Abstract
Loss-of-function mutations in SGCE, which encodes ε-sarcoglycan (ε-SG), cause myoclonus--dystonia syndrome (OMIM159900, DYT11). A “major” ε-SG protein derived from CCDS5637.1 (NM_003919.2) and a “brain-specific” protein, that includes sequence derived from alternative exon 11b (CCDS47642.1, NM_001099400.1), are reportedly localized in post- and pre-synaptic membrane fractions, respectively. Moreover, deficiency of the “brain-specific” isoform and other isoforms derived from exon 11b may be central to the pathogenesis of DYT11. However, no animal model supports this hypothesis. Gene-trapped ES cells (CMHD-GT_148G1-3, intron 9 of NM_011360) were used to generate a novel Sgce mouse model (C57BL/6J background) with markedly reduced expression of isoforms derived from exons 3′ to exon 9 of NM_011360. Among those brain regions analyzed in adult (2 month-old) wild-type (WT) mice, cerebellum showed the highest relative expression of isoforms incorporating exon 11b. Homozygotes (SgceGt(148G1)Cmhd/Gt(148G1)Cmhd or SgceGt/Gt) and paternal heterozygotes (Sgcem+/pGt, m-maternal, p-paternal) showed 60 to 70% reductions in expression of total Sgce. Although expression of the major (NM_011360) and brain-specific (NM_001130189) isoforms was markedly reduced, expression of short isoforms was preserved and relatively small amounts of chimeric ε-SG/β-galactosidase fusion protein was produced by the Sgce gene-trap locus. Immunoaffinity purification followed by mass spectrometry assessments of Sgcem+/pGt mouse brain using pan- or brain-specific ε-SG antibodies revealed significant reductions of ε-SG and other interacting sarcoglycans. Genome-wide gene-expression data using RNA derived from adult Sgcem+/pGt mouse cerebellum showed that the top up-regulated genes were involved in cell cycle, cellular development, cell death and survival, while the top down-regulated genes were associated with protein synthesis, cellular development, and cell death and survival. In comparison to WT littermates, Sgcem+/pGt mice exhibited “tiptoe” gait and stimulus-induced appendicular posturing between Postnatal Days 14 to 16. Abnormalities noted in older Sgcem+/pGt mice included reduced body weight, altered gait dynamics, and reduced open-field activity. Overt spontaneous or stimulus-sensitive myoclonus was not apparent on the C57BL/6J background or mixed C57BL/6J-BALB/c and C57BL/6J-129S2 backgrounds. Our data confirm that mouse Sgce is a maternally imprinted gene and suggests that short Sgce isoforms may compensate, in part, for deficiency of major and brain-specific Sgce isoforms.
Keywords: Sgce, Dystonia, Myoclonus, Sarcoglycans, Gene Trap
1. Introduction
Myoclonus-dystonia syndrome is a clinically and genetically heterogeneous disorder characterized by myoclonic jerks affecting mainly proximal muscles of the upper extremities, neck and trunk. Mutations (missense, nonsense, splicing, insertions, and deletions) in SGCE represent a major cause of the myoclonus-dystonia syndrome (MDS, DYT11, OMIM-159900)(Zimprich et al., 2001, Aichhorn et al., 2006) (Asmus et al., 2005) and are believed to cause disease via loss-of-function mechanisms (Kinugawa et al., 2009, Xiao et al., 2013). The onset of the disorder is usually in the first or second decade of life. Females tend to have earlier onset and greater probability of leg involvement than males (Raymond et al., 2008). SGCE is a maternally imprinted gene and MDS is inherited in an autosomal dominant fashion with reduced penetrance (Zimprich et al., 2001). SGCE encodes the epsilon member of the sarcoglycan (SG) family (ε-SG), single pass transmembrane proteins that are components of the dystrophin-glycoprotein complex (DGC). In cardiac and skeletal muscle, the DGC connects the actin cytoskeleton to the extracellular matrix.
In humans and mice, SGCE/Sgce harbors several alternatively spliced exons (Fig. 1) (Yokoi et al., 2005, Nishiyama et al., 2004, Ritz et al., 2011). In humans, a “brain-specific” isoform that incorporates alternative exon 11b (CCDS47642.1, NM_001099400.1, NP_001123661.1) shows high expression in Purkinje cells and neurons of the cerebellar dentate nucleus with significantly lower levels in the globus pallidus, striatum and substantia nigra (Ritz et al., 2011). In mice, the “major” ε-SG protein derived from NM_011360 (NP_035490.3) and the “brain-specific” isoform are reportedly localized in post- and pre-synaptic membrane fractions, respectively (Nishiyama et al., 2004). Mice may also express another “brain-specific” isoform encoded by an elongated exon 11b (exon 11c) (Yokoi et al., 2005). Proteins derived from transcripts containing either exon 11b or 11c encode proteins with C-terminal PDZ-binding motifs (Yokoi et al., 2005). Therefore, it has been suggested that the brain-specific isoforms may be central to the pathogenesis of MDS. However, although 11b was not screened in several published cohorts, no disease-associated mutations in terminal exons of SGCE (exons 11, 11b and 12) have been reported in patients with MDS (Table 1). Furthermore, the capacity of ε-SG to tolerate deleterious variants encoded by 11b is difficult to estimate given that 11b was not captured in many large whole-exome sequencing projects (Table S1). On the other hand, there are relatively few reported sequence variants in the terminal exons of SGCE (Table S1) which encode the C-terminus PDZ-binding cytoplasmic domain of ε-SG.
Fig. 1.
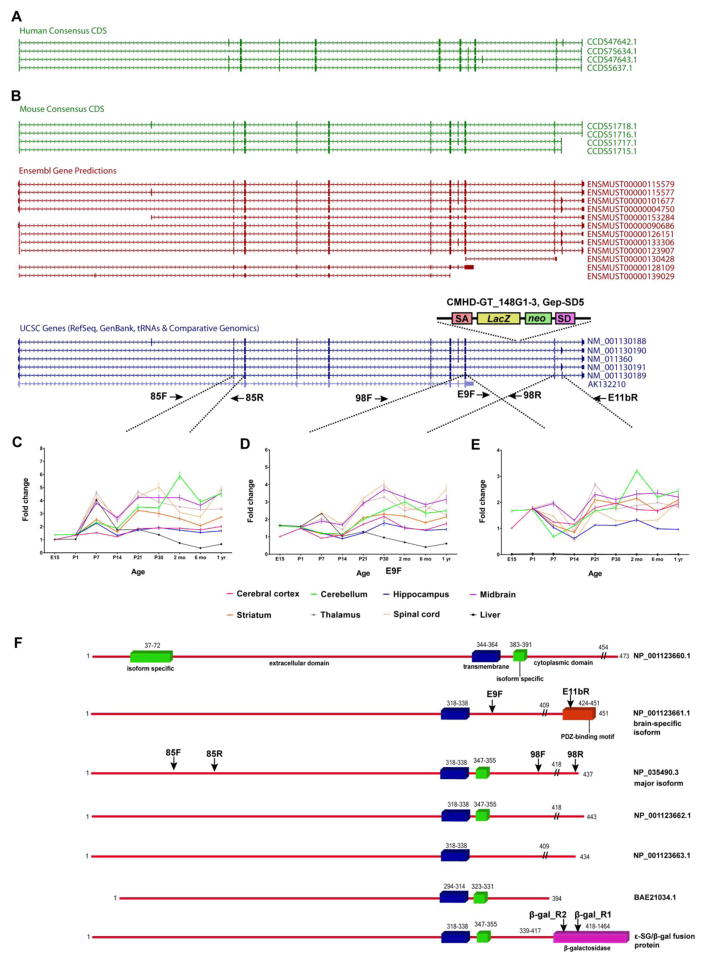
(A) Human SGCE Consensus CDS isoforms. (B) Mouse Sgce isoforms derived from Consensus CDS, Ensembl gene predictions and UCSC genes showing the location of the CMHD-GT_14G1-3 Gep-SD5 gene trap. Three distinct primer pairs (C, 85F/85R; D, 98F/98R; and E, E9F/E11bR) were designed to examine Sgce expression in eight different tissues across nine developmental time points. (F) Protein structures of the major (NP_035490.3), longest (NP_001123660.1), brain-specific (NP_001123661), and shortest (BAE21034.1) mouse isoforms, and predicted chimeric ε-SG/β-gal fusion protein derived from the major isoform. Blue bars, transmembrane domain. Green bars, isoform specific domains. Red bar, protein sequence derived from exon 11b that includes a PDZ-binding motif. Pink bar, β-galactosidase.//, predicted sites of ε-SG/β-gal protein fusion. Also shown are the approximate locations of primers that would amplify the corresponding regions of Sgce and hybrid transcripts. SA, splice acceptor. lacZ, gene encoding β-galactosidase. neo, neomycin resistance gene. SD, splice donor.
Table 1.
SGCE variant screening in myoclonus-dystonia syndrome
| Study | Subjects screened (positive) | Exon 11b screened | Mutation locations |
|---|---|---|---|
| Han et al (2003) | 7 families (3) | No | Exons 3 and 7 |
| Schüle et als (2004) | 10 families (2) | No | Exons 2 and 7 |
| Valente EM et al. (2005) | 58 index patients (6) | No | Exons 3, 4, 5 and 9 |
| Tezenas du Montcel et al. (2006) | 76 index patients (16) | Yes | Exons 2, 3, 4, 6, 7, and 9 |
| Gerrits et al. (2006) | 31 index patients (7) | No | Exons 2, 3, 5, 6, 7 |
| Nardocci N et al. (2008) | 11 families (9) | No | Exons 3, 4, 6, and 7 |
| Ritz K et al. (2009) | 86 index patients (13) | No | Exons 2, 3, 5, 6, and 7 |
| Asmus F et al. (2009) | 23 index patients (7) | Yes | Exons 3, 6, 7, and 10 |
| Carecchio M et al. (2013) | 46 new index patients (8) | No | Exons 3, 4, 6, and 9 |
| Peall KJ et al. (2014) | 89 index patients (19) | Yes | Exons 3, 5, 6, 7 and 9 |
Several mouse models have been used to explore the biology of ε-SG and pathobiology of MDS. Overexpression of wild-type (WT) ε-SG in mice resulted in substitution of ε-SG for α-SG in muscle DGC without overt behavioral or morphological consequences (Imamura et al., 2005). Paternal heterozygous null mice (Sgcem+/p−) were reported to exhibit myoclonus, increased slips on a beam-walking test, hyperactivity, and higher levels of striatal dopamine (DA) and dopamine metabolites 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA) (Yokoi et al., 2006). In contrast, mice with conditional knock-out (KO) of Sgce in Purkinje cells showed subtle motor learning deficits but did not exhibit myoclonus or robust motor abnormalities (Yokoi et al., 2012a). To gain additional insights into ε-SG function and the pathogenesis of MDS due to mutations in SGCE, we developed and characterized a novel Sgce gene-trap mouse model that disrupts expression of brain isoforms derived from exon 11b and other exons 3′ to exon 9.
2. Materials and methods
2.1. Developmental expression of Sgce
All mouse experiments were performed in accordance with the National Institutes of Health’s Guidelines for the Care and Use of Laboratory Animals and approved by our Institutional Animal Care and Use Committee. Expression patterns of Sgce transcripts were established in WT mice with relative quantitative reverse transcriptase-PCR (QRT-PCR) using tissues from cerebral cortex, cerebellum, hippocampus, ventral midbrain, striatum, thalamus, spinal cord, and liver. We examined 6 mice (3 male, 3 female) at 9 developmental time points (E15, P1, P7, P14, P21, P30, 2 mo, 6 mo, and 1 yr). Dissection of all brain regions which were readily identifiable in older mice was not possible at E15 and P1. SYBR Green-based QRT-PCR was performed with Ambion’s RETROscript® Reverse Transcription Kit and a LightCycler® 480 System (Roche, Indianapolis, IN, USA). One primer pair (Table S2, 85F and 85R) was designed to examine expression of all isoforms (Fig. 1). Expression of other isoforms was established by placing primers in exons 9 and 10 (98F and 98R) or exons 9 and 11b (E9F and E11bR) with β-actin as the endogenous control (Fig. 1 and Table S2). Data was normalized to E15 cerebral cortex. Detailed methods are provided in a previous publication from our laboratory (Xiao et al., 2012).
2.2. Generation of Sgce gene-trap mice
One line of ES cells on a 129X1/SvJ × 129S1/Sv background and generated using the Gep-SD5 vector (clone number CW509161 [CMHD-GT_148G1-3], Fig. 1) was obtained from The Center for Modeling Human Disease (CMHD, Ontario, Canada; www.cmhd.ca/genetrap/vectors.html). The Gep-SD5 vector does not contain an internal ribosome entry sequence. Due to the rules of nonsense-mediated decay, the selection transcript is only produced if the insertion site is close to the polyA near the 3′ end of the gene. The polyA sequence of the reporter gene (lacZ) is deleted which results in an unstable trapped transcript leading to hypomorphic mutants. DNA was extracted from cultured ES cells and used to confirm correct targeting of Sgce. Gene-trapped ES cells were injected into the blastocoel cavity of E3.5 C57BL/6 embryos using standard procedures. Male chimeras were bred to C57BL/6J female mice to establish coat color and germline transmission. The insertion site of the gene trap was identified with long-range PCR and confirmed with Sanger sequencing. We established that the terminal exons (3′ to exon 9) of Sgce were disrupted in homozygotes (SgceGt(148G1)Cmhd/Gt(148G1)Cmhd or SgceGt/Gt) and paternal heterozygotes (Sgcem+/pGt, m-maternal, p-paternal). Male Sgcem+/pGt mice were crossed to C57BL/6J female mice for 10 generations. Three primers (Table S2) were used for PCR-based genotyping with one primer located within the gene trap vector (Sgce_KO_V2L) and the two other primers flanking the gene-trap insertion within intron 9 of Sgce (NM_011360). The Mouse Direct PCR kit from Biotool (Houston, TX, USA) was used for genotyping with the following cycling conditions: 95°C for 5 min; 35 cycles at 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s; and final at 72°C for 7 min. The WT allele yields a 552 bp amplicon whereas the gene-trap mutant allele generates a 393 bp amplicon. Sgcem+/pGt mice on the C57BL/6J background were crossed to BALB/c and 129S2 mice for 5 or more generations to determine if the occurrence of myoclonus depended on genetic background.
2.3. Sgce expression in gene-trap mice
Relative levels of mouse Sgce mRNA were determined in the cerebral cortex and cerebellum of 1-month-old WT, Sgcem+/pGt, SgcemGt/p+, and SgcemGt/pGt mice (n= 3/genotype) using 3 primer pairs targeting different regions of Sgce (Fig. 1, Table S2). Sanger sequencing with two reverse primers in lacZ and one forward primer in exon 9 (Sgce_E9F) was used to establish in frame splicing of exon 9 to the Gep-SD5 vector sequence, and expression of Sgce/lacZ fusion transcript(s) was quantified with a forward primer in exon 9 (Sgce_E9F) and reverse primer in lacZ (β-Gal_R2, Table S2, Fig. 1). The predicted molecular weight of a chimeric ε-SG/β-gal fusion protein derived from the major ε-SG isoform and the Gep-SD5 β-galactosidase was calculated at ~159 kDa using two independent algorithms (Stothard, 2000, Bjellqvist et al., 1993). Of note, the chimeric protein harbors 25 amino acids between the end of ε-SG and the first methionine of β-galactosidase (Fig. S1). Mouse β-actin was used as the endogenous control. To ascertain spatial expression patterns, relative levels of mouse Sgce mRNA were determined in 6 brain regions (cerebral cortex, cerebellum, hippocampus, striatum, thalamus, and midbrain), cervical spinal cord, and liver harvested from 8 adult mice (3-month old, 4 males and 4 females) of WT, Sgcem+/pGt, and SgcemGt/pGt genotypes. SYBR Green-based QRT-PCR was performed using 3 primer pairs targeting different regions of Sgce (Fig. 1, Table S2). Mouse β-actin was used as the endogenous control. SgcemGt/p+ mice were not included in region of interest analyses since they showed no significant expression differences from WT mice in cerebral cortex and cerebellum (Table S3).
2.4. Immunoaffinity purification (IAP) and mass spectrometry analyses
Antibodies used for IAP and Western blotting included pan-ε-SG antibody esg3788, pan-ε-SG antibody esg3790, and brain-specific antibody esg2-1358 (Waite et al., 2016). The esg2-1358 antibody was generated against a short peptide (NH2-C-QRFEVNGIPEERKLTEAMSL-COOH) derived exclusively from exon 11b (Waite et al., 2016). The pan-ε-SG antibodies esg3788 and esg3790 were raised in rabbits immunized with a thioredoxin fusion protein containing the entire C-terminal intracellular domain of mouse ε-SG (NP_035490.3) that was used to generate previous ε-SG antibodies (Esapa et al., 2007). The anti-β-dystroglycan monoclonal antibody MANDAG2, developed by G.E. Morris, was obtained through the Developmental Studies Hybridoma Bank. Whole brain tissues from C57BL/6J WT and Sgcem+/pGt (all 3-month-old male) were snap frozen in liquid nitrogen and homogenized in digitonin (Merck Chemicals, Darmstadt, Germany) lysis buffer [1% digitonin (w/v), 150mM NaCl, 50mM Tris pH 8.0, 1mM EGTA, 1mM sodium orthovanadate and cOmplete™ Protease Inhibitor Cocktail (Roche, West Sussex, UK)], clarified and pre-cleared on protein A-agarose beads as previously described (Esapa et al., 2007). Lysates were then incubated with antibody-conjugated protein A-agarose beads overnight at 4°C. Proteins were eluted from the beads via incubation in 2° non-reducing Laemmli buffer at 60°C for 30 minutes followed by boiling at 95°C for 5 minutes. DTT was added to the elution at a final concentration of 50 mM, and the elution was boiled at 95°C for 5 min. An aliquot of each IAP elution was analyzed via Western blot for enrichment of ε-SG and β-dystroglycan. The remaining eluate was resolved on a 4–12% gradient NuPAGE Novex Bis-Tris gel (Invitrogen, Loughborough, UK) for 10 min, and fixed in 50% (v/v) methanol with 4% (v/v) acetic acid for 15 min. The gel was stained using colloidal Coomassie InstantBlue protein stain (Expedeon, Swavesey, UK) for 1 hr, then destained in ultrapure water before each elution was excised in its entirety as two gel plugs. Gel plugs were sent for in-gel tryptic digest followed by Velos Orbitrap mass spectrometry at the Advanced Mass Spectrometry Facility (University of Birmingham-England). Data analysis was carried out using Proteome Discoverer (Thermo Scientific, Loughborough, UK) with the SEQUEST search algorithm.
2.5. X-gal staining
X-gal staining was performed with a LacZ Detection Kit for Tissues (InvivoGen San Diego, CA) following the manufacturer’s protocol. Briefly, adult (3-month old) Sgcem+/pGt mice were transcardially perfused with normal saline followed by 4% paraformaldehyde in 0.1M phosphate buffered saline (PBS) with 2 mM MgCl2 (pH 7.4). Brains were then placed into a cryoprotection solution (30% sucrose in 0.1M phosphate buffer [PB] with 2 mM MgCl2) overnight. Coronal and sagittal cryosections (40 μm) were acquired and rinsed in PBS (2mM MgCl2, pH 7.4). Floating sections were then incubated with the staining solution (6 mM potassium ferricyanide, 6 mM potassium ferrocyanide, 2 mM MgCl2, 0.02% Igepal, 0.01% sodium deoxycholate, and 1 mg/ml X-gal solution in PBS) for 48 hrs at 37°C. Sections then rinsed with PBS, mounted, dehydrated and coverslipped.
2.6. Biogenic monoamines
Striatum was acutely harvested from 3 male and 3 female WT and Sgcem+/pGt mice at 3-months of age. All subsequent processing was completed by the Vanderbilt University Neurochemistry Core. Tissue samples were homogenized with a dismembrator in a solution (pH 3.8) containing 0.1M trichloroacetic acid (TCA), 0.01 M sodium acetate, 0.0001 M EDTA, 5 ng/ml isoproterenol (as internal standard) and 10.5 % methanol. Ten microliters of homogenates were collected for assay of protein concentrations. Samples were then spun in a microcentrifuge at 10,000 g for 20 min and the supernatant was removed for monoamine analysis. Biogenic amines were determined by high-pressure liquid chromatography utilizing an Antec Decade II (oxidation: 0.65) electrochemical detector operated at 33°C. Supernatants (20 μl) were injected using a Water 2707 autosampler onto a Phenomenex Kintex (2.6 u, 100 A) C18 HPLC column (100 × 4.60 mm). Biogenic amines were eluted with a mobile phase consisting of 89.5% 0.1M TCA, 0.01 M sodium acetate, 0.0001 M EDTA and 10.5 % methanol (pH 3.8). Solvents were delivered at 0.6 ml/min using a Waters 515 HPLC pump. Using this HPLC solvent the following biogenic amines were eluted in the following order: noradrenaline, 3, 4-dihydroxyphenylacetic acid (DOPAC); dopamine (DA); 5-hydroxyindoleacetic acid (5-HIAA); homovanillic acid (HVA); 5-hydroxytryptophan (5-HT); and 3-methoxytyramine (3-MT). HPLC control and data acquisition were managed by Empower software.
Protein concentrations were determined with a BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA, USA). Ten microliter volumes of tissue homogenate were distributed onto a 96-well plate and 200 μl of mixed BCA reagent (25 ml of Protein Reagent A is mixed with 500 μl of Protein Reagent B) was added. The plate was then incubated for 2 hrs at room temperature for color development. A BSA standard curve was simultaneously run. Absorbance was measured with a POLARstar Omega plate reader (BMG LABTECH, Cary, NC, USA).
2.7. Behavioral assessments
Adult (3-month-old) Sgcem+/pGt mice and gender-matched WT littermates were subjected to a battery of motor and behavioral examinations including open-field activity, rotarod, vertical rope climbing, raised-beam task, grip strength, gait analysis (DigiGait™), dominance tube, and cross-maze test as described in a recent publication from our laboratory (Xiao et al., 2016). Mice were weighted weekly. Video recordings and righting reflex assays were performed prior to weaning in independent groups of WT and Sgcem+/pGt mice.
WT and Sgce mutant mice were observed for the presence of spontaneous and stimulus-induced (audiogenic [100 dB, 40 ms, 1 Hz]; tail, trunk and limb displacement with a microspatula; snout tap; and tail pinch) abnormal movements (tonus, clonus, tremor, twisting, bobbing, wagging, sustained flexion, and sustained extension; Table S4) on a daily basis from Postnatal Day 1 (P1) through P30 and then 3X weekly through 6 months of age. Although assessments were qualitative, abnormal movements were most apparent between P10 and P20, particularly P12 to P16.
An initial cohort of 10 Postnatal Day 14 (P14) mice (all males, 5 WT and 5 Sgcem+/pGt) was videotaped for 5 min. Each mouse was placed in a clear rectangular arena (18 cm x 30 cm) and subjected to limb, tail and truncal displacements with a microspatula. The videotapes were scored by 3 raters blinded to genotype using an adaption of a previously published rating scale (Raike et al., 2012). This ordinal scale was utilized to score all abnormal movements in five body regions (face, neck, trunk, forelimbs, and hindlimbs): 0 - absent abnormal movement, 1- slight and intermittent, 2 - mild and common or moderate and intermittent, 3 - moderate and common, or 4 - marked and prolonged (Table S4). In addition, specific types of involuntary movements (tonus, clonus, tremor, twisting, bobbing, wagging, sustained extension, and sustained flexion) were rated in binary fashion as absent (0) or present (1).
A second cohort of 20 P16 mice (WT [n = 10] and Sgcem+/pGt [n = 10]), (5 males and 5 females for each genotype) was videotaped for a total of 6 min of open-field behavior acquired in 1-min bins and scored by 3 blinded raters. Mice were placed alone in a clear plastic cage (18 cm × 30 cm) for videotaping and returned to their home cages between video sessions.
2.8. Whole-genome gene-expression analysis
Total RNA from mouse cerebellum was isolated from 6 adult (10 month-old) Sgcem+/pGt mice (3 males and 3 females) and 6 age- and gender-matched WT controls (Ambion™ TRI Reagent®, ThermoFisher Scientific). The quality of total RNA was assessed with a NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies) and RNA integrity was verified with an Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nano kit. Whole-genome gene-expression data was generated with the Affymetrix GeneChip® Mouse Gene 2.0 ST Array (Santa Clara, CA, USA). This array was designed using data from RefSeq (release 51), Ensembl (release 65) and lncRNA db and provides comprehensive coverage of over 30,000 mRNA transcripts and 2000 lincRNA transcripts. The 2.0 ST Array was designed with a median of 22 25-mer probes per transcript. Each array includes background antigenomic probes, poly-A controls and hybridization controls. Target RNA was first reverse transcribed into cDNA, followed by in-vitro transcription to generate biotin-labelled cRNA for subsequent hybridization. Hybridized target cRNA were stained with streptavidin phycoerythrin and scanned using an Affymetrix GeneArray Scanner.
Data were processed using Affymetrix Expression Console software that incorporates the Robust Multi-array Average (RMA) normalization algorithm. Genes were annotated using Affymetrix MoGene 2.0 ST, V.1, release 35 annotation files from NetAffx™ server of Affymetrix. The RMA-normalized .chp files were summarized further with GeneSpring GX® 13.1.1 (Agilent® Technologies, Santa Clara, CA). Scatter plots were used to access the reproducibility of gene expression within genotypes. A t-test statistic was used in identifying significantly dysregulated genes (p ≤ 0.05) between WT and Sgcem+/pGt mice. Differential gene expression was also analyzed using a False Discovery Rate (FDR) corrected p ≤ 0.05 (Benjamini and Heller, 2008). A heat map created using unsupervised hierarchical clustering with average linkage and Euclidean distance and a volcano plot were used to visualize differential gene expression. Using WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) and Ingenuity Pathway Analysis (IPA), we investigated the effects of differentially regulated genes on pathways (KEGG, Kyoto Encyclopedia of Genes and Genomes) and molecular/cellular networks (Zhang et al., 2005).
To validate data obtained with the Affymetrix GeneChip® Mouse Gene 2.0 ST Array, we selected 6 up-regulated and 6 down-regulated genes moderately expressed in cerebellum, and 3 genes related to the DGC, for QRT-PCR. Using TaqMan® probes, QRT-PCR was performed on the Roche LightCycler® 480 system with primers designed using the Roche Universal ProbeLibrary Assay Design Center. A total of 16 cerebellar RNA samples from WT and Sgcem+/pGt mice (8 males and 8 females in each group, including the 6 samples used for whole-genome gene-expression analysis) were employed for validation with β-actin as the endogenous control. Technical triplicates were performed for all samples and median values were utilized for statistical analysis.
2.9. Statistics
ANOVA with post-hoc tests was used to determine the effects of genotype and gender on parametric behavioral measures. The Mann-Whitney test was used to determine the effects of genotype within gender for a non-parametric behavioral measure (slips on the raised beam task). Two-tailed t-tests were used to establish the effects of genotype on monoamine levels in striatum. Fisher’s exact test was used to determine the effects of genotype on the results of dominance tube testing. An alpha (α) of 0.05 was chosen for statistical significance.
3. Results
3.1 Expression of Sgce is developmentally regulated
Overall, brain expression of total (Fig. 1C) and brain-specific (Fig. 1E) Sgce increased with increasing postnatal age (p < 0.0001, for both). There was a significant effect of region on expression of all Sgce isoforms (primers 85F and 85R), “long” isoforms (primers 98F and 98R), and the brain-specific isoform derived, in part, from exon 11b (p < 0.0001, for all). The highest expression levels for all isoforms and the brain-specific isoform were detected in 2-month-old mouse brain (Fig. 1). In general, cerebellar levels of all isoforms and the brain-specific isoform were 2 to 3 fold higher than hippocampus and cerebral cortex. In older adult mice (1 yr), expression of all isoforms and the brain-specific isoform were similar in cerebellum and striatum. The brain-specific isoform was not detected in liver and total Sgce mRNA was very low in this non-neural tissue.
3.2 Sgce is maternally imprinted
Male and female heterozygous and homozygous mice were fertile and pups of all genotypes and both genders were born at normal Mendelian ratios. As seen in Table 2, there were significant effects of gender (F1,59 = 135.2, p < 0.0001) and genotype (F1,59 = 78.1, p < 0.0001) on weights in 3-month-old mice (Table 2). Overall, Sgcem+/pGt mice were 10 to 15% smaller than their WT littermates.
Table 2.
Effects of genotype and gender on weight and behavioral measures in 3-month-old mice. Ambulatory count, the total number of X + Y photo beam breaks while in ambulatory movement status. Stereotypic count, any partial-body movements that occur within the ambulatory box such as grooming, head-weaving or scratching. Vertical count, number of periods of continuous Z photo beam breaks. Jump count, the number of times that the mouse leaves the photo beam array for a period of time. Ambulatory episodes, the number of times the mouse has started moving after the resting delay has expired. Values are means ± SEM except for dominance tube.
| Male | Female | |||
|---|---|---|---|---|
| Sgce+/+ (n=18) | Sgcem+/pGt (n=15) | Sgce+/+ (n=16) | Sgcem+/pGt (n=15) | |
| Weight (g) | 31.2 ± 0.7 | 26.3 ± 0.5* | 24.9 ± 0.4 | 20.2 ± 0.4* |
| Grip strength (g) | 329.2 ± 6.9 | 274.3 ± 5.8* | 276.4 ± 4.5 | 222.1 ± 4.8* |
| Grip strength/weight | 10.6 ± 0.3 | 10.4 ± 0.2 | 11.2 ± 0.2 | 11.0 ± 0.3 |
| Dominance tube | 53.3% | 46.7% | 49.0% | 47.1% |
| Cross maze score (%) | 33.5 ± 2.3 | 25.4 ± 2.0* | 33.1 ± 2.5 | 23.5 ± 2.3* |
| Rope climbing (s) | 5.4 ± 0.5 | 4.5 ± 0.4 | 4.1 ± 0.6 | 4.3 ± 0.4 |
| Rope climbing/weight | 0.17 ± 0.01 | 0.17 ± 0.01 | 0.16 ± 0.02 | 0.21 ± 0.02 |
| Open field activity | ||||
| Distance traveled (cm) | 1858.4 ± 52.0 | 1594.7 ± 30.4* | 2455.7 ± 53.2 | 1798.7 ± 43.3* |
| Ambulatory count | 897.4 ± 32.7 | 782.1 ± 18.5* | 1168.4 ± 31.4 | 904.2 ± 31.3* |
| Stereotypic Count | 2596.8 ± 102.3 | 2245.1 ± 42.1* | 2607.0 ± 49.5 | 2183.5 ± 56.8* |
| Vertical count | 189.9 ± 11.2 | 187.5 ± 15.0 | 135.3 ± 10.7 | 101.4 ± 10.2* |
| Jump count | 35.1 ± 2.9 | 38.3 ± 2.7 | 45.9 ± 4.1 | 29.5 ± 2.4* |
| Average velocity (cm/s) | 36.2 ± 1.5 | 34.4 ± 1.6 | 43.5 ± 1.9 | 45.2 ± 2.1 |
| Ambulatory episodes | 85.4 ± 6.0 | 75.9 ± 6.4 | 112.9 ± 6.2 | 85.2 ± 3.7* |
| DigiGait™ | ||||
| Propel (s) Forelimb | 0.127 ± 0.003 | 0.116 ± 0.003* | 0.116 ± 0.004 | 0.110 ± 0.004 |
| Propel (s) Hindlimb | 0.195 ± 0.003 | 0.168 ± 0.002* | 0.175 ± 0.004 | 0.165 ± 0.002* |
| Stride length (cm) Forelimb | 7.06 ± 0.13 | 6.38 ± 0.10* | 6.56 ± 0.15 | 6.44 ± 0.11 |
| Stride length (cm) Hindlimb | 7.22 ± 0.09 | 6.45 ± 0.08* | 6.75 ± 0.10 | 6.57 ± 0.06 |
| Stride Frequency (steps/s) Forelimb | 2.87 ± 0.04 | 3.29 ± 0.03* | 2.92 ± 0.06 | 3.04 ± 0.05 |
| Stride Frequency (steps/s) Hindlimb | 2.81 ± 0.04 | 3.09 ± 0.04* | 2.86 ± 0.05 | 3.00 ± 0.02* |
| Stance width (cm) Forelimb | 1.58 ± 0.02 | 1.49 ± 0.03* | 1.44 ± 0.03 | 1.40 ± 0.03 |
| Stance width (cm) Hindlimb | 2.60 ± 0.05 | 2.35 ± 0.04* | 2.30 ± 0.02 | 2.21 ± 0.02* |
| Step angle (deg) Forelimb | 64.87 ± 1.83 | 65.81 ± 1.74 | 67.79 ± 1.49 | 65.63 ± 1.66 |
| Step angle (deg) Hindlimb | 54.77 ± 1.62 | 55.93 ± 0.99 | 61.04 ± 1.32 | 59.54 ± 1.33 |
| Paw Area (cm2) Forelimb | 0.23 ± 0.01 | 0.20 ± 0.01* | 0.22 ± 0.01 | 0.19 ± 0.01* |
| Paw Area (cm2) Hindlimb | 0.46 ± 0.01 | 0.38 ± 0.01* | 0.41 ± 0.01 | 0.40 ± 0.01 |
p < 0.05, for effect of genotype within gender.
Overall, long isoforms comprised roughly 70% of all isoforms in Sgce+/+ mice. More exclusively, the brain-specific isoform derived from exon 11b only comprised 10% of total Sgce (Table 3). Total expression of Sgce transcripts was reduced by approximately 60 to 70% in Sgcem+/pGt and SgcemGt/pGt mice and unchanged in SgcemGt/p+ mice (Tables 3 and S3). Sanger sequencing using a forward primer in exon 9 (Sgce_E9F) and two reverse primers in lacZ (β_Gal_R1 and β_Gal_R2) yielded Sgce/lacZ fusion amplicons (716 and 329 bp, respectively). QRT-PCR showed that long isoforms including the brain-specific isoform were not expressed in Sgcem+/pGt and SgcemGt/pGt mice (Table 3). However, expression of short Sgce isoforms and Sgce/lacZ fusion transcript(s) was preserved. Expression levels of fusion transcript(s) detected with random decamers were about twice the levels obtained with oligo(dT) primers (Table 3 and S3).
Table 3.
Sgce expression in 3-month-old gene-trap mice using relative quantitative reverse transcriptase PCR (QRT-PCR). Primers 1 (85F/85R), all isoforms. Primers 2 (98F/98R), long isoforms. Primers 3 (E9F/E11bR), brain-specific isoform. Primers 4 (E9F/β-gal_R2), Sgce/lacZ fusion transcript. Values are referenced to wild-type (Sgce+/+) cerebral cortex and presented as means ± standard error of the mean (SEM) (n = 8 mice/genotype).
| Tissue | Sgce+/+ | Sgcem+/pGt | SgcemGt/pGt | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Primers 1 | Primers 2 | Primers 3 | Primers 4 | Primers 1 | Primers 2 | Primers 3 | Primers 4 | Primers 1 | Primers 2 | Primers 3 | Primers 4 | |
| Cerebral cortex | 1.00 ± 0.02 | 0.70 ± 0.05 | 0.09 ± 0.01 | 0.00 ± 0.00 | 0.33 ± 0.04 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.15 ± 0.04 | 0.32 ± 0.03 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.18 ± 0.03 |
| Cerebellum | 2.70 ± 0.09 | 1.38 ± 0.08 | 0.17 ± 0.03 | 0.00 ± 0.00 | 1.09 ± 0.08 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.55 ± 0.03 | 0.99 ± 0.08 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.53 ± 0.04 |
| Hippocampus | 0.92 ± 0.02 | 0.62 ± 0.05 | 0.07 ± 0.01 | 0.00 ± 0.00 | 0.27 ± 0.04 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.15 ± 0.03 | 0.24 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.14 ± 0.03 |
| Midbrain | 1.77 ± 0.05 | 1.27 ± 0.06 | 0.12 ± 0.01 | 0.00 ± 0.00 | 0.80 ± 0.09 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.38 ± 0.05 | 0.71 ± 0.06 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.30 ± 0.04 |
| Striatum | 1.27 ± 0.02 | 0.86 ± 0.05 | 0.11 ± 0.02 | 0.00 ± 0.00 | 0.46 ± 0.04 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.24 ± 0.03 | 0.39 ± 0.03 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.21 ± 0.03 |
| Thalamus | 1.47 ± 0.04 | 1.05 ± 0.04 | 0.10 ± 0.01 | 0.00 ± 0.00 | 0.60 ± 0.07 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.29 ± 0.03 | 0.57 ± 0.04 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.27 ± 0.04 |
| Spinal cord | 1.38 ± 0.02 | 1.11 ± 0.07 | 0.07 ± 0.00 | 0.00 ± 0.00 | 0.59 ± 0.05 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.25 ± 0.03 | 0.53 ± 0.03 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.23 ± 0.03 |
| Liver | 0.39 ± 0.02 | 0.28 ± 0.03 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.01 ± 0.00 |
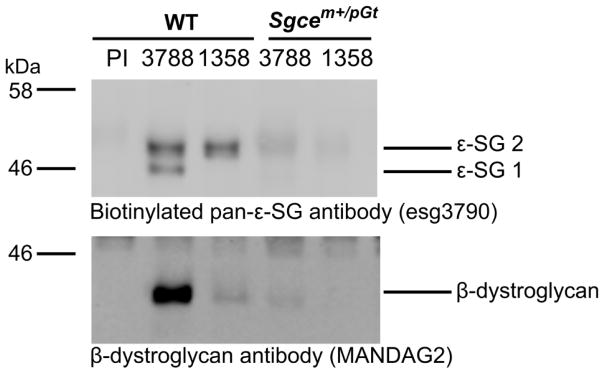
3.3 Sgcem+/pGt mice express reduced levels of ε-SG
ε-SG has been previously shown to form part of dystrophin-associated protein complexes in the brain that contain β-, δ- and ζ-sarcoglycans and β-dystroglycan (Waite et al., 2016). Therefore, we performed IAP and mass spectrometry analyses to investigate whether these complexes were disrupted in the Sgcem+/pGt mice compared to WT littermates. IAPs were performed with two different antibodies to enrich for total ε-SG (esg3788) or the brain-specific ε-SG isoform specifically (esg2-1358). Purification of all ε-SG isoforms was verified by Western blot analysis using an ε-SG polyclonal antibody (Figs. 2 and S2). Both the ubiquitous and brain-specific ε-SG isoforms were enriched in the esg3788 IAP and the brain-specific ε-SG isoforms in the esg2-1358 IAP from WT brain tissue. However, there was a major reduction in detectable ε-SG in both IAPs from the Sgcem+/pGt tissue leaving a residual immunoglobulin-related signal of approximately 50 kDa. Western blot of the IAPs using an anti-β-dystroglycan antibody demonstrated co-purification of the DGC component β-dystroglycan in the esg3788 IAP from WT tissue and to a lesser extent in the esg2-1358 IAP. By contrast, there was only trace detection of β-dystroglycan in the esg3788 IAP from Sgcem+/pGt tissue that was absent in the esg2-1358 IAP. With high exposure, a putative ε-SG/β-gal band was seen at ~159 kDa in the esg3788 IAP from Sgcem+/pGt tissue (Fig. S2).
Fig. 2.
Western blots of immunoaffinity purifications (IAPs) of ε-SG-containing complexes from WT C57BL/6J and Sgcem+/pGt mice. The left three lanes contain proteins isolated from WT brain tissue whereas the right two lanes contain protein from Sgcem+/pGt brain tissue. Lanes are labelled with the antibody used to isolate proteins: PI, preimmune IgG; 3788, esg3788 anti-ε-SG; 1358, esg2-1358 anti-ε-SG brain-specific isoform. The biotinylated anti-ε-SG antibody detects both the major isoform (lower band, ε-SG 1) and brain-specific isoform (upper bands, ε-SG 2) of ε-SG in IAPs using the esg3788 anti-ε-SG antibody, and only the brain-specific isoform in IAPs using the esg2-1358 anti-ε-SG brain-specific isoform antibody. Western blot using the anti-β-dystroglycan antibody MANDAG2 showing co-purifications of β-dystroglycan in the esg3788 and esg-1358 IAPs from WT tissue. β-dystroglycan is severely reduced in the esg3788 IAP from Sgcem+/pGt brain tissue and absent in the esg-1358 IAP.
The remaining immunoaffinity-purified material from all samples was separated by polyacrylamide gel electrophoresis and stained with colloidal Coomassie blue. Each sample was excised as two gel bands and analysed via mass spectrometry to identify the protein constituents. Multiple high confidence ε-, β-, δ- and ζ-sarcoglycan peptides were detected in the esg3788 and esg2-1358 IAPs from WT tissue indicating purification of the entire brain sarcoglycan complex. Only a subset of these peptides was detected in the esg3788 IAP from the Sgcem+/pGt mice (Table 4). Additionally, the spectral counts for each sarcoglycan were reduced in Sgcem+/pGt mice versus WT littermates, reflecting a lower abundance of the sarcoglycans in the esg3788 IAP from Sgcem+/pGt mice (Table S5). However, there was a complete absence of detectable sarcoglycan peptides in the esg2-1358 IAP from Sgcem+/pGt tissue indicating a loss of brain-specific ε-SG isoform containing protein complexes.
Table 4.
Unique sarcoglycan peptides identified in wild-type and Sgcem+/pGt whole brain ε-SG immunoaffinity purifications (IAPs). For each IAP (wild-type versus Sgcem+/pGt tissue, esg3788 antibody versus esg2-1358 antibody), all unique tryptic sarcoglycan peptides detected via mass spectrometry are included in the table. The ε-SG2 brain-specific peptides detected in wild-type tissue are in bold. Abbreviations: IAP, immunoaffinity purification; esg3788, anti-ε-SG antibody 3788; esg2-1358, anti-ε-SG2 antibody 1358.
| Protein | esg3788 IAP | esg2-1358 IAP | ||
|---|---|---|---|---|
|
| ||||
| Wild-type | Sgcem+/pGt | Wild-type | Sgcem+/pGt | |
| ε-SG | TPYSDGVLYGSPTAENVGKPTIIEITAYNRR | None | ||
| QVSTYQEVVR | QVSTYQEVVR | QVSTYQEVVR | ||
| EVENPQNQLR | EVENPQNQLR | |||
| FEVNGIPEER | ||||
| KLTEAmSL | ||||
|
| ||||
| β-SG | RNENLVITGNNQPIVFQQGTTK | RNENLVITGNNQPIVFQQGTTK | RNENLVITGNNQPIVFQQGTTK | None |
| LPSSSSGDQSGSGDWVR | LPSSSSGDQSGSGDWVR | |||
| THNILFSTDYETHEFHLPSGVK | ||||
| TSITSDIGmQFFDPR | TSITSDIGmQFFDPR | TSITSDIGmQFFDPR | ||
| LcMcADGTLFK | LcMcADGTLFK | |||
| GNEGVFIMGK | GNEGVFIMGK | |||
|
| ||||
| δ-SG | GVEINAEAGNMEAIcR | GVEINAEAGNMEAIcR | None | |
| LEGDSEFLQPLYAK | LEGDSEFLQPLYAK | LEGDSEFLQPLYAK | ||
| LLFSADDSEVVVGAER | LLFSADDSEVVVGAER | LLFSADDSEVVVGAER | ||
| VLGAEGTVFPK | VLGAEGTVFPK | VLGAEGTVFPK | ||
| VFEVcVcANGR | VFEVcVcANGR | |||
| VLTQLVTGPK | VLTQLVTGPK | VLTQLVTGPK | ||
| SRPGNALYFK | ||||
| SLVMEAPK | ||||
|
| ||||
| ζ-SG | ELHLQSTEGEIFLNADSIR | ELHLQSTEGEIFLNADSIR | None | |
| VLFSADEDEITIGAEK | VLFSADEDEITIGAEK | |||
| LEGISEFLLPLYVK | LEGISEFLLPLYVK | LEGISEFLLPLYVK | ||
| LGNLPIGSFSSSTSSSNSR | LGNLPIGSFSSSTSSSNSR | |||
| QTVYELcVcPnGK | QTVYELcVcPnGK | |||
| GVQVSAAAGDFK | GVQVSAAAGDFK | GVQVSAAAGDFK | ||
| STDLDIQELK | ||||
| VTGTEGAVFGHSVETPHIR | ||||
Both the Western blot and mass spectrometry data from the IAPs indicate a significant reduction in ε-sarcoglycan protein isoforms in Sgcem+/pGt mice compared with WT littermates with a complete loss of detectable ε-SG isoform 2. However, detection of β-dystroglycan by Western blot and ε-, β-, δ- and ζ-sarcoglycans using mass spectrometry in the esg3788 IAP indicate the existence of residual ε-SG containing sarcoglycan complexes and dystrophin-associated protein complexes in the Sgcem+/pGt mice.
3.4 Sgce is widely expressed in the central nervous system (CNS)
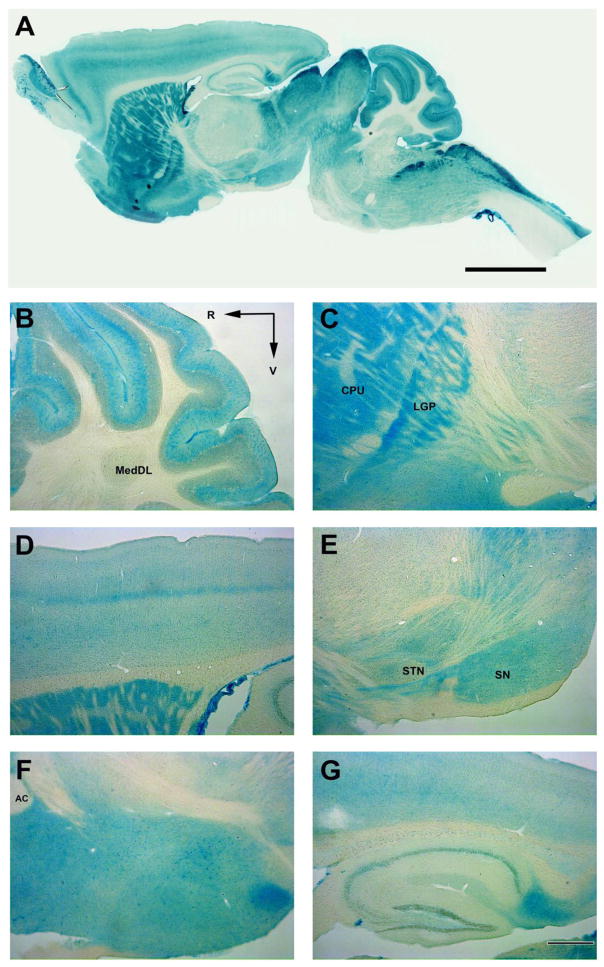
In Sgcem+/pGt mouse brain, X-gal staining was seen throughout the CNS (Fig. 3). In cerebellum, staining was prominent in the molecular layer with weaker expression in the granule cell layer and cerebellar nuclei (Fig. 3B). In the basal ganglia, staining in the caudate-putamen and lateral globus pallidus was more intense than that seen in the subthalamic nucleus and substantia nigra (Fig. 3C). In cerebral cortex, staining was most prominent within Layer V but also stood out in Layers II and III (Figs. 3A & D). In hippocampus, modest staining intensity was seen in the pyramidal cell layer and dentate gyrus (Fig. 3G).
Fig. 3.
X-gal expression in Sgcem+/pGt mice. A parasagittal section (A) of Sgcem+/pGt mouse brain, showing intense reaction in the gray matter of cerebellar cortex, striatum and cerebral cortex. (B) Cerebellum. (C) Striatum. (D) Cerebral cortex. (E) Ventral midbrain. (F) Hypothalamus. (G) Hippocampus. R, rostral; V, ventral; AC, anterior commissure; MedDL, medial cerebellar nucleus, dorsolateral protuberance; CPU, caudate putamen; LGP, lateral globus pallidus; STN, subthalamic nucleus; SN, substantia nigra. Scale bar = 400 μm.
3.5 Sgcem+/pGt mice have normal monoamine levels in the striatum
There were no significant effects of gender or genotype on striatal monoamine levels (Table S6). However, noradrenaline levels trended higher in Sgcem+/pGt mice.
3.6. Abnormal gait and appendicular posturing are seen in preweanling Sgcem+/pGt mice
No evidence of myoclonus or limb dystonia was noted while Sgcem+/pGt mice on the C57BL/6J background or mixed C57BL/6JxBABL/c and C57BL/6Jx129S2 backgrounds were routinely observed in their home cages or during open field behavior from the early postnatal period through 1.5 yrs of age. Moreover, myoclonus was not observed in response to audiogenic or tactile stimuli. However, abnormal hindlimb posturing in response to perturbations and “tiptoe” walking were significantly more common in preweanling Sgcem+/pGt mice (Videos S1 & S2) in comparison with WT littermates. Male and female Sgcem+/pGt mice had significantly higher scores than WT littermates (Table 5) on the rating scale for abnormal movements (Table S4). With the use of perturbations, the identification of abnormal motor phenotypes showed a low false positive rate. False positive and negative rates were higher with ratings of unperturbed open-field behavior (Table 5).
Table 5.
Blinded video analyses of abnormal movements in preweanling Sgcem+/pGt mice and WT littermates at P14 and P16.
| Sex, Genotypes (numbers, age) | Total scorea (mean ± SEM) | Number of genotype-phenotype matchesb | Percent correct |
|---|---|---|---|
| M, WT (n=5, P14) | 1.7 ± 1.2 | 4/5 | 80.0 |
| M, Sgcem+/pGt (n=5, P14) | 8.3 ± 3.3* | 5/5 | 100.0 |
| M, WT (n=10, P16) | 18.7 ± 4.7 | 22/30 | 73.3 |
| M, Sgcem+/pGt (n=10, P16) | 36.0 ± 5.3* | 18/30 | 60.0 |
| F, WT (n=10, P16) | 20.0 ± 8.1 | 23/30 | 76.7 |
| F, Sgcem+/pGt (n=10, P16) | 39.0 ± 10.4* | 22/30 | 73.3 |
Mean total scores for abnormal movements were generated by three blinded raters.
Genotype-phenotype matches were declared if ≥ 2/3 of the raters scored the presence of abnormal movements in Sgcem+/pGt mice or if ≤ 1/3 scored abnormal movements in WT littermates.
p < 0.05.
3.7. Sgcem+/pGt mice show evidence of mild abnormalities on measures of gait, open-field activity and behavior
In the preweanling period, Sgcem+/pGt mice and WT littermates showed no righting time differences (Fig. S3). In adult mice, there were significant effects of gender (F1,60 = 84.1, p < 0.0001) and genotype (F1,60 = 91.0, p < 0.0001) on grip strength (Table 2). Male WT mice exhibited greater grip strength than Sgcem+/pGt littermates (mean difference = 55 g). The effect of genotype was similar in female mice (mean difference = 54 g). However, after normalized by weight, the effect of genotype on grip strength was no longer apparent (p = 0.56). There were no effects of genotype on rope climbing or tube dominance. There was a notable effect of genotype (F1,60 = 15.2, p = 0.0003) but no effect of gender or the genotype-gender interaction on cross-maze scores. Male and female Sgcem+/pGt mice had significantly lower scores than their gender-matched WT littermates on the cross-maze test (p < 0.05, for both).
Sgcem+/pGt mice were less active than their WT littermates on multiple parameters open-field activity (Table 2). There were significant genotype*gender interactions with female mice showing larger effects of genotype on open-field assays of distance traveled, jump count, and ambulatory count (p = 0.0001, 0.0036, and 0.014, respectively). There were large effects of genotype on stereotypic counts (F1,60 = 32.65, p < 0.0001) and ambulatory episodes (F1,60 = 10.89, p = 0.0016) but no significant effects of the genotype*gender interaction.
DigiGait™ analyses showed minor effects of genotype (Table 2) on several gait parameters. Male Sgcem+/pGt mice had shorter forepaw (p = 0.004) and hindpaw (p < 0.0001) stride lengths and higher forepaw and hindpaw stride frequencies (p <0.0001, for both) than WT gender-matched littermates. These differences did not reach statistical significance in female mice. Forepaw areas were smaller in Sgcem+/pGt mice than WT littermates (F1,60 = 17.1, p = 0.0001). Overall, hindpaw areas were also smaller in Sgcem+/pGt mice than WT littermates (F1,60 = 30.6, p < 0.0001) but there was a genotype*gender interaction (F1,60 = 18.8, p < 0.0001) with male mice showing a more marked effect of the mutant allele. There was a small overall effect of genotype on forepaw stance width (F1,60 = 4.1, p = 0.047). Hindpaw stance width was narrower in Sgcem+/pGt mice than WT littermates (F1,60 = 23.33, p < 0.0001) although the difference between Sgcem+/pGt mice and WT females did not reach statistical significance (p = 0.060).
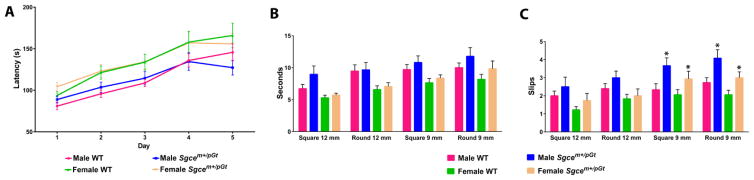
On the rotarod, both Sgcem+/pGt mice and WT littermates showed increased latencies to fall from Day 1 through Day 5 but there was no effect of genotype (Fig. 4A). On the raised-beam task, there were no significant effects of gender or genotype on beam traversal times (Figs. 4B & C). On the 9-mm round beam, male and female Sgcem+/pGt mice had more slips than gender-matched WT littermates (p = 0.016 and p = 0.22, respectively). Male and female Sgcem+/pGt mice also had more slips on the 9-mm square beam than gender-matched WT littermates (p = 0.0003 and p = 0.035, respectively).
Fig. 4.
Performance of Sgcem+/pGt mice (n = 15 males and 15 females) and WT littermates (n = 18 males and 16 females) on the rotarod (A) and raised beam tasks (B and C). Beam traversal times (B) and slips (C) were recorded for 4 different beams. *significant effect of genotype within gender (p<0.05)
3.8. Whole-genome gene expression abnormalities distinguish Sgcem+/pGt mice from WT littermates
The high linear correlations among scatter plots in male and female WT and Sgcem+/pGt mice support the reproducibility of our whole-genome gene expression data (Figs. S4 & S5). Differential gene expression was visualized with a heat map (Fig. S6) and volcano plot (Fig. S7). The symmetric volcano plot also attests to the quality of our gene expression analyses. With FDR ≤ 0.05 and fold change (FC) ≥ 2.0, we ended up with only two genes (Sgce and 1700016P03Rik) that were significantly dysregulated (Table 6, Fig. S7). Without correction for familywise error, we found 6 and 5 genes that were down- or up-regulated by 1.5X, respectively (Table 6). At a 1.25X FC, we found 38 and 31 genes that were significantly up- or down-regulated, respectively. QRT-PCR corroborated the direction and relative magnitude of differential expression for 11/12 genes that were assayed (Table 7).
Table 6.
Genes significantly up- and down-regulated in Sgcem+/pGt mice
| Affymetrix Cluster ID | Gene Symbol | Gene Name | Fold Change ≥ 1.25 & p ≤ 0.05) |
|---|---|---|---|
| 17400862 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | 1.250 |
| 17522876 | Eomes | Eomesodermin homolog (Xenopus laevis) | 1.250 |
| 17347355 | Eif2ak2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | 1.255 |
| 17436237 | Fosl2 | FOS-like antigen 2 | 1.255 |
| 17301615 | Gm21685 | Predicted gene, 21685 | 1.256 |
| 17259078 | Rnf213 | Ring finger protein 213 | 1.256 |
| 17222819 | 9330175M20Rik | RIKEN cDNA 9330175M20 gene | 1.262 |
| 17300591 | Irf9 | Interferon regulatory factor 9 | 1.264 |
| 17473666 | Vmn1r67 | Vomeronasal 1 receptor 67 | 1.266 |
| 17435577 | LOC102638085 | 1.268 | |
| 17234423 | Derl3 | Der1-like domain family, member 3 | 1.274 |
| 17403237 | Gbp3 | Guanylate binding protein 3 | 1.275 |
| 17411147 | Ifi44 | Interferon-induced protein 44 | 1.284 |
| 17547813 | Gm19845 | Predicted gene, 19845 | 1.289 |
| 17350009 | Gm24690 | Predicted gene, 24690 | 1.290 |
| 17487489 | Pvr | Poliovirus receptor | 1.293 |
| 17516383 | Snord14e | Small nucleolar RNA, C/D box 35A | 1.303 |
| 17538425 | Snora35 | Small nucleolar RNA, H/ACA box 35 | 1.305 |
| 17292632 | n-R5s54 | RNA, 5S Ribosomal Pseudogene 212 | 1.307 |
| 17320684 | Gm24647 | Predicted gene, 24647 | 1.318 |
| 17261033 | Gm22753 | Predicted gene, 22753 | 1.327 |
| 17481504 | Olfr713 | Olfactory receptor 713 | 1.341 |
| 17226622 | Cd55 | CD55 antigen | 1.344 |
| 17490622 | Snord35a | Small Nucleolar RNA, C/D Box 35A | 1.359 |
| 17455903 | Asb4 | Ankyrin repeat and SOCS box-containing 4 | 1.360 |
| 17290072 | Gm25985 | Predicted gene, 25985 | 1.373 |
| 17290072 | Traj13 | T Cell Receptor Alpha Joining 13 | 1.389 |
| 17361435 | Npas4 | Neuronal PAS domain protein 4 | 1.394 |
| 17315178 | Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 1.404 |
| 17335467 | Cdkn1a | Cyclin-dependent kinase inhibitor 1A (P21) | 1.410 |
| 17464654 | Pdk4 | Pyruvate dehydrogenase kinase, isoenzyme 4 | 1.426 |
| 17342642 | Dusp1 | Dual specificity phosphatase 1 | 1.453 |
| 17385374 | Nr4a2 | Nuclear receptor subfamily 4, group A, member 2 | 1.494 |
| 17358658 | Gm23426 | Predicted gene, 23426 | 1.518 |
| 17277387 | Fos | FBJ osteosarcoma oncogene | 1.698 |
| 17413945 | Nr4a3 | Nuclear receptor subfamily 4, group A, member 3 | 1.718 |
| 17308165 | Gm21464 | Predicted gene, 21464 | 1.856 |
| 17252875 | 1700016P03Rik | RIKEN cDNA 1700016P03 gene | 2.046 |
| 17464588 | Sgce | Sarcoglycan, epsilon | −2.098 |
| 17546762 | Mid1 | Midline 1 | −1.883 |
| 17532694 | Pisd-ps3 | Phosphatidylserine decarboxylase, pseudogene 3 | −1.805 |
| 17464803 | Ica1 | Islet cell autoantigen 1 | −1.566 |
| 17546212 | G530011O06Rik | RIKEN cDNA G530011O06 gene | −1.564 |
| 17403844 | Gm24373 | Predicted gene, 24373 | −1.551 |
| 17270615 | Gm22743 | Predicted gene, 22743 | −1.478 |
| 17491505 | Gm24966 | Predicted gene, 24966 | −1.457 |
| 17308963 | Gm23926 | Predicted gene, 23926 | −1.398 |
| 17278781 | Gm23347 | Predicted gene, 23347 | −1.349 |
| 17455971 | Mios | Missing oocyte, meiosis regulator, homolog (Drosophila) | −1.334 |
| 17491503 | Gm25499 | Predicted gene, 25499 | −1.333 |
| 17456204 | Capza2 | Capping protein (actin filament) muscle Z-line, alpha 2 | −1.333 |
| 17539189 | Gm23806 | Predicted gene, 23806 | −1.330 |
| 17364365 | Gm24930 | Predicted gene, 24930 | −1.327 |
| 17491527 | Gm25870 | Predicted gene, 25870 | −1.318 |
| 17356739 | Mir194-2 | microRNA 194-2 | −1.297 |
| 17283683 | Gm24334 | Predicted gene, 24334 | −1.288 |
| 17274184 | Socs2 | Suppressor of cytokine signaling 2 | −1.288 |
| 17432577 | Gm436 | Predicted gene 436 | −1.287 |
| 17399769 | S100a13 | S100 calcium binding protein A13 | −1.282 |
| 17491642 | Snord107 | Small Nucleolar RNA, C/D Box 107 | −1.281 |
| 17363082 | Olfr1437 | Olfactory receptor 1437 | −1.278 |
| 17440618 | Mir701 | microRNA 701 | −1.278 |
| 17478956 | Gm7551 | Heterogeneous nuclear ribonucleoprotein A3 pseudogene | −1.277 |
| 17460861 | Slc41a3 | Solute carrier family 41, member 3 | −1.273 |
| 17278840 | Mir376a | microRNA 376a | −1.271 |
| 17288946 | Mir9-2 | microRNA 9-2 | −1.261 |
| 17491507 | Gm22496 | Predicted gene, 22496 | −1.259 |
| 17491515 | Gm25121 | Predicted gene, 25121 | −1.256 |
| 17284609 | Ighv1-62-3 | Immunoglobulin heavy variable 1-62-3 | −1.254 |
Table 7.
Validation of whole-genome gene expression with relative quantitative RT-PCR.
| Gene symbol | Protein | Function | Fold Change (p value) *
|
|
|---|---|---|---|---|
| Microarray | QRT-PCR | |||
| Nr4a3 | Nuclear receptor subfamily 4 group A member 3 | Transcriptional activator | 1.72 ± 0.17 (9.0E-4) | 1.93 ± 0.15 (6.0E-5) |
| Nr4a2 | Nuclear receptor subfamily 4 group A member 2 | Transcriptional activator | 1.49 ± 0.14 (4.0E-3) | 1.29 ± 0.10 (0.010) |
| Dusp1 | Dual specificity protein phosphatase 1 | Regulation of cellular proliferation | 1.45 ± 0.14 (3.0E-4) | 2.17 ± 0.12 (8.0e-7) |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1 | Regulator of cell cycle progression | 1.41 ±0.16 (4.0E-3) | 1.50 ± 0.12 (1,0E-3) |
| Eomes | Eomesodermin | Transcriptional activator | 1.25 ± 0.06 (0.035) | 1.63 ± 0.17 (1.0E-4) |
| Spry4 | Sprouty homolog 4 | Inhibitor of mitogen-activated protein kinase (MAPK) signaling pathway | 1.24 ± 0.09 (0.027) | 1.21 ± 0.08 (0.018) |
| Slc41a3 | Solute carrier family 41 member 3 | Cation transmembrane transporter activity | −1.27 ± 0.09 (0.045) | −1.42 ± 0.07 (3.0E-4) |
| S100a13 | S100 calcium-binding protein A13 | Calcium ion binding and lipid binding | −1.28 ± 0.04 (1.7E-3) | −1.52 ± 0.05 (1.0E-4) |
| Socs2 | Suppressor of cytokine signaling 2 | Suppressor of cytokine signaling (SOCS) | −1.29 ± 0.09 (0.016) | −1.29 ± 0.04 (4.0E-4) |
| Capza2 | F-actin-capping protein subunit alpha-2 | Regulator of the actin filaments | −1.33 ± 0.05 (6.0E-5) | −1.18 ± 0.16 (0.131) |
| Mios | WD repeat-containing protein mio | Inhibitor of the target of rapamycin complex I | −1.33 ± 0.04 (2.0E-4) | −1.22 ± 0.11 (0.049) |
| Ica1 | Islet cell autoantigen 1 | Membrane protein on Golgi complex and immature secretory granules | −1.56 ± 0.04 (6.0E-6) | −1.86 ± 0.04 (3.0E-6) |
| Pomt2 | Protein-O-mannosyltransferase 2 | Modification of the protein alpha-dystroglycan | −1.01 ± 0.02 (0.678) | 1.01 ± 0.07 (0.886) |
| Sgca | Sarcoglycan, alpha | Component of the dystrophin-glycoprotein complex (DGC) | 1.04 ± 0.02 (0.216) | 1.05 ± 0.03 (0.459) |
| Sgcb | Sarcoglycan, beta | Component of DGC | −1.06 ± 0.02 (0.209) | −1.05 ± 0.05 (0.308) |
Means ± SEM (p value)
Genes showing the largest up-regulation (Table 6) included the immediate-early gene Fos and three members of the nuclear receptor subfamily 4 (Nr4a3, Nr4a2 and Nr4a1). Other up-regulated genes encode cell-cycle (Cdkn1a) or complement (Cd55) proteins or small nucleolar RNAs (Snord35a, Snora35, and Snord14e). The top down-regulated genes (Table 6) included those encoding an E3-ubiquitin ligase (Mid1) and islet cell autoantigen 1 (Ica1).
A large collection of genes directly or indirectly associated with the dystrophin-glycoprotein complex were specifically examined for evidence of differential expression between WT and Sgcem+/pGt cerebellum (Table S7). Based on BioGPS (biogps.org) and the Allen Brain Atlas (www.brain-map.org), the majority of these genes show significant expression in cerebellum. The largest, albeit modest, FCs were seen for Stx1A (syntaxin 1A) and Cav3 (caveolin). There was no significant up-regulation of other sarcoglycans (α β, γ, δ, or ζ).
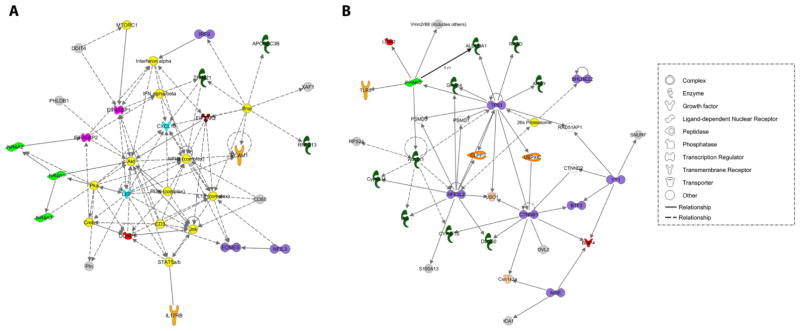
There were few enriched KEGG pathways for up-regulated genes and none for down-regulated genes. The largest collection of significantly up-regulated genes was associated with systemic lupus erythematosus and the ribosome (Table S8). IPA analysis indicated that a relatively large percentage of up- or down-regulated genes are involved in cell cycle, cellular development, and cell death and survival (Table S9). Regulation of eIF4 and p70S6K signaling and mTOR signaling were the top canonical pathway for up-regulated genes. Significant upstream regulators of dysregulated genes are shown in Table S10 and include Ifnar1, Crem, and Creb1 for up-regulated genes, and Tnfrsf4 and Nfe2l1 for down-regulated genes. Interactome analysis identified NF-κB and TP53 as the major hubs for up- and down-regulated genes, respectively (Fig. 5). Akt and NFE2L2 were secondary hubs for up- and down-regulated genes, respectively.
Fig. 5.
Ingenuity interactome analysis of up-regulated (A) and down-regulated (B) genes in Sgcem+/pGt mouse cerebellum.
4. Discussion
4.1 Splice variants of SGCE and Sgce
Human SGCE, located on the reverse strand of Chr. 7q21.3 (CRCh38/hg38), has 4 Consensus CDS isoforms (Fig. 1) and 4 RefSeq isoforms. Mouse Sgce, located on the reverse strand of Chr. 6qA1 (CRCm38/mm10) has 4 Consensus CDS isoforms and 5 RefSeq isoforms. The major isoform (NM_011360) harbors 11 exons with exon 8, but without exons 1a, 11, and 11b; while the brain-specific isoform (NM_001130189) also harbors 11 exons with exon 11b, but without exons 1a and 8. The gene-trap insertion site within intron 9 of NM_011360 is predicted to disrupt all isoforms with the exception of AK132210 (9 exons and without exons 1a, 10, 11, and 12) and ENSMUST00000139029. Total Sgce expression was reduced by 60 to 70% in our mouse model which exhibited mild motor and cognitive abnormalities along with stimulus-induced appendicular posturing. Presumably, residual expression of Sgce was enough to prevent the appearance of overt myoclonus. Amino acids 1 to 317 of mouse and human ε-SG are located in the extracellular space with only 23% of the full-length protein located in the cytoplasmic space (amino acids 339 – 437). The few SGCE mutations causally-associated with DYT11 and localized to the cytoplasmic domain of ε-SG are frameshifts and likely associated with nonsense-mediated decay (Valente et al., 2005, Asmus et al., 2005). However, it is possible that the failure to include all alternative exons, including exon 11b, in screening efforts has contributed to the relatively low reported frequency of SGCE mutations in subjects with MDS (Ritz et al., 2009, Peall et al., 2014, Valente et al., 2005). Moreover, our data suggest that the long isoforms of Sgce are required for entirely normal motor and cognitive functioning, at least in mice.
4.2 Long isoforms of Sgce are widely expressed in the CNS and enriched in cerebellum
Compatible with data reported by BioGPS (BioGPS.org) and Allen Brain Atlas (www.brain-map.org), we showed that Sgce is widely expressed in mouse brain from the early postnatal period through adulthood and modestly enriched in cerebellum in comparison with other sensorimotor structures such as striatum, thalamus and spinal cord. The expression patterns of Sgce identified with X-gal staining in our mouse model are similar to those reported in rats with in situ hybridization (Xiao and LeDoux, 2003). For the data described herein, β-gal staining was derived from a chimeric ε-SG/β-gal fusion protein and appears to represent expression of the major isoform and/or other protein isoforms that are derived from exon 10 and exons 3′ to exon 10. The β-gal staining does not incorporate expression of the shortest Sgce isoforms. IAP-mass spectrometry and QRT-PCR showed that the major and brain-specific isoforms of Sgce were eliminated in Sgcem+/pGt mice. However, short isoforms terminating 5′ to intron 9 and small amounts of a chimeric ε-SG/β-gal fusion protein were preserved and may have compensated, at least in part, for absence of the major and brain-specific isoforms.
4.3 Sgcem+/pGt mice exhibit mild behavioral and motor abnormalities
As reported previously in humans and Sgce null mice, our Sgce mutant mice generated with gene-trap technology showed clear-cut maternal imprinting such that Sgce expression from both SgcemGt/pGt and Sgcem+/pGt mice was absent (Yokoi et al., 2005). Male and female Sgcem+/pGt mice were smaller than WT littermates and showed several abnormalities on behavioral and motor testing. Male and female Sgcem+/pGt mice showed reduced scores on the cross-maze test. Based on the results of rope climbing, rotarod and open-field activity, it is unlikely that the significantly lower scores on the cross-maze test were due to motor deficits. Instead, the cross-maze scores are a likely manifestation of increased anxiety in Sgcem+/pGt mice although contributions from cognitive deficits cannot be excluded (Salimov et al., 1995, Salimov et al., 1996). Anxiety is a well-known psychiatric co-morbidity in human myoclonus-syndrome due to mutations in SGCE (Peall et al., 2013, Peall et al., 2016). Accordingly, it is possible that deleterious sequence variants localized to terminal exons of SGCE could be risk factors for human anxiety even if not associated with dystonia and/or myoclonus.
Dystonia or dystonic-like posturing was not described in previously published murine models of DYT11 (Yokoi et al., 2005, Yokoi et al., 2006, Yokoi et al., 2012a, Yokoi et al., 2012b). Stimulus-induced flexion posturing was seen in our preweanling Sgcem+/pGt mice mainly between P14 and P16, a key period for development of cerebellar Purkinje cells and cerebellar cortex (Kano and Hashimoto, 2012). In theory, Purkinje cells and/or local area networks could be unstable until additional maturation has been completed. In this regard, stimulus-induced dystonia, seen in humans with paroxysmal kinesigenic dyskinesias due to mutation in PRRT2, often becomes manifest during childhood (Erro et al., 2014), and transient dystonia of infancy usually manifests between 5 and 10 months of age and then gradually disappears after 3 months to 5 years (Bonnet et al., 2010). Given that dystonia remains an inexact clinical diagnosis, both in mice and men, we have restricted our description to flexion posturing. We could not illicit abnormal posturing in postweanling mice or extensor posturing in preweanling mice. Sgcem+/pGt mice also exhibited a “tiptoe” gait with raised caudal trunk and narrowed hindlimb stance widths. The narrowed hindlimb stance widths persisted into adulthood (Table 2). Tiptoe walking is not specific to dystonia or Sgcem+/pGt mice and has also been described in Yoshimura mice, a model of chronic cervical myelopathy (Wang et al., 2014). However, early gait abnormalities may be common in humans with SGCE mutations (Asmus et al., 2005). In one study, for example, all SGCE mutations carriers had a gait disorder, unsteadiness or frequent falls before 18 months of age (Asmus et al., 2005).
Our gene-trap Sgce mouse model showed some similarities and a few important differences from previously reported Sgce null and conditional KO mice, as well as Tor1a-Sgce double null mice. All Sgce models have shown impaired performance on the raised-beam task with increased numbers of slips (Yokoi et al., 2006, Yokoi et al., 2012a, Yokoi et al., 2012b, Yokoi et al., 2010). Sgce null mice had increased vertical open-field motor activity, anxiety-like behavior, myoclonus and increased striatal dopamine (Yokoi et al., 2006). In comparison, our gene-trap model also showed evidence of anxiety but exhibited reduced vertical open-field motor activity, had no evidence of myoclonus and striatal dopamine levels were normal. In addition, slips on the raised-beam task are not specific to Sgce models and have been reported in Tor1a, Thap1, and Ciz1 models of dystonia (Yokoi et al., 2015, Ruiz et al., 2015, Xiao et al., 2016, Zhao et al., 2008). Increased slips on the raised beam task have been reported in heterozygous Tor1a null mice and hMT1 transgenic mice that overexpress mutant human torsinA (Zhao et al., 2008, Dang et al., 2005, Song et al., 2014).
4.4 Gene expression abnormalities in Sgcem+/pGt mice
IAP-mass spectrometry showed that brain-specific ε-SG interacts with β-SG, δ-SG, and ζ-SG. Gene-expression data did not show evidence for their compensatory up-regulation in Sgcem+/pGt mice. Furthermore, there was no apparent up-regulation for other components of the DGC at the transcript level. Given the modest phenotypes in our Sgcem+/pGt mice, it seems that short isoforms of ε-SG were able to partially compensate for loss of the major and brain-specific isoforms and other long isoforms.
Based on IPA analysis, it appears that up- and/or down-regulation of genes in various cell cycle, cellular development, and cell death and survival pathways served to compensate for ε-SG deficiency in our mouse model. However, we cannot exclude the possibility that some of these changes in gene expression were not due to novel effects of mutant ε-SG/β-gal fusion protein(s) that resulted from insertion of the Gep-SD5 gene-trap vector into intron 9 of Sgce. Although a single-pass transmembrane cell-surface protein seemingly involved in synaptic transmission, our gene expression data suggests that ε-SG shows functional overlap with other dystonia proteins such as TAF1, CIZ1, torsinA, THAP1, and Gα(olf) which play direct or indirect roles in cell cycle control, DNA repair and neurodevelopment (LeDoux et al., 2013, Xiao et al., 2016).
Several relatively distinct groups of genes showed up-regulation (Nr4a3, Fos, Nr4a2, Nr4a1, Cdkn1a, Cd55, Snord35a, Snora35 and Snord14e) or down-regulation (Mid1 and Ica1). Intermittent subclinical involuntary movements and/or anxiety could be responsible for differential up-regulation of the immediate-early genes Fos, Nr4a3, Nr4a2, and Nr4a1 in Sgcem+/pGt mice (Huguet et al., 2016). CDKN1A is an essential cell-cycle protein that interacts with the dystonia-associated protein CIZ1 (LeDoux et al., 2013, Xiao et al., 2012) and CD55 regulates the complement system, an important player in neuronal differentiation (Stephan et al., 2012). Snord35a, Snora35 and Snord14e encode small nucleolar RNAs, a class of RNA molecules that contribute to modifications of rRNA and tRNAs such as methylation and pseudouridylation. MID1 functions as a ubiquitin ligase and regulator of mTOR signaling and plays an important role in neurodevelopment and neurodegeneration. Lack of MID1 causes abnormal development of the cerebellum (Lancioni et al., 2010).
Top up-regulated canonical pathways (eIF4, p7056K, mTOR) and IPA nodes (NF-kB, Akt) are related to intracellular pathways regulating the cell cycle. The Akt/mTOR component of this intracellular network has been shown to regulate neurite outgrowth in cerebellar granule cells (Okada et al., 2011) and the morphology of Purkinje cells (Thomanetz et al., 2013). The major hubs for down-regulated genes, TP53 and NFE2L2, can also be readily linked to cell cycle control and neurodevelopment. In cerebellum, elevation of NFE2L2 (alias NRF2) levels in beneficial in a mouse model of Alexander disease caused by autosomal mutation in GFAP (LaPash Daniels et al., 2012). TP53 regulates CDKN1A at the G1/S cell-cycle checkpoint. In aggregate, gene expression studies suggest that compensatory up- or down-regulation of genes related to neurodevelopment and neuronal morphology may compensate for loss of major and brain-specific ε-SG isoforms.
5. Conclusions
Sgcem+/pGt mice show germline absence of major and brain-specific isoforms harboring exons 3′ to intron 9. Sgce was maternally-imprinted in our mouse model. Sgcem+/pGt mice exhibited tiptoe walking and occasional stimulus-induced appendicular flexion posturing in the preweanling period from P10 to P20. Adult Sgcem+/pGt mice showed increased slips on a raised-beam task, anxiety-like behavioral abnormalities, and narrowed hindlimb stance widths but no myoclonus or differences in striatal monoamines in comparison with WT littermates. Overall, adult Sgcem+/pGt mice were less active than WT littermates. There was no evidence for up-regulation of other sarcoglycans in Sgcem+/pGt mice. Genes differentially expressed between Sgcem+/pGt mice and WT littermates were involved in cell cycle, cellular development, and cell death and survival. Our gene-trap mice provide novel insights into the biology and pathobiology of ε-SG.
Supplementary Material
Highlights.
We generated a gene-trap mouse model with reduced expression of ε-sarcoglycan.
Sgce mutant mice exhibit a tiptoe gait and appendicular posturing.
Sgce mutant mice have altered gait dynamics and reduced open-field activity.
In the cerebellum, dysregulated genes were involved in cell cycle and development.
Short Sgce isoforms compensate for deficiency of major and brain-specific isoforms.
Acknowledgments
This study was supported by the Neuroscience Institute at the University of Tennessee Health Science Center, Dystonia Medical Research Foundation, Dorothy/Daniel Gerwin Parkinson’s Research Fund, and National Institutes of Health grants R03 NS050185, R01 NS082296 and R01 NS069936.
Footnotes
Supplementary data to the article can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AICHHORN W, WHITWORTH AB, WEISS EM, MARKSTEINER J. Second-generation antipsychotics: is there evidence for sex differences in pharmacokinetic and adverse effect profiles? Drug Saf. 2006;29:587–98. doi: 10.2165/00002018-200629070-00004. [DOI] [PubMed] [Google Scholar]
- ASMUS F, SALIH F, HJERMIND LE, OSTERGAARD K, MUNZ M, KUHN AA, DUPONT E, KUPSCH A, GASSER T. Myoclonus-dystonia due to genomic deletions in the epsilon-sarcoglycan gene. Ann Neurol. 2005;58:792–7. doi: 10.1002/ana.20661. [DOI] [PubMed] [Google Scholar]
- BENJAMINI Y, HELLER R. Screening for partial conjunction hypotheses. Biometrics. 2008;64:1215–22. doi: 10.1111/j.1541-0420.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- BJELLQVIST B, HUGHES GJ, PASQUALI C, PAQUET N, RAVIER F, SANCHEZ JC, FRUTIGER S, HOCHSTRASSER D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–31. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- BONNET C, ROUBERTIE A, DOUMMAR D, BAHI-BUISSON N, COCHEN DE COCK V, ROZE E. Developmental and benign movement disorders in childhood. Mov Disord. 2010;25:1317–34. doi: 10.1002/mds.22944. [DOI] [PubMed] [Google Scholar]
- DANG MT, YOKOI F, MCNAUGHT KS, JENGELLEY TA, JACKSON T, LI J, LI Y. Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset dystonia. Exp Neurol. 2005;196:452–63. doi: 10.1016/j.expneurol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- ERRO R, SHEERIN UM, BHATIA KP. Paroxysmal dyskinesias revisited: a review of 500 genetically proven cases and a new classification. Mov Disord. 2014;29:1108–16. doi: 10.1002/mds.25933. [DOI] [PubMed] [Google Scholar]
- ESAPA CT, WAITE A, LOCKE M, BENSON MA, KRAUS M, MCILHINNEY RA, SILLITOE RV, BEESLEY PW, BLAKE DJ. SGCE missense mutations that cause myoclonus-dystonia syndrome impair epsilon-sarcoglycan trafficking to the plasma membrane: modulation by ubiquitination and torsinA. Hum Mol Genet. 2007;16:327–42. doi: 10.1093/hmg/ddl472. [DOI] [PubMed] [Google Scholar]
- HUGUET G, KADAR E, TEMEL Y, LIM LW. Electrical Stimulation Normalizes c-Fos Expression in the Deep Cerebellar Nuclei of Depressive-like Rats: Implication of Antidepressant Activity. Cerebellum. 2016 doi: 10.1007/s12311-016-0812-y. [DOI] [PubMed] [Google Scholar]
- IMAMURA M, MOCHIZUKI Y, ENGVALL E, TAKEDA S. Epsilon-sarcoglycan compensates for lack of alpha-sarcoglycan in a mouse model of limb-girdle muscular dystrophy. Hum Mol Genet. 2005;14:775–83. doi: 10.1093/hmg/ddi072. [DOI] [PubMed] [Google Scholar]
- KANO M, HASHIMOTO K. Activity-dependent maturation of climbing fiber to Purkinje cell synapses during postnatal cerebellar development. Cerebellum. 2012;11:449–50. doi: 10.1007/s12311-011-0337-3. [DOI] [PubMed] [Google Scholar]
- KINUGAWA K, VIDAILHET M, CLOT F, APARTIS E, GRABLI D, ROZE E. Myoclonus-dystonia: an update. Mov Disord. 2009;24:479–89. doi: 10.1002/mds.22425. [DOI] [PubMed] [Google Scholar]
- LANCIONI A, PIZZO M, FONTANELLA B, FERRENTINO R, NAPOLITANO LM, DE LEONIBUS E, MERONI G. Lack of Mid1, the mouse ortholog of the Opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J Neurosci. 2010;30:2880–7. doi: 10.1523/JNEUROSCI.4196-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAPASH DANIELS CM, AUSTIN EV, ROCKNEY DE, JACKA EM, HAGEMANN TL, JOHNSON DA, JOHNSON JA, MESSING A. Beneficial effects of Nrf2 overexpression in a mouse model of Alexander disease. J Neurosci. 2012;32:10507–15. doi: 10.1523/JNEUROSCI.1494-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEDOUX MS, DAUER WT, WARNER TT. Emerging common molecular pathways for primary dystonia. Mov Disord. 2013;28:968–81. doi: 10.1002/mds.25547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NISHIYAMA A, ENDO T, TAKEDA S, IMAMURA M. Identification and characterization of epsilon-sarcoglycans in the central nervous system. Brain Res Mol Brain Res. 2004;125:1–12. doi: 10.1016/j.molbrainres.2004.01.012. [DOI] [PubMed] [Google Scholar]
- OKADA K, TANAKA H, TEMPORIN K, OKAMOTO M, KURODA Y, MORITOMO H, MURASE T, YOSHIKAWA H. Akt/mammalian target of rapamycin signaling pathway regulates neurite outgrowth in cerebellar granule neurons stimulated by methylcobalamin. Neurosci Lett. 2011;495:201–4. doi: 10.1016/j.neulet.2011.03.065. [DOI] [PubMed] [Google Scholar]
- PEALL KJ, DIJK JM, SAUNDERS-PULLMAN R, DREISSEN YE, VAN LOON I, CATH D, KURIAN MA, OWEN MJ, FONCKE EM, MORRIS HR, GASSER T, BRESSMAN S, ASMUS F, TIJSSEN MA. Psychiatric disorders, myoclonus dystonia and SGCE: an international study. Ann Clin Transl Neurol. 2016;3:4–11. doi: 10.1002/acn3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEALL KJ, KURIAN MA, WARDLE M, WAITE AJ, HEDDERLY T, LIN JP, SMITH M, WHONE A, PALL H, WHITE C, LUX A, JARDINE PE, LYNCH B, KIROV G, O’RIORDAN S, SAMUEL M, LYNCH T, KING MD, CHINNERY PF, WARNER TT, BLAKE DJ, OWEN MJ, MORRIS HR. SGCE and myoclonus dystonia: motor characteristics, diagnostic criteria and clinical predictors of genotype. J Neurol. 2014;261:2296–304. doi: 10.1007/s00415-014-7488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEALL KJ, SMITH DJ, KURIAN MA, WARDLE M, WAITE AJ, HEDDERLY T, LIN JP, SMITH M, WHONE A, PALL H, WHITE C, LUX A, JARDINE P, BAJAJ N, LYNCH B, KIROV G, O’RIORDAN S, SAMUEL M, LYNCH T, KING MD, CHINNERY PF, WARNER TT, BLAKE DJ, OWEN MJ, MORRIS HR. SGCE mutations cause psychiatric disorders: clinical and genetic characterization. Brain. 2013;136:294–303. doi: 10.1093/brain/aws308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAIKE RS, PIZOLI CE, WEISZ C, VAN DEN MAAGDENBERG AM, JINNAH HA, HESS EJ. Limited regional cerebellar dysfunction induces focal dystonia in mice. Neurobiol Dis. 2012;49C:200–210. doi: 10.1016/j.nbd.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAYMOND D, SAUNDERS-PULLMAN R, DE CARVALHO AGUIAR P, SCHULE B, KOCK N, FRIEDMAN J, HARRIS J, FORD B, FRUCHT S, HEIMAN GA, JENNINGS D, DOHENY D, BRIN MF, DE LEON BRIN D, MULTHAUPT-BUELL T, LANG AE, KURLAN R, KLEIN C, OZELIUS L, BRESSMAN S. Phenotypic spectrum and sex effects in eleven myoclonus-dystonia families with epsilon-sarcoglycan mutations. Mov Disord. 2008;23:588–92. doi: 10.1002/mds.21785. [DOI] [PubMed] [Google Scholar]
- RITZ K, GERRITS MC, FONCKE EM, VAN RUISSEN F, VAN DER LINDEN C, VERGOUWEN MD, BLOEM BR, VANDENBERGHE W, CROLS R, SPEELMAN JD, BAAS F, TIJSSEN MA. Myoclonus-dystonia: clinical and genetic evaluation of a large cohort. J Neurol Neurosurg Psychiatry. 2009;80:653–8. doi: 10.1136/jnnp.2008.162099. [DOI] [PubMed] [Google Scholar]
- RITZ K, VAN SCHAIK BD, JAKOBS ME, VAN KAMPEN AH, ARONICA E, TIJSSEN MA, BAAS F. SGCE isoform characterization and expression in human brain: implications for myoclonus-dystonia pathogenesis? Eur J Hum Genet. 2011;19:438–44. doi: 10.1038/ejhg.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUIZ M, PEREZ-GARCIA G, ORTIZ-VIRUMBRALES M, MENERET A, MORANT A, KOTTWITZ J, FUCHS T, BONET J, GONZALEZ-ALEGRE P, HOF PR, OZELIUS LJ, EHRLICH ME. Abnormalities of motor function, transcription and cerebellar structure in mouse models of THAP1 dystonia. Hum Mol Genet. 2015;24:7159–70. doi: 10.1093/hmg/ddv384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALIMOV R, SALIMOVA N, SHVETS L, SHVETS N. Effect of chronic piracetam on age-related changes of cross-maze exploration in mice. Pharmacol Biochem Behav. 1995;52:637–40. doi: 10.1016/0091-3057(95)00179-z. [DOI] [PubMed] [Google Scholar]
- SALIMOV RM, MCBRIDE WJ, MCKINZIE DL, LUMENG L, LI TK. Effects of ethanol consumption by adolescent alcohol-preferring P rats on subsequent behavioral performance in the cross-maze and slip funnel tests. Alcohol. 1996;13:297–300. doi: 10.1016/0741-8329(95)02060-8. [DOI] [PubMed] [Google Scholar]
- SONG CH, BERNHARD D, HESS EJ, JINNAH HA. Subtle microstructural changes of the cerebellum in a knock-in mouse model of DYT1 dystonia. Neurobiol Dis. 2014;62:372–80. doi: 10.1016/j.nbd.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEPHAN AH, BARRES BA, STEVENS B. The complement system: an unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012;35:369–89. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- STOTHARD P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102, 1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- THOMANETZ V, ANGLIKER N, CLOETTA D, LUSTENBERGER RM, SCHWEIGHAUSER M, OLIVERI F, SUZUKI N, RUEGG MA. Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J Cell Biol. 2013;201:293–308. doi: 10.1083/jcb.201205030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VALENTE EM, EDWARDS MJ, MIR P, DIGIORGIO A, SALVI S, DAVIS M, RUSSO N, BOZI M, KIM HT, PENNISI G, QUINN N, DALLAPICCOLA B, BHATIA KP. The epsilon-sarcoglycan gene in myoclonic syndromes. Neurology. 2005;64:737–9. doi: 10.1212/01.WNL.0000151979.68010.9B. [DOI] [PubMed] [Google Scholar]
- WAITE AJ, CARLISLE FA, CHAN YM, BLAKE DJ. Myoclonus dystonia and muscular dystrophy: epsilon-sarcoglycan is part of the dystrophin-associated protein complex in brain. Mov Disord. 2016 doi: 10.1002/mds.26738. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG J, WANG X, RONG W, LV J, WEI F, LIU Z. Alteration in chondroitin sulfate proteoglycan expression at the epicenter of spinal cord is associated with the loss of behavioral function in Tiptoe walking Yoshimura mice. Neurochem Res. 2014;39:2394–406. doi: 10.1007/s11064-014-1442-8. [DOI] [PubMed] [Google Scholar]
- XIAO J, LEDOUX MS. Cloning, developmental regulation and neural localization of rat epsilon-sarcoglycan. Brain Res Mol Brain Res. 2003;119:132–43. doi: 10.1016/j.molbrainres.2003.09.004. [DOI] [PubMed] [Google Scholar]
- XIAO J, NANCE MA, LEDOUX MS. Incomplete nonsense-mediated decay facilitates detection of a multi-exonic deletion mutation in SGCE. Clin Genet. 2013;84:276–80. doi: 10.1111/cge.12059. [DOI] [PubMed] [Google Scholar]
- XIAO J, UITTI RJ, ZHAO Y, VEMULA SR, PERLMUTTER JS, WSZOLEK ZK, MARAGANORE DM, AUBURGER G, LEUBE B, LEHNHOFF K, LEDOUX MS. Mutations in CIZ1 cause adult onset primary cervical dystonia. Ann Neurol. 2012;71:458–69. doi: 10.1002/ana.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIAO J, VEMULA SR, XUE Y, KHAN MM, KURUVILLA KP, MARQUEZ-LONA EM, COBB MR, LEDOUX MS. Motor phenotypes and molecular networks associated with germline deficiency of Ciz1. Exp Neurol. 2016;283:110–120. doi: 10.1016/j.expneurol.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOI F, CHEN HX, DANG MT, CHEETHAM CC, CAMPBELL SL, ROPER SN, SWEATT JD, LI Y. Behavioral and electrophysiological characterization of Dyt1 heterozygous knockout mice. PLoS One. 2015;10:e0120916. doi: 10.1371/journal.pone.0120916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOI F, DANG MT, LI J, LI Y. Myoclonus, motor deficits, alterations in emotional responses and monoamine metabolism in epsilon-sarcoglycan deficient mice. J Biochem. 2006;140:141–6. doi: 10.1093/jb/mvj138. [DOI] [PubMed] [Google Scholar]
- YOKOI F, DANG MT, MITSUI S, LI Y. Exclusive paternal expression and novel alternatively spliced variants of epsilon-sarcoglycan mRNA in mouse brain. FEBS Lett. 2005;579:4822–8. doi: 10.1016/j.febslet.2005.07.065. [DOI] [PubMed] [Google Scholar]
- YOKOI F, DANG MT, YANG G, LI J, DOROODCHI A, ZHOU T, LI Y. Abnormal nuclear envelope in the cerebellar Purkinje cells and impaired motor learning in DYT11 myoclonus-dystonia mouse models. Behav Brain Res. 2012a;227:12–20. doi: 10.1016/j.bbr.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOI F, DANG MT, ZHOU T, LI Y. Abnormal nuclear envelopes in the striatum and motor deficits in DYT11 myoclonus-dystonia mouse models. Hum Mol Genet. 2012b;21:916–25. doi: 10.1093/hmg/ddr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOI F, YANG G, LI J, DEANDRADE MP, ZHOU T, LI Y. Earlier onset of motor deficits in mice with double mutations in Dyt1 and Sgce. J Biochem. 2010;148:459–66. doi: 10.1093/jb/mvq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG B, KIROV S, SNODDY J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO Y, DECUYPERE M, LEDOUX MS. Abnormal motor function and dopamine neurotransmission in DYT1 DeltaGAG transgenic mice. Exp Neurol. 2008;210:719–30. doi: 10.1016/j.expneurol.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIMPRICH A, GRABOWSKI M, ASMUS F, NAUMANN M, BERG D, BERTRAM M, SCHEIDTMANN K, KERN P, WINKELMANN J, MULLER-MYHSOK B, RIEDEL L, BAUER M, MULLER T, CASTRO M, MEITINGER T, STROM TM, GASSER T. Mutations in the gene encoding epsilon-sarcoglycan cause myoclonus-dystonia syndrome. Nat Genet. 2001;29:66–9. doi: 10.1038/ng709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.