Cy5) were used as donor and acceptor, respectively. After preparation and characterization of the qRAS (Figure S1), we investigated its fluorescence activation in response to a disulfide-reducing agent, tris(2-carboxyethyl)phosphine (TCEP). Before TCEP addition, fluorescence spectrum showed nearly extinguished emission at 575 nm for the donor TMR dye (Figure 1B). The FRET quenching efficiency was calculated to be >97%. Upon addition of 5 mM TCEP, a dramatic increase of the emission intensity at 575 nm was observed, which was accompanied by a decrease in the emission intensity at 665 nm. These results are consistent with the cleavage of the disulfide bond of qRAS, which abolishes the FRET from TMR to Cy5. Plot of F575/F665 as a function of time showed a short half-activation time (t1/2) at 3 mins and an impressive 70-fold activation ratio over the initial state (Figure 1C). In contrast, cleavage of TMR has much less pronounced effect (<1.4 fold) on the emission intensity of acceptor Cy5 (F665) when irradiated at 640 nm (Figure 1C). The relatively redox-insensitive acceptor signal is valuable to track labeled macromolecules during cell uptake studies. Similar FRET quenching and redox activation ratios were obtained when the donor and acceptor positions were switched (Figure S2).
Besides TMR/Cy5, we also extended qRAS design to additional donor/acceptor pairs, including 7-diethylaminocoumarin/QSY35, 7-hydroxycoumarin/dabcyl, and BODIPY-493/BHQ-1 quencher.[10] All exhibited efficient fluorescence activation in response to a reducing agent (Figure S3). In addition, 7-hydroxycoumarin/dabcyl pair also took advantage of the pH sensitivity of the donor 7-hydroxycoumarin, where the fluorescence signal is further suppressed in the acidic environment such as endocytic organelles (Figure S4).[11] The dual sensitivity design has the potential to further enhance the fluorescence signal in cell cytosol over endocytic organelles. The demonstrated chemical versatility allows a custom-made qRAS probe from a broad selection of fluorophores for different detection platforms (e.g., fluorescence microscopy, plate reader, flow cytometry).
Next, we examined whether qRAS-conjugated proteins are able to maintain the redox activation property. Ovalbumin was used as model protein and labeled by qRAS (OVAqRAS, Figure 2A). As shown in Figure 2B, OVAqRAS itself was almost nonfluorescent in the TMR channel as a result of efficient FRET quenching. After incubation with GSH (5 mM) for 1 h, a substantial increase of emission intensity of TMR was observed (30-fold). Fluorescent images of OVAqRAS solution with and without GSH addition demonstrated conspicuous on/off output in the donor channel (Figure 2C). Similar fluorescence activation profile was detected when treating OVAqRAS with dithiothreitol (DTT), a commonly used reducing agent for the disulfide bond (Figure S5). The achieved fluorescence activation ratio (30-fold) offers a superior imaging window over reported FRET-based protein sensors.[12] Quantification of the kinetics of redox activation showed a half time (t1/2) of 15 mins (Figure 2D). In addition, the TMR/Cy5 ratio remained constant across a broad pH range from 4.0 to 8.0 (Figure S6), which indicates that pH alone did not activate qRAS along the endocytic path.
Figure 2.
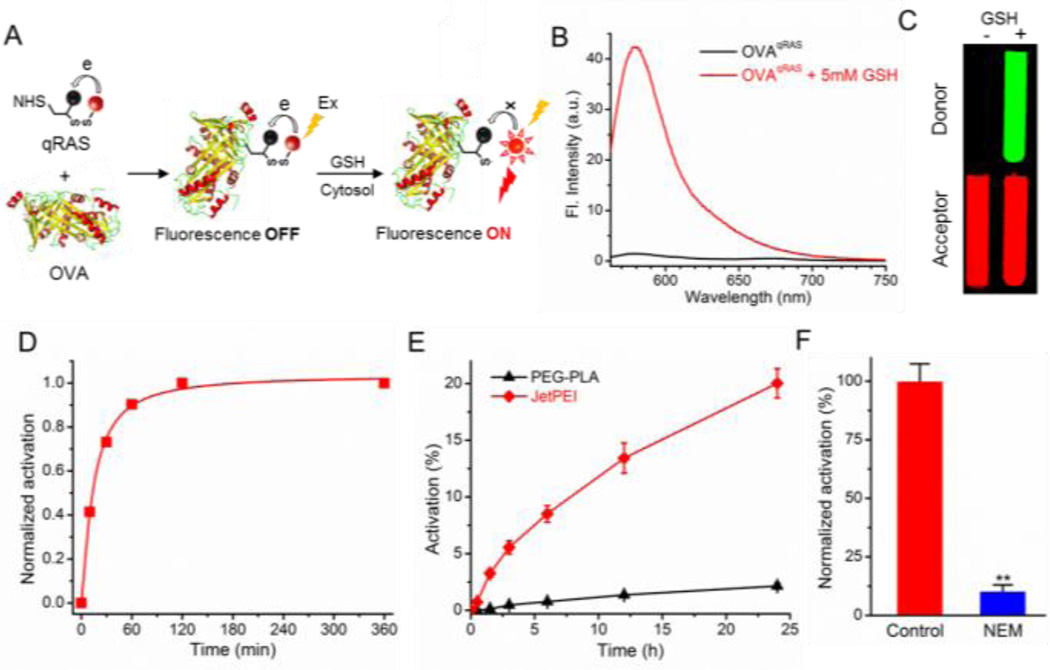
A) Schematic of qRAS conjugation to OVA and redox activation by GSH in the cytosol. B) Emission spectra of qRAS-labeled OVA (OVAqRAS) before and after the addition of GSH (5 mM) in PBS. Samples were excited at 550 nm. C) Fluorescent images of OVAqRAS solution with and without GSH by a Maestro Imaging system. D) Normalized fluorescence activation of OVAqRAS over time in response to a redox stimulus. E) Real-time monitoring of cytosolic delivery efficiency of OVAqRAS by JetPEI and PEG-PLA in the A549 lung cancer cells (n = 3). F) A549 cells were pretreated with NEM before incubation with JetPEI and OVAqRAS. The activation percentage is normalized to the control cells without NEM treatment (n = 3; **: p < 0.01).
To examine the capability of OVAqRAS for quantification of cytosolic delivery in living cells, we firstly demonstarted the redox activation of OVAqRAS (Figure S7) and a linear corrleation between detected fluorescence signal and OVAqRAS concentration on a plate reader (Figure S8). Polyethylenimine (PEI) has been known to mediate endosomal escape of nucleic acids through the “proton sponge effect”.[13] A commercially available JetPEI was used in this study. PEG-b-poly(D, L-lactic acid) (PEG-PLA) was employed as a negative control due to its low transfection efficiency. Following co-incubation with OVAqRAS, the fluorescence activation was monitored for 24 h. Results showed that JetPEI led to significantly higher signal of activated OVAqRAS over 24 h when compared to PEG-PLA (Figure 2E), consistent with the ability of JetPET for disrupting the endolysosomal membranes for cytosolic delivery. In addition, when replacing OVAqRAS with a cytotoxic protein, ribonuclease A, JetPEI exhibited significantly stronger inhibitory effect on the proliferation of cancer cells over the PEG-PLA control (Figure S9). To further demonstrate the GSH-specific activation of OVAqRAS, we pretreated the A549 cells with N-ethylmaleimide (NEM), an irreversible and membrane-permeable sulfhydryl blocker. Data showed significant suppression of fluorescence signals (Figure 2F), suggesting the redox activatable mechanism of OVAqRAS. The ability to quantify cytosolic delivery efficiency on a plate reader offers a simple and high-throughput method for real-time monitoring of endolysosomal escape and cytosolic delivery of biomacromolecules.
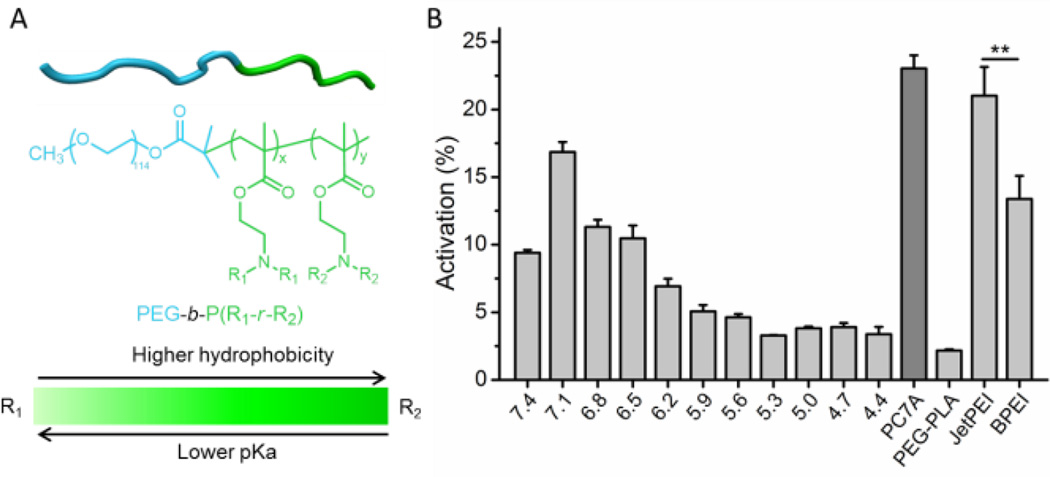
Previously, we synthesized a library of ultra-pH-sensitive (UPS) copolymers with ionizable tertiary amine side chains. The library covers an entire physiologic range of endocytic pH (4.0−7.4) with 0.3 pH increments (Figure 3A and S10).[14] In this study, we screened the library of UPS copolymers and evaluated their ability for cytosolic delivery of proteins. For the UPS copolymers with linear aliphatic side chains, higher pKa resulted in stronger activation signal of OVAqRAS (Figure 3B). We previously reported the pH-specific strong buffer effect of UPS copolymers at their pKa values.[15] Results from this study indicate a pH selectivity where buffering the organelle pH at early stage of endocytic maturation is more effective for cytosolic delivery. In addition, we observed that a UPS copolymer with a cyclic 7-membered ring on the side chain (PC7A, pKa = 7.0) rendered the most efficient cytosolic delivery of proteins with minimal cytotoxicity (Figure S11). Our data also showed that JetPEI induced higher level of cytosol delivery of protein over branched PEI (BPEI, Figure 3B), consistent with previous reports.[16] Similar structural dependence of cytosolic delivery of UPS copolymers was also observed when immunoglobulin G was used as a model protein (Figure S12), demonstrating the versatility of the qRAS probe for the labeling of different macromolecules to assess their cytosolic delivery in a high-throughput setting.
Figure 3.
High-throughput screening of cytosolic delivery of OVAqRAS by UPS polymeric nanoparticles on a plate reader. A) Chemical structures of UPS copolymers PEG-b-P(R1-r-R2) with finely tunable hydrophobicity and pKa. B) Cytosolic delivery efficiency of the polymers at 24 h after co-incubation with OVAqRAS in A549 cells for 40 min (n = 3; **: p < 0.01).
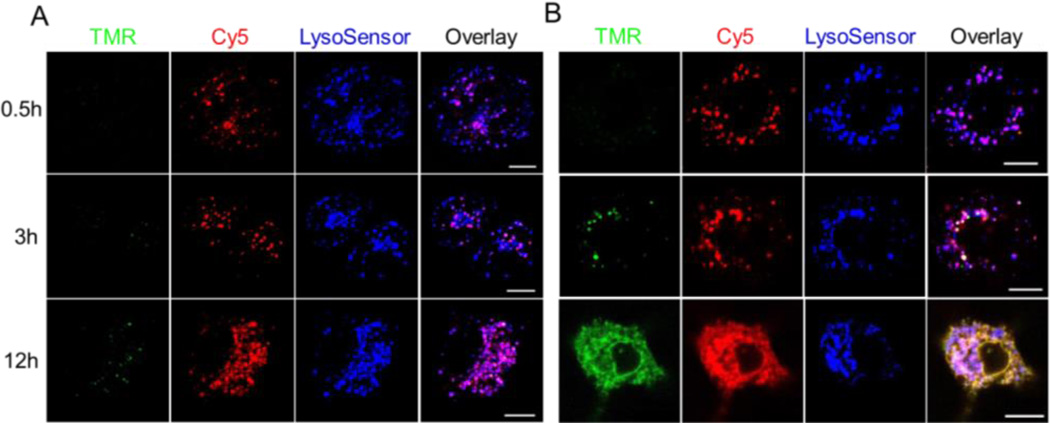
To evaluate whether qRAS is able to improve the accuracy of microscopic imaging of cytosolic delivery of macromolecules, we performed confocal microscopy analysis of A549 cells incubated with IgGqRAS. PC7A and UPS4.4 copolymers with strong and weak cytosolic delivery efficiency, respectively, were employed for comparison (Figure S13). Cells treated with UPS4.4 showed a nominal donor TMR signal up to 12 h indicative of minimal activation. In contrast, punctate distribution of acceptor Cy5 signal fully colocalized with lysosensor probe confirming endolysosomal entrapment of the protein (Figure 4A and S14A). For the cells treated with PC7A, a dynamic pattern of endolysosomal disruption and cytosolic delivery of IgG was observed in PC7A treated cells. Emission intensity in the donor TMR channel was dramatically increased over time (Figure 4B and S14B). At 0.5 h, no TMR signal was observed demonstrating the advantage of qRAS design in suppressing the signal from dose accumulation in the endocytic vesicles (IgG can still be tracked by the acceptor Cy5 signal). At 3 h, a punctate distribution of TMR signal was observed, which indicates that at early times, PC7A may disrupt endolysosomal membranes for GSH entry and activation of IgGqRAS inside the vesicles. A subset of TMR and Cy5 signals was separated from the lysosensor signals. At 12 h, endolysosomal escape of IgG proteins was pervasive as evidenced by broad TMR and Cy5 fluorescence spread throughout the cytoplasm and decreased colocalization level with lysosensor (Figure 4B). These data suggest that the qRAS design is able to overcome the challenge in cytosolic dilution and high signal intensity in endocytic vesicles for microscopic imaging of macromolecules using “always on” labels, leading to improved accuracy to study endolysosomal escape of macromolecules into the cell cytosol.
Figure 4.
Confocal microscopy analysis of endolysosomal escape and cytosolic delivery of IgG in live cells. A549 cancer cells were co-incubated with IgGqRAS and UPS4.4 (A) or PC7A (B) for 40 min. LysoSensor DND-189 was used to image the lysosomes (Blue). Both the donor TMR (Green) and acceptor Cy5 (Red) signals of qRAS were recorded over time. Scale bars = 10 µm.
In conclusion, we report the design and development of a versatile redox-activatable sensor for real-time monitoring and high-throughput quantification of cytosolic delivery of macromolecules. The qRAS probe can be easily synthesized and conjugated to multiple classes of macromolecules (OVA, IgG, and additional proteins and polymers, Figure S15). Macromolecules labeled with qRAS remain silent in the extracellular environment and intact endocytic vesicles, but can be dramatically activated in response to reducing environment of cytosol. The highly sensitive and specific cytosolic activation of the qRAS conjugates enables the quantitative assay of endolysosomal disruption on a microtiter plate. This high-throughput capability offers an attractive sensing platform to mechanistically investigate key physicochemical parameters of existing nanocarriers as well as discover new compositions for cytosolic delivery of multiple classes of macromolecules. These efforts will help accelerate the clinical translation of macromolecular therapeutics for a variety of disease indications.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01EB013149). We thank Z. Zeng for help with cell culture. We also thank the Simmons Cancer Center Support Grant (P30 CA142543) for support of nanomedicine core facility.
References
- 1.a) Kong HJ, Mooney DJ. Nat. Rev. Drug Discov. 2007;6:455–463. doi: 10.1038/nrd2309. [DOI] [PubMed] [Google Scholar]; b) Lachelt U, Wagner E. Chem. Rev. 2015;115:11043–11078. doi: 10.1021/cr5006793. [DOI] [PubMed] [Google Scholar]; c) Castanotto D, Rossi JJ. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Bareford LM, Swaan PW. Adv. Drug Deliv. Rev. 2007;59:748–758. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Pack DW, Hoffman AS, Pun S, Stayton PS. Nat. Rev. Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]; b) Gu Z, Biswas A, Zhao M, Tang Y. Chem. Soc. Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]; c) Jiang Y, Tang R, Duncan B, Jiang Z, Yan B, Mout R, Rotello VM. Angew. Chem. Int. Ed. 2015;54:506–510. doi: 10.1002/anie.201409161. Angew. Chem.2015, 127, 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang XL, Ramusovic S, Nguyen T, Lu ZR. Bioconjug. Chem. 2007;18:2169–2177. doi: 10.1021/bc700285q. [DOI] [PubMed] [Google Scholar]
- 4.a) Holub JM, Larochelle JR, Appelbaum JS, Schepartz A. Biochemistry. 2013;52:9036–9046. doi: 10.1021/bi401069g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Deng ZJ, Morton SW, Bonner DK, Gu L, Ow H, Hammond PT. Biomaterials. 2015;51:250–256. doi: 10.1016/j.biomaterials.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]; b) Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Rietsch A, Beckwith J. Annu. Rev. Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]; b) Schafer FQ, Buettner GR. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 7.a) Feener EP, Shen WC, Ryser HJ. J. Biol. Chem. 1990;265:18780–18785. [PubMed] [Google Scholar]; b) Go YM, Jones DP. Biochim. Biophys. Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Santra S, Kaittanis C, Santiesteban OJ, Perez JM. J. Am. Chem. Soc. 2011;133:16680–16688. doi: 10.1021/ja207463b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Maiti S, Park N, Han JH, Jeon HM, Lee JH, Bhuniya S, Kang C, Kim JS. J. Am. Chem. Soc. 2013;135:4567–4572. doi: 10.1021/ja401350x. [DOI] [PubMed] [Google Scholar]; b) Lee MH, Kim JY, Han JH, Bhuniya S, Sessler JL, Kang C, Kim JS. J. Am. Chem. Soc. 2012;134:12668–12674. doi: 10.1021/ja303998y. [DOI] [PubMed] [Google Scholar]; c) Onyango JO, Chung MS, Eng CH, Klees LM, Langenbacher R, Yao L, An M. Angew. Chem. Int. Ed. 2015;54:3658–3663. doi: 10.1002/anie.201409770. Angew. Chem.2015, 127, 3729–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Austin CD, Wen X, Gazzard L, Nelson C, Scheller RH, Scales SJ. Proc. Natl. Acad. Sci. USA. 2005;102:17987–17992. doi: 10.1073/pnas.0509035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy R, Hohng S, Ha T. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansson MK. Methods Mol. Biol. 2006;335:17–29. doi: 10.1385/1-59745-069-3:17. [DOI] [PubMed] [Google Scholar]
- 11.Lee M, Gubernator NG, Sulzer D, Sames D. J. Am. Chem. Soc. 2010;132:8828–8830. doi: 10.1021/ja101740k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Xiong H, Zhou Z, Zhu M, Lv X, Li A, Li S, Li L, Yang T, Wang S, Yang Z, Xu T, Luo Q, Gong H, Zeng S. Nat. Commun. 2014;5:3992. doi: 10.1038/ncomms4992. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) George Abraham B, Sarkisyan KS, Mishin AS, Santala V, Tkachenko NV, Karp M. PLoS One. 2015;10:e0134436. doi: 10.1371/journal.pone.0134436. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Xue L, Prifti E, Johnsson K. J. Am. Chem. Soc. 2016;138:5258–5261. doi: 10.1021/jacs.6b03034. [DOI] [PubMed] [Google Scholar]; d) Ogawa M, Kosaka N, Choyke PL, Kobayashi H. ACS Chem. Biol. 2009;4:535–546. doi: 10.1021/cb900089j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. Mol. Ther. 2013;21:149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 14.Ma X, Wang Y, Zhao T, Li Y, Su LC, Wang Z, Huang G, Sumer BD, Gao J. J. Am. Chem. Soc. 2014;136:11085–11092. doi: 10.1021/ja5053158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Wang Y, Li Y, Bodemann B, Zhao T, Ma X, Huang G, Hu Z, DeBerardinis RJ, White MA, Gao J. Nat. Commun. 2015;6:8524. doi: 10.1038/ncomms9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Breunig M, Lungwitz U, Liebl R, Goepferich A. Proc. Natl. Acad. Sci. USA. 2007;104:14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wiseman JW, Goddard CA, McLelland D, Colledge WH. Gene Ther. 2003;10:1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]; c) Derouazi M, Girard P, Van Tilborgh F, Iglesias K, Muller N, Bertschinger M, Wurm FM. Biotechnol. Bioeng. 2004;87:537–545. doi: 10.1002/bit.20161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.