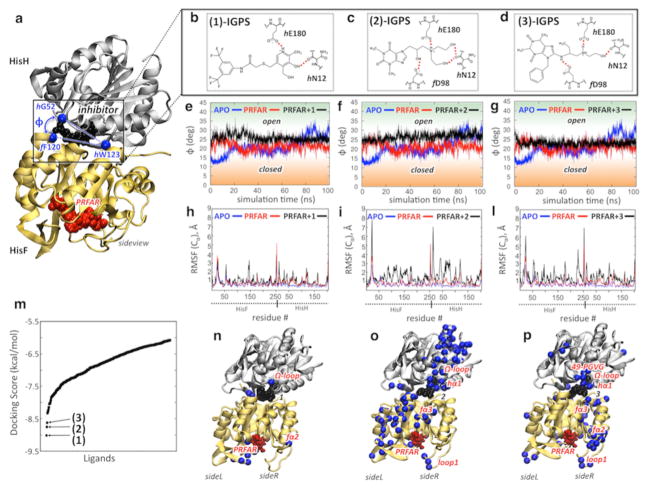
Figure 3.
Docking of allosteric inhibitors 1–3 and their effects on IGPS motions. (a) Inhibitors (black) dock at the HisF–HisH interface, affecting the motion of HisH relative to HisF (breathing motion). The breathing motion is measured by the angle ϕ defined by the α-carbon atoms (Cα) of f F120, hW123, and hG52. (b–d) Docking poses of allosteric inhibitors 1–3. (e–g) Evolution of ϕ during the MD simulation of the apo complex (blue), the binary PRFAR-bound complex (red), and the ternary complex with inhibitors 1–3 (black). (h, i, and l) The average root-mean-square fluctuations (RMSF) of Cα atoms, relative to average configurations, are reported for the apo complex (blue), the binary PRFAR-bound complex (red), and the ternary complex with inhibitors 1–3 (black). (m) Inhibitors 1–3 were selected according to the docking scores of the top 400 molecules screened by Glide from the 83766 heterocyclic small molecules of the Maybridge library. (n–p) The three inhibitors affect IGPS motions differently. The Cα atoms of the amino acid residues showing the highest differences in the RMSF average values (ΔRMSF > 1.1 Å) between the ternary and binary complexes (i.e., the RMSF of those residues that have been largely affected by the presence of potential inhibitors) are mapped onto the structures of the ternary IGPS complexes.