Abstract
In contrast to the traditional relational semiotics, biosemiotics decisively deviates towards dynamical aspects of signs at the evolutionary and developmental time scales. The analysis of sign dynamics requires constructivism (in a broad sense) to explain how new components such as subagents, sensors, effectors, and interpretation networks are produced by developing and evolving organisms. Semiotic networks that include signs, tools, and subagents are multilevel, and this feature supports the plasticity, robustness, and evolvability of organisms. The origin of life is described here as the emergence of simple self-constructing semiotic networks that progressively increased the diversity of their components and relations. Primitive organisms have no capacity to classify and track objects; thus, we need to admit the existence of proto-signs that directly regulate activities of agents without being associated with objects. However, object recognition and handling became possible in eukaryotic species with the development of extensive rewritable epigenetic memory as well as sensorial and effector capacities. Semiotic networks are based on sequential and recursive construction, where each step produces components (i.e., agents, scaffolds, signs, and resources) that are needed for the following steps of construction. Construction is not limited to repair and reproduction of what already exists or is unambiguously encoded, it also includes production of new components and behaviors via learning and evolution. A special case is the emergence of new levels of organization known as metasystem transition. Multilevel semiotic networks reshape the phenotype of organisms by combining a mosaic of features developed via learning and evolution of cooperating and/or conflicting subagents.
Keywords: Evolutionary semiotics, constructivism, semiotic network, evolvability, metasystem transition, constraints on learning
1. Introduction: Biosemiotics requires constructivism
Traditional semiotics is focused on the relations between sign vehicles, objects, and interpretants (i.e., thoughts or actions that follow the interpretation of signs), whereas questions about the origin and evolution of sign relations are mostly ignored. This is natural for a discipline that is strongly integrated with logic and linguistics because logic and language are stable within the human life span. But recently semiotics has expanded into biology, where the new discipline of biosemiotics attempts to apply the notions of sign and meaning to all organisms (Sharov 1992; Hoffmeyer 1996, 2008; Barbieri 2008). Although biosemiotics has strong connections with traditional relational semiotics (Deely 1992), it decisively deviates towards dynamical aspects of signs at the evolutionary and developmental time scales (Sharov 1992; Cariani 1998). The main feature of this approach in biosemiotics can be formulated as constructivism in a broad sense. Everything has to be constructed: sense organs – to detect signals; networks – to integrate and analyze signals, effector organs – to respond; memory – to store information; subagents – to perform downstream tasks including lower-level construction; body – to integrate all functional units; niche – to live in; tools and resources – to increase functional efficiency; and signs – to support communication between parts of an organism and with other organisms.
The term constructivism1 generally denotes a theory of human knowledge that emphasizes the importance of active involvement in knowledge-building and rejects the idea that knowledge comes via passive imprinting or copying (Tobias and Duffy 2009; Riegler 2006). In systems science, constructivism is used to describe agents that actively modify the world in contrast to passive observers and predictors (Klir 1991). In philosophy, this term is often used to emphasize the subjective component of behaviors, which are guided not directly by the real world but by previously constructed internal representations of reality (Liu and Matthews 2005). This aspect may be erroneously misunderstood as relativism in a broadest sense, where internal representations are not constrained by reality. However, if we accept the notion of a unity between mind and body, evolution and cognition, and individual and social, as emphasized by Vygotsky (Liu and Matthews 2005) and Piaget (Piaget and Garcia 1989), then internal representations appear strongly constrained (but not determined) by various real interactions in the past (both physical and cultural). In this way, constructivism is closely linked with the philosophy of pragmatism (James 1954; Dewey 1998).
Construction should not be confused with computation, although it can be modeled or controlled computationally. For example, cellular-automata models generate versatile dynamic patterns (von Neumann 1966; Gardner 1970). In 3D printers, construction is controlled by a computer but the result is non-digital because glue drops used for printing slightly vary in size and shape, and their deposition depends on the presence of neighboring structures. Living processes are not fully digital and not computable although some of them resemble computation processes (e.g., DNA replication or polypeptide synthesis). It is debatable if artificial life (AL) can be designed on computation alone, but pure computational systems are not likely to have evolutionary potential and robustness similar to real organisms due to the absence of non-digital self-organization.
The roots of constructivism can be traced back to James Baldwin (Baldwin 1896), who developed “genetic epistemology” and proposed a model of evolution where animal behaviors are both products and factors of evolution (this effect was named after Baldwin). The theory of meaning developed by Jacob von Uexküll is also related to pragmatism and constructivism (Sharov 2001). According to Uexküll, every animal develops its subjective model of the environment, called Umwelt, where objects and perceptions are associated with certain values (food items, sex partners, or orientation marks) or threats (predators) (Uexküll 1982). Ideas of constructivism in relation to biology were further developed by Von Foerster and Bateson (Riegler 2006). Waddington proposed an epigenetic model of sign interpretation, where infinitely small signals become amplified at bifurcation unstable points of embryo development and trigger larger downstream phenotypic effects (Waddington 1968). Potential trajectories of embryo development form an epigenetic landscape where valleys represent stable types of embryo development or cell differentiation. The role of genes in this model is to reshape the epigenetic landscape by pull-and-stretch actions. In this way, genes can support heredity without determining the phenotype (Sharov 2014).
Another important idea of constructivism is the notion of self-construction. Self-construction was initially explored using the formalism of cellular automata (von Neumann 1966; Langton 1984). However, this approach is over-simplified because it is based on discrete states and ignores non-digital epigenetic self-organization processes comparable to protein folding or embryo development. In later theories, mathematics was used mostly for developing concepts rather than for computing. These include the category theory (Rosen 1991), autopoiesis (Maturana and Varela 1980), and eigenbehaviors (Cariani 1998). Because self-construction is recursive (Bickhard 2005), it is possible to explore the long-term dynamics in a sequence of recursive construction acts. It is reasonable to expect the existence of meta-stable states (i.e., eigenstates and eigenbehaviors) in the self-construction dynamics, which explains the phenomenon of heredity without an assumption of determinism. Also, recursive construction allows the emergence of new meta-stable structures and behaviors that represent evolution and learning. Cariani used eigenbehaviors as a guiding principle for developing evolutionary aspects of semiotics and explored the change of internal models of the outer world in artificial and natural agents (Cariani 1998). The theory of code biology also attempts to link construction with semiotics (Barbieri 2003; Barbieri 2008). In particular, Barbieri considers the synthesis of polypeptides based on the genetic code as the construction of meanings. In addition to the genetic code, he considered other codes in living cells, such as signal transduction and splicing codes (Barbieri 2003). However, the theory of code biology is focused on individual coding processes and does not attempt to integrate all functions of organisms into a multi-level network of self-construction.
In this paper I use principles of constructivism to explain the emergence of multi-level semiotic networks in organisms. Multi-levelness appears essential to support the plasticity, robustness, and evolvability of living systems. The origin of life is described here as the emergence of simple self-constructing semiotic networks that progressively increased their complexity. Semiotic networks are based on sequential and recursive construction, where each step produces components that are needed for the following steps of construction. Construction is not limited to repair and reproduction of what already exists or is unambiguously encoded, but also includes production of new components and behaviors via learning and evolution. Because subagents are partially independent in their learning and evolution, the phenotype of organisms appears to be a mosaic of features developed by cooperating and/or conflicting subagents.
2. Signs from the Evolutionary Perspective
There is no consensus on the definition of sign in biosemiotics. Some scholars consider that Peirce’s definition of sign as a triadic relation between representamen, object, and interpretant is universal and applicable to all levels of semiosis from cellular processes to human cognition (Hoffmeyer and Emmeche 1991; Bruni 2008). Similarly, Hoffmeyer and Stjernfelt (2016) argued that biosemiosis at all levels is based on proto-propositions with a dual Subject-Predicate (S-P) structure. Others view molecular signaling, DNA copying, mRNA synthesis, and protein synthesis guided by mRNA as a more primitive kind of sign processes referred to as organic code (Barbieri 2003), vegetative semiosis (Kull 2009) or protosemiosis (Prodi 1988; Sharov and Vehkavaara 2015). The latter point of view is consistent with the general evolutionary principle that functions of organisms, including semiotic functions, evolved from simple to more complex and this change was not just quantitative but also qualitative. It also helps to explain the origin of life because simple signs are more likely to emerge in primordial living systems (Sharov 2009), whereas complex cognitive signs of Peirce’s type require at least minimal mental capacities that did not exist in primordial systems.
According to Sharov and Vehkavaara (Sharov and Vehkavaara 2015), molecular proto-signs are not associated with objects because they are processed by cellular subagents (e.g., ribosomes) that have no capacity to classify and track objects. Instead, proto-signs are linked to actions of agents either directly or via simple logical gates. It seems natural to associate a triplet of nucleotides in the mRNA with an amino acid as an object. However, a ribosome has no internal representation of an amino acid as object and it does not “know” that it makes proteins. Instead, a ribosome detects if a triplet of nucleotides in the mRNA matches to the anticodon sequence of the incoming tRNA molecule loaded with an amino acid and then makes a peptide bond. Humans (e.g., biologists) know the chemical structure of these components and understand the details of their interaction, but a ribosome simply gets a signal that indicates readiness for the reaction and then uses the catalyst tool to finish the action. In other words, proto-propositions with S-P structure do not exist in protosemiosis because primitive organisms and cellular subagents cannot perceive objects and their properties (i.e., subjects and predicates, or S-P). Instead these agents use proto-signs (e.g., signals) to initiate or modify their actions. Thus, I disagree with Hoffmeyer and Stjernfelt (2016) that proto-propositions with S-P structure are universal at all levels of semiosis.
According to the theory of Charles Peirce (Peirce 1976), semiotics is intrinsically linked with logic. Following this tradition, Hoffmeyer and Stjernfelt (2016) wrote that “even very simple sign processes always are truth related”. I agree with this statement if truth is understood as a pragmatic relation2, following William James (1954). Indeed, the correspondence between proto-signs and actions tends to be beneficial for the survival and reproduction of organisms. Here I use mathematical logic to explain the difference between protosemiosis and advanced sign processes (eusemiosis). In short, protosemiosis can be modeled with propositional logic, whereas eusemiosis requires predicate logic (also known as first-order and second-order logic). In propositional logic, propositions are atomic and do not describe any objects, similar to protosemiosis. Such unstructured propositions are rare in human language and can be exemplified by sentences “it’s dark” or “it’s raining”. These propositions should not be confused with “proto-propositions” with S-D structure, as defined by Hoffmeyer and Stjernfelt, which belong to predicate logic that describes objects, their properties, and relations. Predicate logic is substantially more complex than propositional logic and appears more relevant for human communication. Thus, I assume that primitive agents can handle only the most simple atomic propositions, whereas the use of predicates requires additional semiotic capacities such as recognition of objects and their properties, which presumably appeared later in evolution.
In relation to life, a sign is something that repeatedly and consistently regulates or guides the actions of organisms or their subagents (e.g., cells or molecular complexes) in a useful way. In this respect, signs are similar to tools or resources, which are also needed for activities of organisms and cells. But molecular tools and resources are not signs per se because they do not always regulate cellular functions (if sufficiently abundant). If some molecular function is halted or slowed down due to the lack of resources, this effect is forced (i.e., it is purely physical), and thus, cannot be viewed as sign-dependent. However, tools and resources may also serve as signs if they happen to modulate certain signaling pathways in addition to their main job as tools and resources. For example, the depletion of glucose in the environment is detected by bacterial cells and results in a sign-dependent activation of alternative metabolic pathways (Lodish et al. 2000). Signs are both material and ideal; materially they are represented by sign vehicles, and ideally – by relationships with agents (i.e., via the capacity of agents to produce, perceive, and interpret signs), which are reproducible through generations and are potentially immortal (Sharov 2016b).
As the number of proto-signs increased in evolution, they became connected via logical gates. However, these connections were still fixed genetically and could not be modified within the life span of an organism even if they failed to produce beneficial effects. To overcome this limitation, organisms developed epigenetic mechanisms to modify logical gates on demand. These mechanisms can support rewritable memory within cells and even adaptive learning (Sharov 2010). Eventually organisms developed complex sense organs and acquired a capacity to integrate incoming signals into meaningful categories representing real objects and situations (e.g., food items, partner agents, or enemies) and predict events using models. This capacity may have emerged in single-cell organisms but became fully developed in multicellular organisms with a nervous system. It marks an evolutionary transition from protosemiosis to eusemiosis (although protosemiosis still persists at the molecular level) where knowledge about objects becomes possible (Sharov 2016b). Following the terminology of Uexküll, the knowledge about internal parts and functions is the Innenwelt of an organism, whereas the knowledge about external objects and processes is the Umwelt (Uexküll 1982). Signs processed at the eusemiotic level are not necessarily followed by physical actions of organisms; but they may involve mental changes (e.g., accumulation of knowledge) and may affect future actions. This preparedness has been called a disposition to respond (Morris 1964).
3. Life Requires Multilevel Networks of Signs
Organisms use signs to establish relations between their functional components and the environment (both external and internal), and thus, signs are always connected into semiotic networks. The minimum network, known as a functional cycle, includes a receptor and effector (Uexküll 1982: 32); however this network is too small to support heredity, functional plasticity, robustness, and evolvability of signs. Heredity requires at least two levels of interacting components that have digital and analog features, respectively (Hoffmeyer and Emmeche 1991). The quantum nature of small molecules (e.g., nucleic bases) allows them to keep digital identity in a sequence of recursive construction, and therefore they are ideal as heritable signs at the lower level (Schrödinger 1940). In addition, whole organisms represent the higher-level and support self-organization of the analog type. Their complexity is above the quantum threshold where full identity and physical entailment is possible (Kauffman 2014). Nevertheless, whole organisms can reliably reproduce their phenotypes in a sequence of generations due to the meta-stability of developmental pathways and guidance from heritable molecules at the lower level, as follows from Waddington’s model of the epigenetic landscape (see section 1).
Both plasticity and robustness in organisms require multiple alternative signaling pathways to switch to in the case of malfunction, as well as additional compensatory mechanisms to ameliorate the negative effects of external and internal disturbances. Thus, these features cannot be implemented in very simple systems with just a few components. As a result, selection favored organisms with expanded semiotic networks that had more components and relations between them. These complex networks also increase the evolutionary potential of organisms because there are more network connections that can be rewired. However, it appears that the complexity of semiotic networks cannot increase without modularity, as explained below, and therefore, plasticity, robustness, and evolvability require multi-levelness.
Modules are discrete functional self-regulated units that accomplish some useful work (e.g., construction) and are protected from external disturbances via isolation and/or self-repair (Schlosser and Wagner 2004). Thus, modules can combine responsiveness to external signals with enhanced persistence and stable function. As a result, each module can evolve without affecting the function of other modules in the organism (Wagner 1996). In other words, the main advantage of modularity is that it adds freedom and flexibility to semiotic networks. The second advantage is that modules are reusable: (1) they can be recruited by different subsystems and/or (2) duplicated and modified for slightly different jobs. For example, DNA topoisomerase I is used to unwind double-stranded DNA for both transcription and replication, whereas topoisomerase II resolves DNA knots and protects telomeres. Higher-level modules include multiple interconnected genes that regulate developmental pathways, such as limb patterning and growth (this module is reused for each limb). Finally, the third advantage is that modules are adaptable and tend to provide efficient and simple interfaces for communication with higher-level systems. Thus, they can be characterized by the term simplexity (Berthoz 2012), which stands for “[…] the combination of simplicity and complexity within the context of a dynamic relationship between means and ends” (Compain 2003). On one hand, making a module (e.g., a ribosome) is a more complex task than direct construction of a single final product (protein), which means making simple things in a complex way. On the other hand, the module simplifies operations by providing a “user-friendly” interface with standard signaling functions. Thus, operating of a module is a simpler task than repeated direct construction of final products. As an example of an interface, let’s consider ribosomes, which are programmable constructors of proteins. A ribosome receives input in the form of a messenger RNA (mRNA). After binding to the mRNA, the ribosome matches triplets of nucleotides in the mRNA with a reverse-complementary triple of nucleotides in transport RNAs that carry specific individual amino acids used for protein synthesis. Besides appending an amino acids to the protein chain, ribosomes can process several additional signals: they terminate the protein synthesis after encountering a stop-codon (UAG, UAA, or UGA), and may initiate mRNA degradation if a stop-codon is found before the last exon junction. The latter mechanism is important for nonsense mediated decay of improperly synthesized mRNA molecules (Yamasaki et al. 2007). Normal mRNAs have no stop-codons before the last exon junction; but if a nucleotide was erroneously skipped or inserted during mRNA synthesis, then stop-codons may easily appear downstream of the error but before the last exon junction. This feature is utilized as a signal for mRNA destruction to prevent wasteful protein synthesis and potential toxic effects of erroneously synthesized proteins.
Organisms use multilevel networks to outsource routine tasks to their subagents, such as organs, cells, molecular complexes, or symbionts. Moreover, they can outsource adaptation by allowing subagents to solve functional problems on their own via learning and evolution (see section 6). Obviously, some kind of memory or heredity is needed for learning, and thus, not all subagents can learn or evolve. Mitochondria and chloroplasts are organelles within eukaryotic cells, which originated from symbiotic bacteria; they carry their own genome and therefore are capable of adaptive evolution that is partially independent from the evolution of their master organisms. Individual cells can learn and anticipate future events (Ginsburg and Jablonka 2009; Pershin et al. 2009). Simple models show that epigenetic mechanisms can support associative learning by cells (Sharov 2013: 353).
The integrity of networks, the degree of signaling plasticity, and the number of hierarchical levels increased in evolution as new forms of interaction emerged. Molecular networks in prokaryotes are simple and have limited flexibility. Genes involved in the same cellular function are physically integrated into one operon, and thus, they are regulated and transcribed as a group called “operon” (Lodish et al. 2000). Most genes in prokaryotes have one functional domain, which limits their functional repertoire. Bacteria have limited plasticity and adaptability because they lack rewritable epigenetic memory3. In eukaryotes, genes are regulated and transcribed individually, which considerably enriches the flexibility of gene networks. Additional cellular compartments (e.g., nucleus, cytoplasm, mitochondria, Golgi, endoplasmic reticulum, which are absent in bacteria) provide an opportunity to establish context-dependent interactions of signaling molecules, which are different in each compartment. Transport of molecules and organelles between cell compartments adds a new type of relation to signaling networks. Most eukaryotic genes combine multiple functional domains that allow their protein products to participate in complex cellular interactions. The nucleus represents a hub of signaling connections within a eukaryotic cell and can be viewed as a mini-brain. Thanks to the rewritable epigenetic memory, eukaryotic cells can adjust their functions according to the environment or cellular needs and even pass this acquired information through generations. The next step in the evolution of network complexity is the emergence of multicellular organisms, where each cell type and each organ has its unique network of signaling interactions. Multicellular organisms mastered the use of non-coding RNA (e.g., micro-RNA and lnc-RNA) for enriching the plasticity of regulatory networks. Finally, animals developed neural signaling which supports fast and versatile distant communications between cells and organs. The top level of interconnectedness is observed in the brain of animals, but our understanding of brain function is still very limited.
Let us summarize the advantages of multilevel organization of living systems. First, multi-level networks integrate organism functions at a wide range of spatial scales from molecules (~10−9 m) to large organisms such as whales (30 m). Life requires small molecules to support heredity because of their digital properties, whereas larger scales are needed for unique patterns of self-organization. And second, life requires plasticity, robustness, and evolvability, which are all supported by modularity. Modules represent intermediate levels within multilevel semiotic networks.
4. Origin of the First Networks of Signs
Because life and semiosis are generally viewed as coextensive (Anderson et al. 1984; Sharov 1992), the origin of signs should be associated with the origin of life4. Thus, we need to discuss how the first sign networks appeared in primordial living systems. Kauffman suggested that rich networks of interacting components existed from the very beginning of life (Kauffman 1986). In particular, he proposed that living systems originated from autocatalytic sets of molecules, where each kind of molecule (e.g., peptide, according to Kauffman) is synthesized with the help (i.e., catalysis) of some other kinds of molecules. Models show that such systems can indeed persist and propagate if supplied with necessary resources (e.g., amino acids). Catalysis within stable self-organizing systems is certainly a predecessor of a sign relation because catalysts regulate processes that contribute to the stability of the whole system, and therefore appear “useful” in relation to this system. It was shown experimentally that simple autocatalytic sets of replicating RNA molecules can persist in artificial conditions (Vaidya et al. 2012). However, such autocatalytic sets cannot persist in natural environments that provide neither a sufficient amount of resources such as nucleic bases or amino acids, nor enclosure to prevent the dissipation.
More realistic models of the origin of life5 include surface metabolism (Wächtershäuser 1988), and coenzyme world (Sharov 2009, 2016a). These models assume that primordial living systems started with a single function and added more components sequentially. For example, the coenzyme world model assumes that coenzyme-like molecules can establish their own autocatalysis by attachment to the surface of oil microspheres (i.e., hydrocarbons of abiotic origin) and changing surface properties via oxidation (Fig 1A). Changing surface properties (the first function) may benefit coenzyme-like molecules to multiply via autocatalysis mediated by modified oil microspheres, and then colonize other oil microspheres. In such a system, it can be said that coenzyme-like molecules are signs that encode surface properties of oil microspheres (Sharov 2009). This is a 2-level network that includes coenzyme-like molecules at the lower level, and whole microspheres at the upper level. This simple system can evolve via adding new kinds of coenzymes with novel functions (e.g., those that help to capture and store energy and other resources).
Fig. 1.

Model of the origin of life on oil microspheres. (A) Coenzyme-like molecules can attach to the oil microsphere via rare fatty acid molecules; after attachment they start oxidizing hydrocarbons to fatty acids, which in turn provide additional anchoring sites for other coenzyme-like molecules; accumulation of fatty acids increases the chance of a microsphere to split into smaller ones, and small microspheres can infect other oil microspheres (i.e., capture new oil resource). (B) Transition from surface metabolism on oil microspheres to cell-like systems with a bilayer membrane and internal metabolism.
The advantage of this model is that it can explain the origin of coenzymes, nucleic acids, template-based replication, cell membranes, and transition from external to internal metabolism as follows. Polymerization of coenzyme-like molecules may strengthen the surface of oil microspheres and provide a scaffold for making other molecules. At some point of subsequent evolution, we can expect the emergence of template-based synthesis of sign-carrying polymers, which corresponds to the beginning of the RNA-world primordial systems (Sharov 2009). The cell membrane may have appeared via engulfing water inside oil microspheres (Fig. 1B) (Sharov 2016a). Such “bubble microspheres” are easily generated by agitating an emulsion of liquid hydrocarbons in water but they are not stable. Thus, the outer membrane has to be strengthened to sustain mechanical disturbances, which requires the synthesis of glycerol-like molecules to make lipids and phospholipids. Emergence of a heritable metabolism for making glycerol might have been the major evolutionary achievement at that time6. In summary, there are realistic scenarios for the origin of first small networks of signs at the origin of life, and these networks included two levels: the level of functional molecules and the level of proto-organisms, such as oil microspheres with enhanced surface properties.
5. Time Scales and Levels of Construction
The notion of construction in relation to organisms implies that living processes (e.g., metabolism and development) have a certain similarity to human activities such as the construction of homes and machines. Indeed there are many common features between construction processes in organisms and in human life: (1) construction follows certain rules that were developed and tested in the past (e.g., blueprints are used by humans, genetic and epigenetic signs are used by organisms); (2) each action requires certain resources, tools, scaffolds, and subagents which have to be created, acquired, or recruited beforehand; (3) construction is adjusted to the environment or local context; (4) the product of construction is further modified to compensate for imbalances or mistakes and to improve its functions; and (5) the rules of construction are updated based on experience. However there are also some important differences. First, human rules of construction can be updated without delay, whereas the genome is not updated during the life span of organisms (although it can be re-interpreted). But the pool of genomes in a population changes every generation due to selective survival and reproduction. Second, organisms (except humans) are not capable of true engineering, which includes generating new rules of construction from scratch based on mathematical models. And third, humans are still not able to make self-constructing and self-repairing autonomous systems7.
The notion of construction is primarily associated with material objects, but it can be expanded into the ideal sphere when we talk about the construction of knowledge. Let’s clarify the meaning of the term construction when it is applied to signs and sign relations, which have both material and ideal aspects. First, signs are always represented by material sign vehicles; thus, agents have to physically make sign vehicles in order to communicate. But not all sign vehicles are constructed; some of them exist naturally (e.g., the sun and moon are used by organisms for navigation or coordinating physiological processes). Other sign vehicles are produced by organisms but not for communication purposes. For example, gypsy moth males fly towards tree trunks to find females but tree trunks were not made for the purpose of sending signals to gypsy moth males. In this case, gypsy moths reuse construction processes in trees for their own semiosis. The second meaning of the term construction as applied to signs is that organisms have to make all the material tools for executing the sign relation. In particular, organisms produce a set of tools during their development, which include (1) sensors or sense organs to detect or perceive signs, (2) information-processing organs such as signal-transduction pathways, nerves, and brains, and (3) effector organs that execute actions after the processing of signs. Finally, the third meaning of the term construction as applied to signs is the replication and/or modification of memory or heredity that supports the repeated production of sign vehicles and sign-processing tools within the life span of organisms and/or in subsequent generations. The hereditary mechanisms include replication of the genome, copying of epigenetic signs, and creative interpretation of hereditary signs such as compensation and coordination of various processes if they become unbalanced due to mutations, epigenetic modifications, or changes of the environment.
When living cells produce various subagents (e.g., ribosomes, DNA-polymerases, or chromatin-remodeling complexes), they construct or remodel a network of sign relations supported by these subagents. Indeed, subagents are sign-processing devices: ribosomes use mRNA as programs for protein synthesis; DNA-polymerases use parental DNA strand for template synthesis of the reverse-complementary DNA strand; and chromatin-remodeling complexes sense existing chromatin modifications and either extend or modify chromatin properties as guided by transcription factors and other signaling molecules such as non-coding RNA or insulators. Thus, construction of molecular signs and subagents is essential for preserving and modifying sign relations in living cells.
Construction can be studied at various time scales. At short times, we observe the replenishment of cell components, remodeling of cell structures, cell proliferation and differentiation. But it is more interesting to analyze construction processes at longer time scales during development and evolution. Multicellular organisms start their development from a fertilized egg, which is a single cell with a genome, epigenetic signs, and a minimal set of subagents to initiate the construction of the body. Each step of construction expands the semiotic capacities of the growing embryo. New receptors, effector organs, and signaling pathways make new sign relations that can be utilized in the next round of construction. The word “new” in this sentence refers to the ontogenetic novelty for a given organism rather than for a lineage, because these structures are made repeatedly in each generation. Obviously, the construction of these components is well tested in the ancestral generations.
The process of embryo development may include elements of learning at the level of individual cells, and this idea is supported by observations of learning-like behaviors in single-cell organisms (Hennessey 1979; Armus et al. 2006; Saigusa et al. 2008). Cells may actively search for potential differentiation paths based on their position in the embryo and interaction with other cells. Developing organs start functioning very early, and apparently they also learn how to function. Learning extends into the adult stage of organisms and it is most elaborate in adult animals with brains. The main advantage of learning as compared to innate regulation of development and behavior is in the increased semiotic freedom (Hoffmeyer 2010). In particular, organisms can try various algorithms of activity, select (i.e., memorize) the most productive one, and then reproduce it automatically in similar conditions. Learning always generates ontogenetically novel patterns of activity, but these patterns are not necessarily novel within the evolutionary lineage. In fact, most of the learning is reliably repeated in each generation, as supported by heritable capacities to learn (e.g., by sense organs, effector organs, and neural networks). However, individual learning may occasionally produce really novel behaviors that did not exist in previous generations.
Construction at the evolutionary time scale includes the emergence of phylogenetically-new signs, sign relations, and agents. By phylogenetic novelty I mean new features that have not been present in ancestral organisms. However, it appears that every phylogenetic novelty is constructed mostly with the help of old components, such as subagents, sign relations, tools, and resources. Every new protein is constructed by the same ribosomes and the same genetic code as any other protein. Moreover, almost every new gene appears to be a slightly modified copy of already existing genes. Duplication of genes occurs regularly either from errors during DNA replication or from the action of transposable elements or viruses that are often present in the genome.
Identical gene copies are usually not favored by selection because some functions of cells may be affected negatively by the double amount of gene products. Thus, new copies of genes persist only if they become sufficiently different from parental genes and support functions that are not adequately covered by parental genes. Considering that each gene is a part of a gene regulatory network, new gene copies survive only if they modify their relations within the network (Fig. 2). For example, a new gene may become activated in a different tissue or at a different phase of the cell cycle; or the encoded protein may start interacting with another kind of molecules.
Fig. 2.
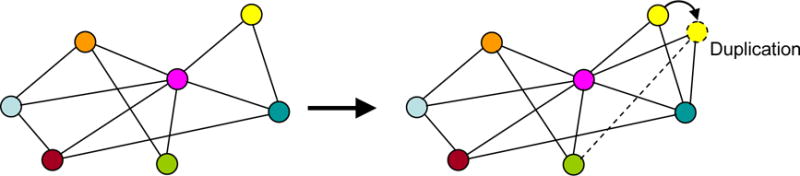
New nodes in a gene regulatory network are retained in evolution only if they modify their relations (dashed line) with other nodes.
Naturally, this prompts another question: how do genes modify their relations in a reasonably short time? Although we cannot get an answer for every gene, it appears that many genes have a hidden internal capacity to establish relations with new partners in the network, and therefore, modifications of gene regulatory networks are not that rare and belong to the category of adjacent possible (Kauffman 2014). Here are a few possible explanations of these capacities. First, molecular interactions are not 100% specific: receptors can be activated by several different ligands, and many signaling molecules may successfully bind several kinds of receptors. Second, organisms and cells often include paralogs of molecular signs and subagents that originated via earlier gene duplication events. Switching relations from one sign or agent to its paralog is presumably more likely to happen in evolution because of the structural and functional similarity of paralogs. Third, some relations within gene regulatory networks may have existed in the past and just need to be restored, which is an easier task that developing them anew. And fourth, due to the high redundancy of regulatory channels, each functional change can be achieved via thousands of potential mutations, and thus, evolution does not have to “wait” for a specific mutation to modify the gene regulatory network (Sharov 2014, 2016b).
This can be illustrated by the color change in the peppered moth Biston betularia in England, which is the best documented case of selection in natural populations (True 2003). Light-colored wings with dark speckles help peppered moths to hide from predators (birds) on the white bark of birches. When birch trunks turned black due to increased industrial pollution, a rapid spread of a dark-colored form of the moth was observed. Obviously, moths had a capacity to produce dark scales on the wings even before birches turned dark, but dark scales were restricted to small speckles. In particular, all biochemical pathways necessary for producing the dark pigment melanin, such genes as yellow and ebony (Wittkopp et al. 2002), were present beforehand. Thus, apparently, a small genetic change was sufficient to redirect melanin synthesis to the entire surface of the wings. Indeed, recent analysis did not reveal association of any known melanin-producing genes to the dark form of the peppered moth (van’t Hof and Saccheri 2010). Authors hypothesized that a high-level unknown developmental factor may regulate the spatial expression of one or more genes related to melanin production. Considering that the ability to change color is beneficial for many moth species, it is reasonable to assume that the evolutionary event with the peppered moth was well tested in the ancestral species.
A special case of evolutionary construction is cooperation between organisms or subagents that eventually may lead to a deep integration indicating the emergence of a new super-agency. In effect, this process adds a new hierarchical level of organization and was called metasystem transition by Valentin Turchin (1977). Examples of metasystem transitions include the emergence of multicellular organisms, multi-segment organisms (e.g., worms or insects), and colonies of social insects with centralized reproduction (Fig. 3). Integration of neurons into a network and finally into a brain is an example of metasystem transition below the organism level. The sequence of events that leads to a metasystem transition is the following: (1) duplication of components without full separation, (2) establishment of cooperation between components, (3) division of labor and specialization, and (4) establishment of central control over components (Turchin 1977). Central control targets all functions of components including reproduction and survival, it suppresses antagonistic relations and promotes cooperation and differentiation of components.
Fig. 3.
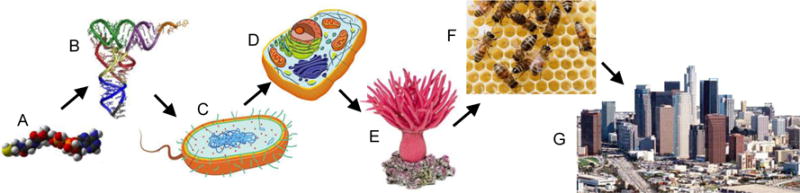
Major metasystem transitions in the evolution of life: A – coenzyme-like molecules; B – replicating polymers; C – prokaryotes; D – eukaryotic single-cell organisms; E – multicellular eukaryotes; F – social organisms; and G – civilization.
Turchin did not discuss symbiogenesis as a pathway to a new hierarchical level of systems, although symbiosis certainly satisfies the definition of metasystem transition. In the case of symbiosis (e.g., during the origin of eukaryotic cells or lichen), cooperating partners are different from the very beginning, and thus there is no need for specialization. Human civilization can be seen as the top level of multi-level integration that includes various organizations, businesses, agriculture, and animal farming (the two latter components are symbiotic).
6. Mosaic Identity in Evolving Multi-Level Agents
Multi-level organization brings enormous advantages to evolving living beings. In particular, it supports the division of labor between subagents and ensures the plasticity, robustness, and evolvability of sign regulatory networks (see section 4). But multi-levelness also brings a problem of coordination between dynamic subagents. Many subagents have enough autonomy for independent learning and evolution, and thus the phenotype of an organism combines a mosaic of features developed by its subagents. For example, individual genes have their own phylogenetic trees, which only approximately match the phylogeny of whole organisms (Puigbo et al. 2013). Horizontal gene transfer mediated by viral infection or symbiosis may result in a rapid change of phenotype in recipient species (Koonin and Galperin 2003). For example, the capacity for photosynthesis appeared in a mosaic pattern in unrelated lineages of bacteria, and it is supported by similar genes, indicating multiple events of horizontal gene transfer in the past (Villareal 2009: 131).
Eukaryotic organisms carry a multitude of various parasites and symbionts, such as transposable elements and viruses integrated in the genome, intracellular parasitic bacteria (e.g., Wolbachia), protozoan latent infections (e.g., Toxoplasma), and gut microbiota. Some types of cancer cells can get transmitted between dogs during copulation and thus behave as independent parasitic species (Murchison et al. 2014).
There is evidence that symbionts can switch between different host species (Bright and Bulgheresi 2010), which supports the notion of their independent evolution. The physiology of the human mind indicates that there is no central decision-making element in the brain; instead there is a “society of mind” composed of many subagents (possibly neurons) that come to “agreement” via a kind of voting system (Minsky 1986). Thus, the phenotype and behavior of organisms is a product of interactions between subagents integrated by a semiotic network.
Evolutionary (or learning) independence of multiple coexisting subagents often leads to internal conflicts, especially in cases when one subagent takes control over others. For example, viruses recruit host ribosomes to produce viral proteins and eventually may kill the cell. Some parasites and symbionts change the phenotype or behavior of the host organism for their own benefit. For example, mice infected with Toxoplasma gondii become attracted to cat’s urine (Ingram et al. 2013). This response is beneficial for the parasite because mice with the altered behavior have a higher chance to be eaten by cats that are definitive hosts of this parasite (i.e., suitable for sexual reproduction). Another example is the parasitic fly Apocephalus borealis which infects honeybees. Larvae of this parasite move to the brain of bees and reprogram it to unusual dispersal activities, which helps the parasite to spread around (Core et al. 2012).
An alternative strategy for parasites is not to harm their host organisms but to reproduce together with them in a latent phase. In this case, the parasite and host become integrated into a kind of semi-symbiotic system where subagents do not attempt to get full control over each other. Interestingly, latent virus can prevent their hosts from developing antiviral mechanisms by selective activation of two viral genes that encode a toxin protein and an antidote to this toxin, which are both synthesized by host ribosomes (Villareal 2009: 37). If a bacterial cell succeeds in removing or inactivating the virus, then the unstable antidote protein quickly disappears but stable toxin persists and kills the bacterial cell. In this case, the virus blocks a certain pathway of evolution in host cells. Mutual constraints on evolutionary and learning pathways between subagents are probably very common in semiotic networks. For example, there is evidence that the immune system selectively eliminates mutant cells that may cause cancer (Corthay 2014), and therefore any genetic changes towards malignancy are disrupted early.
Multi-agent semiotic networks have intrinsic uncertainty in their evolutionary future, which can be compared to quantum uncertainty. A bacterium with a latent virus infection has three potential outcomes: (1) it can recover by killing or inactivating the virus, (2) bacterial cells may die releasing viral particles, and (3) bacteria and virus may continue coexisting. In the latter case, the virus can bring certain advantages to the bacterium, such as immunity against other viruses (Villareal 2009). The existence of multiple evolutionary outcomes may support the balancing selection in many genes whose function depends on the outcome. For example, alleles with a strong anti-viral effect are beneficial for bacteria in scenario #1 but not in scenario #3. As a result, such alleles will persist at some intermediate frequency. In this way, multi-agent semiotic networks contribute to preserving genetic variability, which may appear useful during catastrophic environmental changes that require fast adaptations to new conditions.
Considering potential antagonism and selfish behavior of subagents, what are the requirements for the higher-level agency? Obviously, higher-level agents need sufficient power to channel up the changes of subagents into directions that are beneficial for the whole system. For example, individual genes may occasionally appear “selfish” because of their capacity to replicate and invade other genomes (Dawkins 1976). But cells have established tight constraints on the evolution of genes and do not allow them to evolve towards selfish behaviors. The major restrictive mechanism is the control of gene copy number: only one copy of a gene (or two copies in diploid cells) is transferred to each daughter cell during cell division. Restriction of selfish tendencies of subagents seems to be the major challenge in multi-level semiotic networks. But top-down control should not be too strict because subagents need freedom to solve their local problems via learning and evolution. Thus, higher-level agents need a balance between control and freedom, although we still don’t know the criteria for optimizing these strategies. This principle of combining control and freedom seems to be applicable not just to biology but also to cooperating groups of humans such as families or enterprises.
7. Conclusions
Constructivism is a valuable addition to biosemiotics because it emphasizes the activity of agents in self-construction, self-reproduction, and development of sign relations. New sign relations emerge as modifications of older sign relations and employ already available tools, resources, and subagents. New levels of semiosis emerge via functional integration of interacting agents (meta-system transition). Multilevel semiotic networks are needed to support the plasticity, robustness, and evolvability of organisms. They coordinate the appearance of features developed via learning and evolution of cooperating and/or conflicting subagents. Principles of multilevel semiosis may appear useful not just in biology but also for managing cooperating activities of humans.
Acknowledgments
This paper was supported entirely by the Intramural Research Program of the National Institute on Aging (NIA/NIH), project Z01 AG000656-13. The content of the paper is not endorsed by the funding organization.
Footnotes
It is closely related to constructionism (Noss and Clayson 2015) and evolutionary epistemology (Riegler 2006).
However, the statement would be wrong if truth is interpreted in metaphysical terms, because meaningful sign processes are possible even without true understanding of states-of-affairs (e.g., cooking recipes do not require any knowledge of thermodynamics).
Bacteria have no real histones. However, they change DNA methylation to control their virulence and the cell cycle.
Note, that relational semiotics assumes the existence of signs even in the physical world devoid of life (Deely 1992).
I do not discuss scenarios based on self-replicating nucleic acids, such as RNA-world (Gilbert 1986), because naturally-synthesized nucleotides are too rare and unstable to support self-replication (Sharov and Gordon 2013).
Recent discovery of alcohol and sugar on the comet Lovejoy (Biver et al. 2015) is interesting, but it does not prove that primordial organisms used carbohydrates of abiotic origin as resources. It is very unlikely that life originated on a small comet. And if a comet lands on a planet, organic chemicals would immediately degrade or become diluted.
Here I do not consider products of synthetic biology because all artificial living systems were not engineered from scratch but copied from natural organisms.
References
- Anderson M, Deely J, Krampen M, Ransdell J, Sebeok TA, Uexküll Tv. A semiotic perspective on the sciences: Steps toward a new paradigm. Semiotica. 1984;52(1/2):7–47. [Google Scholar]
- Armus HL, Montgomery AR, Gurney RL. Discrimination learning and extinction in paramecia (P. caudatum) Psychol Rep. 2006;98(3):705–711. doi: 10.2466/pr0.98.3.705-711. [DOI] [PubMed] [Google Scholar]
- Baldwin MJ. A new factor in evolution. American Naturalist. 1896;30:441–451. [Google Scholar]
- Barbieri M. The organic codes: An introduction to semantic biology. Cambridge, New York: Cambridge University Press; 2003. [Google Scholar]
- Barbieri M. Biosemiotics: a new understanding of life. Die Naturwissenschaften. 2008;95(7):577–599. doi: 10.1007/s00114-008-0368-x. [DOI] [PubMed] [Google Scholar]
- Berthoz A. In: Simplexity: Simplifying priciples for a complex world. Weiss G, translator. New Haven, London: Yale University Press; 2012. [Google Scholar]
- Bickhard MH. Functional scaffolding and self-scaffolding. New Ideas in Psychology. 2005;23:166–173. [Google Scholar]
- Biver N, Bockelée-Morvan D, Moreno R, Crovisier J, Colom P, Lis DC, et al. Ethyl alcohol and sugar in comet C/2014 Q2 (Lovejoy) Science Advances. 2015;1(9):e1500863. doi: 10.1126/sciadv.1500863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8(3):218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni LE. Cellular semiotics and signal transduction. In: Barbieri M, editor. Introduction to biosemiotics. The new biological synthesis. Dordrecht: Springer; 2008. pp. 365–407. [Google Scholar]
- Cariani P. Towards an evolutionary semiotics: The emergence of new sign-functions in organisms and devices. In: Van de Vijver SSG, Delpos M, editors. Evolutionary Systems. Dordrecht, Holland: Kluwer; 1998. pp. 359–377. [Google Scholar]
- Compain P. Le pari de la simplexité. Le simple et le complexe en synthèse organique. L’Actualité Chimique. 2003;(4–5):129–134. [Google Scholar]
- Core A, Runckel C, Ivers J, Quock C, Siapno T, Denault S, et al. A new threat to honey bees, the parasitic phorid fly Apocephalus borealis. PLoS One. 2012;7(1):e29639. doi: 10.1371/journal.pone.0029639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthay A. Does the immune system naturally protect against cancer? Front Immunol. 2014;5:197. doi: 10.3389/fimmu.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R. The selfish gene. Oxford: Oxford University Press; 1976. [Google Scholar]
- Deely J. Semiotics and biosemiotics: Are sign-science and life-science coextensive? In: Sebeok TA, Umiker-Sebeok J, editors. Biosemiotics. The semiotic web 1991. New York: Mouton de Gruyter; 1992. pp. 45–75. [Google Scholar]
- Dewey J. The development of American pragmatism. In: Hickman TMALA, editor. The essential Dewey. Vol. 1. Bloomengton: Indiana Univ. Press; 1998. [Google Scholar]
- Gardner M. Mathematical games – The fantastic combinations of John Conway’s new solitaire game “life”. Scientific American. 1970;223:120–123. [Google Scholar]
- Gilbert W. Origin of life: the RNA world. Nature. 1986;319(6055):618. [Google Scholar]
- Ginsburg S, Jablonka E. Epigenetic learning in non-neural organisms. J Biosci. 2009;34(4):633–646. doi: 10.1007/s12038-009-0081-8. [DOI] [PubMed] [Google Scholar]
- Hennessey T. Classical conditioning in paramecia. Anim Learn Behav. 1979;7:419–423. [Google Scholar]
- Hoffmeyer J. Signs of meaning in the universe. Bloomington, IN: Indiana University Press; 1996. [Google Scholar]
- Hoffmeyer J. Biosemiotics: An examination into the signs of life and the life of signs. Scranton, PA: University of Scranton Press; 2008. [Google Scholar]
- Hoffmeyer J. Semiotics of nature. In: Cobley P, editor. The Routledge companion to semiotics. London: Routledge; 2010. pp. 29–42. [Google Scholar]
- Hoffmeyer J, Emmeche C. Code-duality and the semiotics of nature. In: Anderson M, Merrell F, editors. On semiotic modeling. Berlin, New York: Mouton de Gruyter; 1991. pp. 117–166. [Google Scholar]
- Hoffmeyer J, Stjernfelt F. The Great Chain of Semiosis. Investigating the Steps in the Evolution of Semiotic Competence,” Jesper Hoffmeyer and Frederik Stjernfelt. Biosemiotics. 2016;9(1) doi: 10.1007/s12304-015-9247-y. [DOI] [Google Scholar]
- Ingram WM, Goodrich LM, Robey EA, Eisen MB. Mice infected with low-virulence strains of Toxoplasma gondii lose their innate aversion to cat urine, even after extensive parasite clearance. PLoS One. 2013;8(9):e75246. doi: 10.1371/journal.pone.0075246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James W. Essays in pragmatism. New York: Hafner Pub. Co; 1954. [Google Scholar]
- Kauffman SA. Autocatalytic sets of proteins. J Theor Biol. 1986;119(1):1–24. doi: 10.1016/s0022-5193(86)80047-9. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. Prolegomenon to patterns in evolution. Biosystems. 2014;123:3–8. doi: 10.1016/j.biosystems.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Klir GJ. Facets of systems science. Vol. 7. New York, London: Springer; 1991. (IFSR International Series on Systems Science and Engineering). [Google Scholar]
- Koonin EV, Galperin MY. Sequence – evolution – function: Computational approaches in comparative genomics. Boston: Kluwer Academic; 2003. [PubMed] [Google Scholar]
- Kull K. Vegetative, animal, and cultural semiosis: the semiotic threshold zones. Cognitive Semiotics. 2009;4:8–27. [Google Scholar]
- Langton CG. Self-reproduction in cellular automata. Physica D. 1984;10:135–144. [Google Scholar]
- Liu CH, Matthews Vygotsky’s philosophy: Constructivism and its criticisms examined. International Education Journal. 2005;6(3):386–399. [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Darnell J. Molecular cell biology. 4th. New York: W. H. Freeman and Co; 2000. [Google Scholar]
- Maturana H, Varela F. Autopoiesis and cognition: The realization of the living. Vol. 42. Dordecht: D. Reidel Publishing Co; 1980. (Boston Studies in the Philosophy of Science). [Google Scholar]
- Minsky M. The society of mind. New York: Simon and Schuster; 1986. [Google Scholar]
- Morris CW. Signification and significance: A study of the relations of signs and values. Cambridge, MA: MIT Press; 1964. [Google Scholar]
- Murchison EP, Wedge DC, Alexandrov LB, Fu B, Martincorena I, Ning Z, et al. Transmissible dog cancer genome reveals the origin and history of an ancient cell lineage. Science. 2014;343(6169):437–440. doi: 10.1126/science.1247167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noss R, Clayson J. Reconstructing constructionism. Constructivist Foundations. 2015;10(3):285–288. [Google Scholar]
- Peirce CS. The new elements of mathematics. Atlantic Highlands, N.J.: Humanities Press; 1976. [Google Scholar]
- Pershin YV, La Fontaine S, Di Ventra M. Memristive model of amoeba learning. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80(2 Pt 1):021926. doi: 10.1103/PhysRevE.80.021926. [DOI] [PubMed] [Google Scholar]
- Piaget J, Garcia R. Psychogenesis and the history of science. New York: Columbia University Press; 1989. [Google Scholar]
- Prodi G. Material bases of signification. Semiotica. 1988;69(3/4):191–241. [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV. Seeing the Tree of Life behind the phylogenetic forest. BMC Biol. 2013;11:46. doi: 10.1186/1741-7007-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler A. Like cats and dogs: Radical constructivism and evolutionary epistemology. In: Gontier JPvBN, Aerts D., editors. Evolutionary epistemology, language and culture. Dordrecht, Netherlands: Springer; 2006. pp. 47–65. [Google Scholar]
- Rosen R. Life itself: a comprehensive inquiry into the nature, origin, and fabrication of life. New York: Columbia University Press; 1991. [Google Scholar]
- Saigusa T, Tero A, Nakagaki T, Kuramoto Y. Amoebae anticipate periodic events. Phys Rev Lett. 2008;100(1):018101. doi: 10.1103/PhysRevLett.100.018101. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Wagner GP. Introduction: The modularity concept in developmental and evolutionary biology. In: Schlosser G, Wagner GP, editors. Modularity in development and evolution. Chicago, London: University of Chicago Press; 2004. pp. 1–11. [Google Scholar]
- Schrödinger E. What is life? The physical aspect of the living cell. Cambridge: Cambridge University Press; 1940. [Google Scholar]
- Sharov AA. Biosemiotics: functional-evolutionary approach to the problem of the sense of information. In: Sebeok TA, Umiker-Sebeok J, editors. Biosemiotics. The semiotic web 1991. New York: Mouton de Gruyter; 1992. pp. 345–373. [Google Scholar]
- Sharov AA. Umwelt theory and pragmatism. Semiotica. 2001;134:211–228. [Google Scholar]
- Sharov AA. Coenzyme autocatalytic network on the surface of oil microspheres as a model for the origin of life. [Article] International Journal of Molecular Sciences. 2009;10(4):1838–1852. doi: 10.3390/ijms10041838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA. Functional information: Towards synthesis of biosemiotics and cybernetics. Entropy. 2010;12(5):1050–1070. doi: 10.3390/e12051050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA. Minimal mind. In: Swan L, editor. Origins of mind. Dordrecht: Springer; 2013. pp. 343–360. [Google Scholar]
- Sharov AA. Evolutionary constraints or opportunities? Biosystems. 2014;123:9–18. doi: 10.1016/j.biosystems.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Sharov AA. Coenzyme world model of the origin of life. Biosystems. 2016a;144:8–17. doi: 10.1016/j.biosystems.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA. Evolution of natural agents: Preservation, advance, and emergence of functional information. Biosemiotics. 2016b;8 doi: 10.1007/s12304-015-9250-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Gordon R. Life before earth. 2013 http://arxiv.org/ftp/arxiv/papers/1304/1304.3381.pdf2013.
- Sharov AA, Vehkavaara T. Protosemiosis: agency with reduced representation capacity. Biosemiotics. 2015;8(1):103–123. doi: 10.1007/s12304-014-9219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias S, Duffy TM. Constructivist instruction: Success or failure? New York: Taylor & Francis; 2009. [Google Scholar]
- True JR. Insect melanism: the molecules matter. Trends in Ecology and Evolution. 2003;18(12):640–647. [Google Scholar]
- Turchin VF. The phenomenon of science. New York: Columbia University Press; 1977. [Google Scholar]
- Uexküll Jv. The theory of meaning. Semiotica. 1982;42(1):25–82. [Google Scholar]
- van’t Hof AE, Saccheri IJ. Industrial melanism in the peppered moth is not associated with genetic variation in canonical melanisation gene candidates. PLoS One. 2010;5(5):e10889. doi: 10.1371/journal.pone.0010889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal LP. Origin of group identity Viruses, addiction and cooperation. New York: Springer; 2009. [Google Scholar]
- von Neumann J. Theory of self-reproducing automata. Urbana: University of Illinois Press; 1966. [Google Scholar]
- Wächtershäuser G. Before enzymes and templates: theory of surface metabolism. Microbiol Rev. 1988;52(4):452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Towards a theoretical biology. Nature. 1968;218(5141):525–527. doi: 10.1038/218525a0. [DOI] [PubMed] [Google Scholar]
- Wagner GP. Homologues, Natural Kinds and the Evolution of Modularity. American Zoologist. 1996;36:36–43. [Google Scholar]
- Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002;129(8):1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Stoecklin G, Kedersha N, Simarro M, Anderson P. T-cell intracellular antigen-1 (TIA-1)-induced translational silencing promotes the decay of selected mRNAs. J Biol Chem. 2007;282(41):30070–30077. doi: 10.1074/jbc.M706273200. [DOI] [PubMed] [Google Scholar]


