Abstract
Puccinia horiana Hennings, the causal agent of chrysanthemum white rust, is a worldwide quarantine organism and one of the most important fungal pathogens of Chrysanthemum × morifolium cultivars, which are used for cut flowers and as potted plants in commercial production regions of the world. It was previously reported to be controlled by Lecanicillium lecanii, Cladosporium sphaerospermum, C. uredinicola and Aphanocladium album, due to their antagonistic and hyperparasitic effects. We report novel antagonist species on Puccinia horiana. Fungi isolated from rust pustules in a commercial greenhouse from Villa Guerrero, México, were identified as Cladosporium cladosporioides and Cladosporium pseudocladosporioides based upon molecular analysis and morphological characters. The antagonism of C. cladosporioides and C. pseudocladosporioides on chrysanthemum white rust was studied using light and electron microscopy in vitro at the host/parasite interface. Cladosporium cladosporioides and C. pseudocladosporioides grew towards the white rust teliospores and colonized the sporogenous cells, but no direct penetration of teliospores was observed; however, the structure and cytoplasm of teliospores were altered. The two Cladosporium spp. were able to grow on media containing laminarin, but not when chitin was used as the sole carbon source; these results suggest that they are able to produce glucanases. Results from the study indicate that both Cladosporium species had potential as biological control agents of chrysanthemum white rust.
Introduction
Chrysanthemum white rust (CWR), caused by Puccinia horiana Hennings is one of the most destructive diseases of Chrysanthemum × morifolium worldwide [1,2,3]. Due to its economic importance to floriculture, it is classed as a quarantine disease by the European and Mediterranean Plant Protection Organization (EPPO), Inter-African Phytosanitary Council (IAPSC), Andean Community (CAN) and North American Plant Protection Organization (NAPPO), and is also listed as a regulated pest by the International Plant Protection Convention (IPPC). Currently it is present in most chrysanthemum producing areas throughout the world and can cause important losses [3,4,5].
Puccinia horiana is an obligate biotrophic fungus and microcyclic rust, affecting ten different Chrysanthemum species [6,7,8,9]. It produces teliospores, which germinate without a period of dormancy and release basidiospores as the infective propagules. The basidiospores are easily spread by wind and can infect neighboring plants in conditions of high humidity and cool temperatures [10, 11]. On the lower surface of the leaves, these teliospores form yellowish to pinkish immature pustules that become the characteristic white color once they mature [6,4,10].
When disease symptoms appear, they often do so suddenly and on a large scale in greenhouses, leaving little or no time for the farmers to take control actions. This leads to an ecologically and economically suboptimal timetable for the preventive application of fungicides [1,4,12]. Reports of fungicide-resistant strains show that preventive application with some fungicides is not effective [13,14], and this has prompted a search for alternative management solutions such as biological control [4,15,16,17].
Cladosporium Link is one of the most common genera of fungi occurring on various substrates and includes species with diverse lifestyles. Nowadays, after several taxonomic revisions, most saprobe lifestyles are included in the genus [18]. Besides saprophytic behavior, antagonism to pathogenic fungal species has been described. Some of the most common examples come from the relationship between Cladosporium spp. and rust pathogens [19,20], such as: C. uredinicola parasitizing Puccinia violae, P. puta and Cronartium fusiforme [21,22,23]; C. aecidicola on Melampsora medusae [24]; C. tenuissimum parasitizing Uromyces appendiculatus, Cronartium flaccidum and Peridermium pini [25,26,27]; C. gallicola on Endocronartium harknessii [28]; and C. cladosporioides parasitizing Venturia inequalis and Puccinia striiformis f.sp. tritici [29,30]. On Puccinia horiana only C. uredinicola, C. sphaerospermum and Cladosporium sp. have been previously reported [15, 17].
In the present study, we found novel species potentially antagonistic towards P. horiana. Based on morphological characteristics and molecular data, two species not previously reported on P. horiana were identified. Additionally, we investigated in detail the in vitro interaction between the two antagonists and CWR through light and scanning electron microscopy. Our results indicate that the isolates identified had potential as biological control agents of chrysanthemum white rust.
Materials and methods
Isolation, purification and morphological determination
Leaves from 30 different 30-day-old chrysanthemum plants with pustules of P. horiana, extensively colonized by a gray fungus, were collected from a commercial greenhouse with permission and collaboration of the owner in Villa Guerrero, Estado de México, México, in January 2014. Specific permission was not required, based on the epidemiological status of the chrysanthemum white rust in México. Also, this study did not involve endangered or protected species.
Gray dusty mycelia were removed from P. horiana teliospores with a sterilized needle and transferred to synthetic PDA medium (Bioxon, Mexico). After incubation at 24°C for 5 days, a spore suspension was prepared with sterilized water and transferred to water-agar medium. After incubation for 48h at 24°C, single-spore mycelia were picked off and transferred to PDA medium to obtain pure cultures.
Morphological characteristics were determined following the standardized methodology of Schubert et al. [31] for identification of the Cladosporium species. Colonies grown, for 5 days at 24°C in the dark, on synthetic-nutrient-limited media (SNA) plates, were used for morphological and morphometric observations of conidia, ramoconidia and conidiophores, using a photomicroscope Provis AX70 (Olympus, USA). Colony characteristics were determined after growing on PDA (Synthetic Potato-Dextrose-Agar; Bioxon, Mexico), MEA (Malt-Extract Agar; Bioxon, Mexico) and OA (Oat Agar), for 14 days at 24°C in the dark.
Molecular determination
DNA extraction and PCR amplification
DNA was extracted from mycelium and spores taken from PDA cultures using the method described by Falcon and Valera [32]. DNA concentration was determined using a NanoDrop N100 spectrophotometer. For PCR amplification, the stock solution was diluted to 90 ng/mL. Partial gene sequences were amplified as described by Bensh et al. [18] for internal transcribed spacers (ITS), actin (ACT) and translation elongation factor (EF-1α), using ITS1 (5´-TCCGTAGGTGAACCTGCGG-3´) and ITS4 (5´-GCTGCGTTCTTCATCGATGC-3´) from White et al. [33], ACT-512F (5´-ATGTGCAAGGCCGGTTTCGC-3´) and ACT-83R (5´-TACGAGTCCTTCTGGCCAT-3´), EF1-728F (5´-CATCGAGAAGTTCGAGAAGG-3´) and EF1-986R (5´-TACTTGAAGGAACCCTTACC-3´) from Carbone and Cohn [34]. The primers were synthesized by Instituto de Biotecnología, UNAM (Cuernavaca, México). Thermal cycle conditions and PCR mixtures for PCR amplification were those reported by Bensh et al. [18], using thermal cycler TC3000 (Techne, USA) and Taq Polimerase (Biotechmol, Mexico). Five mL of the PCR product was electrophoresed on a 1.5% agarose gel in 1% TBE buffer (0.089 M Tris-borate, 0.089 M boric acid and 0.002 M EDTA) for 45 min at 90 V and stained with ethidium bromide. Bands were detected under UV light in a Gel Documentation and Image Analysis System Geldoc 2000 (BioRad, USA). The PCR products were purified by Wizard SV gel and a PCR clean-up system Kit (Promega, USA), and sequenced at Instituto de Biotecnología, UNAM (Cuernavaca, México).
Sequence analysis
The three gene sequences from each antagonist, Cladosporium cladosporioides and C. pseudocladosporioides, were aligned with the sequences available in GenBank (NCBI, USA). Our sequences were manually edited by CLCbio (Qiagen, USA). Sequence data obtained from Bensh et al. [18,35] were used as reference data for the alignments (Table 1). Multiple alignments were performed by ClustalW software and best nucleotide model determinate by jModelTest v. 2.1.7 [36] using BIC criteria for each locus and then incorporating it in the analysis. A Bayesian phylogenetic inference tree was generated based on data from each partition sequence of the three genes on BEAST v.1.8.1 [37] and Markov Chain Monte Carlo analysis, from four chains started from random tree topology and taken over 80 000 000 generations. Three independent runs were combined by LogCombiner v.1.8.1. Trees were saved each 1 000 generations, resulting in 80 001 saved trees. Using Tracer v.1.6, burn-in was set at 15 000 000 generations, after which the likelihood values were stationary. The coalescent algorithm with GTR+G+I substitution model and a lognormal uncorrelated relaxed clock was selected for the data. Maximum clade credibility tree was visualized by Fig Tree v. 1.4.2. For the stability and robustness of each species, Neighboring-Joining analysis was performed for each data partition, using MEGA 6.0 [38] and 1000 replications using bootstrap. The ITS region has limited resolution for species in Cladosporium, therefore results for the ACT and EF-1α regions were used for comparison of clade stability (S1 Fig).
Table 1. Cladosporium isolates included in the sequence analysis.
| Species | Accession number | GenBank numbers (ITS, EF-1α, ACT) | Substrate | Country | Reference | ||
|---|---|---|---|---|---|---|---|
| C. cladosporioides | DETSC1A | KT877404 | KT887880 | KT721703 | Puccinia horiana | Mexico | This work |
| DETSC1B | KT877405 | KT887881 | KT721704 | Puccinia horiana | Mexico | This work | |
| CBS113738 | HM148004 | HM148245 | HM148491 | Grape blossom | USA | [35] | |
| CBS143.35 | HM148011 | HM148252 | HM148498 | Pisum sativum | South Africa | [35] | |
| CBS674.82; CBS 320.87 |
HM148014 | HM148255 | HM148501 | Gossypium seeds | Israel | [35] | |
| CBS11398 | HM148024 | HM148265 | HM148511 | Phragmidium griseum on Rubus crataegifolius | South Korea | [35] | |
| CPC12762 | HM148030 | HM148271 | HM148517 | Spinacia oleracea seeds | USA | [35] | |
| CPC12852 | HM148033 | HM148274 | HM148520 | Eucalyptus sp. | Australia | [35] | |
| CPC14705 | HM148050 | HM148291 | HM148537 | Phyllactinia sp. Chaesmothecia on Fraxinus rhynchophylla | South Korea | [35] | |
| CPC13220 | HM148054 | HM148296 | HM148541 | Lychen on Acer platanoides | Germany | [35] | |
| CPC14238 | HM148055 | HM148297 | HM148542 | Sambucus nigra fruits | Netherlands | [35] | |
| CBS306.84 | HM148057 | HM148299 | HM148544 | Puccinia allii | UK | [35] | |
| CPC13867 | HM148059 | HM148301 | HM148546 | Leptosphaeria sp. | South Africa | [35] | |
| CBS113746 | HM148061 | HM148303 | HM148548 | Cherry fruits | USA | [35] | |
| CPC13362 | HM148063 | HM148305 | HM148550 | Paeonia obovata | Germany | [35] | |
| CBS1123881 | HM148003 | HM148244 | HM148490 | Indoor air | Germany | [18,35] | |
| C. pseudocladosporioides | DETSC03 | KT877407 | KT887883 | KT887879 | Puccinia horiana | Mexico | This work |
| CBS117134 | HM148156 | HM148400 | HM148645 | Cloud | - | [35] | |
| CBS117153 | HM148157 | HM148401 | HM148646 | Paeonia sp. leaves | Germany | [35] | |
| CBS 1259932 | HM148158 | HM148402 | HM148647 | Outdoor air | Netherlands | [18,35] | |
| CBS 176.82 | HM148162 | HM148406 | HM148651 | Pteridium aquilinum | Romania | [35] | |
| CBS 574.78A | HM148163 | HM148407 | HM148652 | Melampsoporidium betulae | Russia | [35] | |
| CBS 574.78B | HM148164 | HM148408 | HM148653 | Melampsoporidium betulae | Russia | [35] | |
| CPC 11392 | HM148166 | HM148410 | HM148655 | Chrysanthemum coronarium var. spatiosum | South Korea | [35] | |
| CPC 11841 | HM148168 | HM148412 | HM148657 | Phalaris aquatica leaves | New Zeland | [35] | |
| CPC 12850 | HM148169 | HM148413 | HM148658 | Rotten wood | USA | [35] | |
| CPC 13488 | HM148171 | HM148415 | HM148660 | Vernonia sp. | Brazil | [35] | |
| CPC 14295 | HM148188 | HM148432 | HM148677 | Soil | Chile | [35] | |
| CPC 14357 | HM148189 | HM148433 | HM148678 | Food, Coffee leaves | Uganda | [35] | |
| CPC 14992 | HM148192 | HM148436 | HM148681 | Eucalyptus sp. | Indonesia | [35] | |
| CPC 13992 | HM148174 | HM148418 | HM148663 | Coffee tree | USA | [35] | |
| C. delicatulum | DETSC02 | KT877406 | KT887882 | KT887878 | Puccinia horiana | Mexico | This work |
| CBS 126344; CPC 113893 |
HM148081 | HM148325 | HM148570 | Tilia cordata | Germany | [18,35] | |
| C. sphaerospermum | CBS193.54 | DQ780343 | EU570261 | EU570269 | - | - | [39] |
| C. herbarum | CBS 121621; CPC 12177 |
EF679363 | EF679440 | EF679516 | - | - | [31] |
| C. tenuissimum | CBS125995; CPC14253 |
HM148197 | HM148442 | HM148687 | - | - | [18] |
| Cercospora beticola | CBS116456 | AY840527 | AY840494 | AY840458 | - | - | [18,35] |
ACT: partial actin gene, EFα1: partial translation elongation 1-α gene, ITS: internal transcribed spacer with 5.8 rRNA gene.
1 Ex-type from neotype;
2 Ex-type from holotype;
3 Reference strain
Antagonism assay
For the antagonism test, leaves infected (severity up 30%) and non-infected by chrysanthemum white rust, were collected from 20 different 30-day-old Chrysanthemum × morifolium cv. Polaris plants in a commercial greenhouse at Texcoco, México. The leaves were disinfested by immersion in sodium hypochlorite 3% for 3 min and then triple washed with sterilized water. There were five treatments: 1) antagonist 1 vs P. horiana (conidia were applied on pustules); 2) antagonist 2 vs P. horiana; 3) antagonist 1, conidia applied on healthy chrysanthemum leaves; 4) antagonist 2, conidia applied on healthy chrysanthemum leaves; and 5) control, P. horiana infected leaves treated with sterile water. Ten leaves per treatment were put into a humid chamber, each leaf representinged a repetition. A spore suspension (2×105 conidia mL-1) of each antagonist was sprayed onto pustules on diseased leaves and onto healthy chrysanthemum leaves. All treatments were incubated at 24°C and 12 h light/dark. When signs of antagonists appeared, a sample of the fungus was cultured on synthetic PDA (Bioxon, Mexico) to confirm that it was the fungus originally inoculated.
After 96 h of incubation, antagonism percentages, measured as the proportion of pustules of P. horiana colonized by C. cladosporioides and C. pseudocladosporioides, were recorded. Differences in percentage were statistically tested by one-way ANOVA. To meet ANOVA assumptions, normal distribution was assessed by a Shapiro-Wilk test [P>0.05] and homogeneity of variance was evaluated by Levene’s test [P>0.05]. The differences among treatments were tested by post hoc Ryan-Einot-Gabriel-Welch based on an F test (REGW-F; P = 0.05). All statistical analyses were carried out using SPSS Statistics 21.0.
Microscope observations
Leaves collected from Chrysanthemum × morifolium, infected and non-infected with chrysanthemum white rust, were treated under the same conditions and subjected to the same treatments as in the antagonism assay described above. After 36 h of incubation, 0.5 × 0.5 mm samples of the leaves were fixed in glutaraldehyde/paraformaldehyde 3:1 (in 0.2 M phosphate buffer, pH 6.8) overnight. Fixed leaf samples were washed four times with phosphate buffer for 15 min each, dehydrated through an ethanol series (30–100%, 1 h each) and infiltrated with LR-White [40]. Sections of 1μm thickness were cut on an ultramicrotome Reichert Jung Ultra E (USA) and stained with toluidine blue 1%. For SEM, pustules were dehydrated in a CO2 vacuum, mounted on carbon tape and coated with gold. The samples were observed on a JEOL JSM6360LV low vacuum SEM (JEOL, USA).
Glucanase and chitinase production
Glucanase production of both isolates was evaluated on growth medium consisting of an agar synthetic medium (NaNO3, 0.2; KH2PO4, 0.1; MgSO47H2O, 0.05; KCl, 0.05; agar 15 g/L and deionized water) supplemented with laminarin 1% (laminarin from Laminaria digitata, Sigma Aldrich, México) as a sole carbon source. Chitinase production was evaluated on the same synthetic basal medium but supplemented with colloidal chitin 1% rather than laminarin. Colloidal chitin was prepared using 10 g of purified crab shell chitin (Sigma Aldrich, México) suspended in 100 mL concentrated HCl for 2.5 h at 4°C and then was washed with cold deionized water and NaOH overnight at 4°C, followed by re-washing with cold deionized water at approximately pH 7.0. Mycelia discs (4 mm) were removed from PDA purified cultures with a sterilized needle and transferred to specific carbon-source media. After incubation at 24°C for 15 days, growth was evaluated either by measurement of the diameter of the developing colonies in comparison with a negative control (basal medium without carbon source), or by comparison with a positive control (basal medium supplemented with D-glucose 1%) in threefold replication.
Results
Morphological determination of Cladosporium spp.
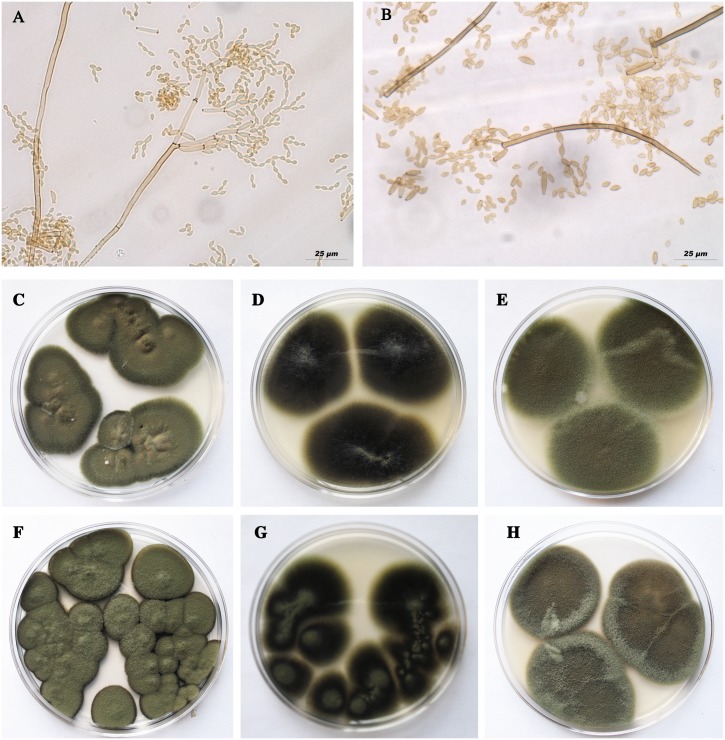
Our collections from chrysanthemum plants yielded two different strains of fungus resembling the Cladosporium genus (Cladosporiaceae, Capnodiales) [18]. Based on the morphological characteristics, one of the isolated fungi was identified as Cladosporium cladosporioides (Fresen.) GA de Vries, and the second isolated fungus was identified as Cladosporium pseudocladosporioides Bensh, Braun & Crous (Fig 1). Their morphological characteristics are summarized in Table 2.
Fig 1. Morphological characteristics of Cladosporium spp. associated with Puccinia horiana.
Light microscopy of conidiophores, ramoconidia and conidia on SNA medium. (A) Cladosporium cladosporioides, (B) C. pseudocladosporioides. Morphology of C. cladosporioides colonies on three different media: PDA (C), MEA (D), OA (E); C. pseudocladosporioides colonies on PDA (F), MEA (G), OA (H).
Table 2. Morphological characteristics of two different Cladosporium species associated with Puccinia horiana.
Conidiophore, ramoconidia and conidia characteristics were from colonies grown on SNA medium for 5 days at 24°C in the dark.
| Cladosporium cladosporioides | Cladosporium pseudocladosporioides | |
|---|---|---|
| REPRODUCTIVE STRUCTURES | ||
| Conidiophore | Straight, solitary, unbranched, terminal or lateral and without nodules. | Straight, solitary, unbranched, terminal or lateral and without nodules. |
| 3.63–3.1–2.77 μm | 4.57–3.44–2.21 μm | |
| Ramoconidia | Usually in groups of three or four, at the tip of conidiophores, straight, cylindrical-oblong in shape. | Usually in groups of three, at the tip of conidiophores, cylindrical-oblong in shape. |
| 5.47–9.27–18.15 × 2.43–2.96–3.83 μm | 7.37–13.94–38.23 × 2.43–3.57–4.76 μm | |
| Conidia | Numerous, in chains of up to nine conidia. They are limoniform, ovoid, obovoid to subglobose, aseptate, light brown, hila conspicuous. | Numerous, in chains of up to six conidia. They are limoniform, obovoid, ovoid to ellipsoidal in shape, aseptate, light brown, hila conspicuous. |
| 2.94–4.08–5.02 × 1.77–2.18–2.94 μm | 4.08–5.64–7.59 × 1.82–2.76–3.97 μm | |
| COLONIES ON DIFFERENT GROWING MEDIA | ||
| On PDA medium | Olivaceous with aerial mycelia diffuse, floccose-felty, reverse olive-black color. | Velvety, brown-reddish with dark margins, reverse brown-reddish color. |
| On MEA medium | Olivaceous-black and floccose, reverse olivaceous-black color. | Floccose olivaceous-black, reverse olivaceous-black color. |
| On OA medium | Velvety olivaceous to grey-olivaceous color with aerial mycelia, reverse grey-olivaceous color. | Velvety grey to grey-brown, reverse greybrown color. |
Sequence analysis of C. cladosporioides and C. pseudocladosporioides, ITS, ACT & EF-1α
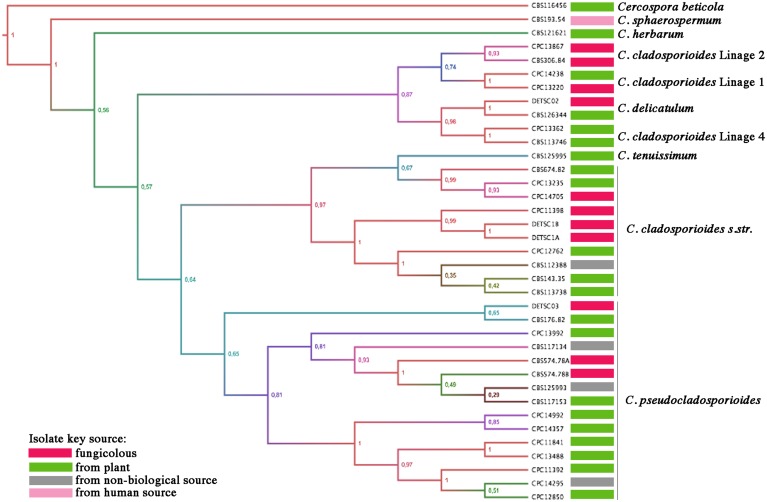
The analysis of the genomic sequences of internal transcribed spacer (ITS) region, actin (ACT) and translation elongation factor-1α (EF-1α) supported the morphological identification of the fungal isolates as C. pseudocladosporioides and C. cladosporioides. In order to determine inter- and intra-species phylogenetic similarities, based on sequence analysis and the origin of isolates, we performed a Bayesian analysis (Fig 2). All sequences were clustered into the Cladosporium genus, within the C. cladosporioides complex with strong support probabilities. Cladosporium cladosporioides, was clustered with CPC-11398 (from Phragmidium griseum) in one of three well defined sub-clades at the species core (Fig 2), consistently a member of C. cladosporioides. This species is composed of one of three well defined clades clustered with Cladosporium delicatulum, so in consequence is polyphyletic. Cladosporium pseudocladosporioides was clustered with CBS-176.82 (from Pteridium aquilinum) in one well defined sub-clade (Fig 2); all isolates were clustered together, so corresponding to a monophyletic lineage and consequently representing a true member of C. pseudocladosporioides.
Fig 2. Consensus phylogram from 80 0001 trees resulting from Bayesian analysis of 43 isolates in a combined ITS, ACT & EF-1α alignment.
Bayesian posterior probabilities on the tree are marked on the nodes. Isolate sources are color coded on branch tips, as indicated in the legend. The tree was rooted to sequences of Cercospora beticola strain CPC 11557, Cladosporium herbarum and Cladosporium sphaerospermum, representing the other two well defined phylogenetic groups within the genus. Also were included C. tenuissimum as support clade and C. delicatulum due to its presence on P. horiana pustules, but without antagonism effect.
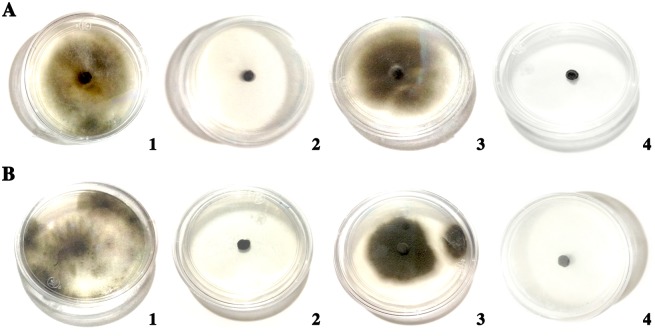
Antagonism assay
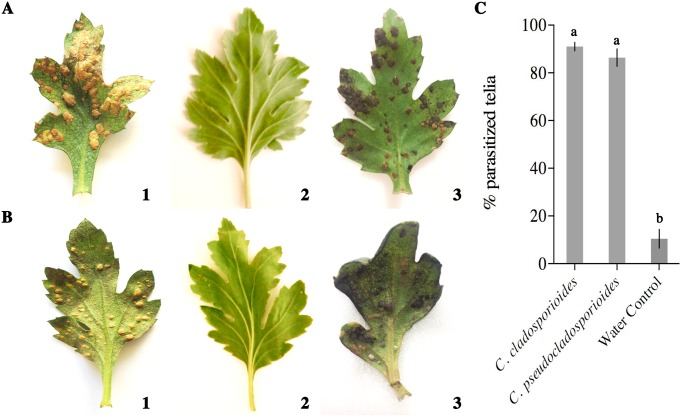
The telia of P. horiana treated only with water showed no alteration and had normal appearance (Fig 3A1 and 3B1). No C. cladosporioides or C. pseudocladosporioides conidia or hyphae were observed on inoculated leaf surfaces in areas devoid of P. horiana pustules (Fig 3A2 and 3B2). In contrast, 96 h after inoculation with Cladosporium isolates, pustules showed an appearance similar to that observed on leaves from diseased plants collected in the commercial greenhouse (Figs 3A3 to 3B3 and 4). The two Cladosporium isolates showed significant (P<0.05) parasitism on P. horiana pustules (Fig 3C). Pure cultures obtained from the parasitized pustules inoculated with the two antagonists exhibited the same morphological characteristics as the original isolates (data not shown).
Fig 3. Antagonism of Cladosporium cladosporioides and C. pseudocladosporioides on P. horiana telia.
(A) Assay with C. cladosporioides. (B) Assay with C. pseudocladosporioides. (1): Chrysanthemum leaves infected with P. horiana, without the antagonists; (2): Leaves without P. horiana and antagonist isolates applied; (3): Leaves with P. horiana and treated with the antagonists. (C) Percentage of P. horiana pustules parasitized by Cladosporium spp. Bars with different letters indicate significant differences at P<0.05 (REGW-F test). The line in each bar represents the standard error.
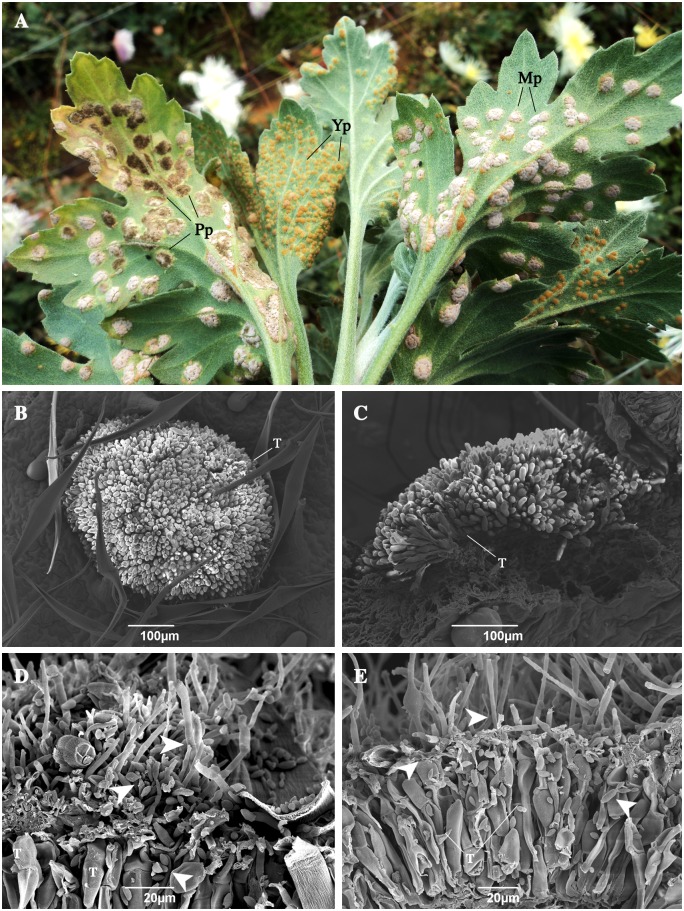
Fig 4. Telia of Puccinia horiana parasitized by fungi in the field.
(A) Leaves collected from field. Pp: parasitized pustule; Yp: young pustule; Mp: mature pustule. (B-C) Undamaged pustules. (D-E) Damaged pustules with fungus morphologically resembling the Cladosporium genus. Arrowheads indicates structures resembling Cladosporium sp. T: teliospores of P. horiana.
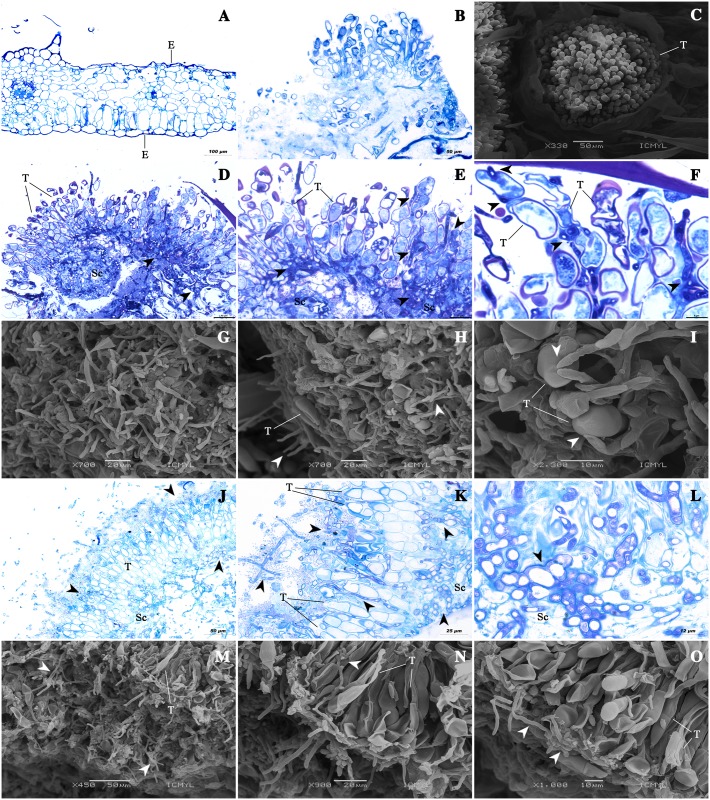
Interaction of Cladosporium isolates and P. horiana examined under SEM and light microscopy
The morphology of Puccinia horiana corresponded to the recently available descriptions [6, 9, 41], and almost all plant tissue was colonized by the CWR (Fig 5B and 5C). The healthy chrysanthemum leaves were not parasitized by the Cladosporium spp. isolates (Figs 3 and 5A). Once Cladosporium cladosporioides and C. pseudocladosporioides conidia germinated, they began to develop an intimate and active physical association with P. horiana. Teliospores from pustules colonized by C. cladosporioides (Fig 5D–5I) and C. pseudocladosporioides (Fig 5J–5O) were collapsed but no evidence of direct penetration was observed. Their cytoplasm was disrupted showing a vacuolated appearance and the wall structures were slimmed and collapsed causing the deformation of teliospores (Fig 5F and 5G for C. cladosporioides and Fig 5K, 5N and 5O for C. pseudocladosporioides). Further invasion and damage on P. horiana sporogenous cells occurred, and conidiophores of the antagonists were observed protruding from the pustules (Fig 5D–5O).
Fig 5. SEM and light microscopy observations of Puccinia horiana telia parasitized by two Cladosporium spp. isolates.
(A) Chrysanthemum leaves without P. horiana and antagonist isolates applied; (B-C) Leaves infected with P. horiana, without the antagonists; (D-F) C. cladosporioides in interaction with P. horiana pustules, teliospores and sporogenous cells under light microscopy; (G-I) C. cladosporioides and P. horiana teliospores under SEM, showing the colonized pustule surface; (J-L) C. pseudocladosporioides in interaction with P. horiana pustules, teliospores and sporogenous cells under light microscopy; (M-O) C. pseudocladosporioides and P. horiana teliospores under SEM, showing the surface of the colonized pustule. Arrowheads indicates structures of the antagonist Cladosporium spp. E: leaf epidermis, and T: P. horiana teliospores, Sc: P. horiana sporogenous cells.
Growth in vitro on glucanase and chitinase media
On the specific nutritional carbon-source media, Cladosporium cladosporioides grew 49.66 ± 0.47 mm on laminarin 1%, 44.66 ± 0.47 mm on glucose 1% and has no growth on colloidal chitin 1%, in case of Cladosporium pseudocladosporioides grew 32.66 ± 3.29 mm on laminarin 1%, 36.00 ± 4.32 mm on glucose 1% and has no growth on colloidal chitin 1%. Both Cladosporium isolates grew on the medium with laminarin as a sole carbon source, but not on that prepared with colloidal chitin (Fig 6).
Fig 6. Growth of two Cladosporium spp. isolates on media with different carbon sources.
(A) C. cladosporioides. (B) C. pseudocladosporioides. (1) laminarin 1%; (2) colloidal chitin 1%; (3) glucose 1%; (4) medium without carbon source.
Discussion
Morphological identification of Cladosporium spp. has been a difficult subject. Conidiophore and conidia size and shape are important characters, but usually dimensions overlap among species in the genus. However, molecular analysis has been a useful approach for the identification of Cladosporium species. Discrimination between C. cladosporioides, C. pseudocladosporioides and other taxa in the C. cladosporioides complex was made by Bensh et al., until 2010 [35], based on molecular phylogeny using ITS, ACT and EF-1α regions. These regions were used by Bensh et al. [18] to explain diversity and evolutionary trends in the Cladosporium genus. ITS alone does not give good species resolution [39] but ACT and EF-1α, in contrast, demonstrate a high degree of divergence among species [35, 42]. In the present study, we used an integrated approach based on the analysis of both molecular and morphological characters to determine the mycoparasitic species isolates as C. cladosporioides and C. pseudocladosporioides. Regarding intra-species relationships, our C. cladosporioides isolate showed some degree of diversification, as it clustered with another rust fungicolous isolate in a clearly supported clade inside core C. cladosporioides; likewise, there was strong clade support in C. pseudocladosporioides and C. cladosporioides lineages. Our results are consistent with Bensh et al. [35], and support the possible presence of cryptic species complexes on C. pseudocladosporioides and C. cladosporioides lineages. Both species are widely distributed and well adapted to various environments [18,35]. Cladosporium cladosporioides has already been reported parasitizing other fungi, such as Venturia inaequalis [29], Erisyphe cichoracearum [43], Botrytis fabae [44], Sclerotinia sclerotiorum [45] and rust fungi such as Puccinia graminis f.sp. tritici [30]. To our knowledge, C. pseudocladosporioides has not previously been reported parasitizing another fungus. This is the first report of both species potentially parasitizing P. horiana telia, and they occur naturally on this rust.
The association of both Cladosporium spp. on the sporogenous cells, without direct penetration of spores, was previously reported in Cladosporium sp. parasitizing Exobasidium camelliae var. gracilis [46], C. phylophillum on Taphrina sp., C. exobasidii on Exobasidium vaccinii and Exobasidium warmingii, and C. epichloës on Epichloë typhina [47]. This kind of relationship, without direct penetration, was probably due to differences between the teliospores and sporogenous cells of P. horiana, such as wall structural complexity between the stroma and spores, as well as different arrangement and proportion of chitin, glucans, glycoproteins, melanin and some other structural chemical compounds between different structures, as reported on Puccinia graminis and some other Pucciniales [48, 49, 50, 51], and it has been hypothesized that β-1,3 glucanases could be determinant for this Cladosporium spp. nutritional and spatial association [25]. Although there was evidence that C. cladosporioides and C. pseudocladosporioides were able to grow on laminarin media as the sole carbon source, we are not certain if these fungi excrete glucanases to parasitize P. horiana; however, the involvement of this enzyme in mycoparasitic Cladosporium relationships was previously reported for C. tenuissimum against Uromyces appendiculatus [25]. It is also possible that antibiotic mechanisms were affecting teliospore morphology and had repercussions probably in terms of potential loss of viability of teliospores, as previously reported in almost all mycoparasitic relationships of strains of C. tenuissimum [25, 26, 27], C. uredinicola [21, 22, 23], C. gallicola [28], C. aecidiicola [24] and Cladosporium sp. [17, 52]. Various antifungal compounds have been reported and isolated from some C. cladosporioides strains [53, 54] and C. pseudocladosporioides has shown some antibiotic activity [55].
Currently, control of chrysanthemum white rust in greenhouses and semi-covered growing systems is focused mainly on fungicides and some resistant cultivars. The potential resistance of P. horiana to fungicides represents a challenge for the development of new schemes to reduce damage by chrysanthemum white rust. Although efforts have been made to apply biological control against this disease using Verticillium lecanii, Aphanocladium album or Cladosporium spp. [4, 15, 16, 17], so far none of them are widely used. Since C. cladosporioides and C. pseudocladosporioides isolates altered the morphology of teliospores, and possibly reduced both viability and production, they might have potential for chrysanthemum white rust management in an integrated disease management scheme. Further studies must be carried out as, among other things, it is necessary to know the disease’s biology, ecology and mycoparasitism under controlled and commercial conditions, and to ascertain the antagonistic activity of the hyperparasites and their role in nature. In addition, it must be confirmed that these isolates do not harm other crops and that they have no adverse effects on humans or other animals. The present study provides a basis for such further studies.
Supporting information
(A) EFα1 partition; (B) ACT partition. (Using the same sequences as in Table 1).
(TIF)
Acknowledgments
The first author (David Eduardo Torres) expresses his appreciation to CONACyT for his Master´s scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank M.C. Mónica Pérez Pacheco from Laboratorio de Desarrollo en Plantas, Departamento de Biología Comparada, Facultad de Ciencias, UNAM and Biol. Yolanda Hornelas Cruces from Laboratorio de Microscopia Electrónica, Instituto de Ciencias del Mar y Limnología, and UNAM for the facilities for processing material for LR-White and SEM images.
Data Availability
All DNA sequences are available from the GenBank database (accession numbers: KT877404, KT887880, KT721703, KT877405, KT887881, KT721704, KT877407, KT887883, KT887879, KT877406, KT887882, KT887878).
Funding Statement
The first author (David Eduardo Torres) expresses his appreciation to CONACyT for his Master´s scholarship. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alaei H, Baeyen S, Maes M, Höfte M, Heungens K. Molecular detection of Puccinia horiana in Chrysanthemum × morifolium through conventional and real-time PCR. J Microbiol Methods. 2009; 76(2): 136–145. doi: 10.1016/j.mimet.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 2.Alaei H, De Backer M, Nuytinck J, Maes M, Höfte M, Heungens K. Phylogenetic relationships of Puccinia horiana and other rust pathogens of Chrysanthemum × morifolium based on rDNA ITS sequence analysis. Microbiol Res. 2009; 113(6): 668–683. [DOI] [PubMed] [Google Scholar]
- 3.European and Mediterranean Plant Protection Organization. Puccinia horiana. Bulletin of the European and Mediterranean Plant Protection Organization (EPPO). 2004; 34(1): 209–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whipps MJ. A review of white rust (Puccinia horiana Henn.) disease on chrysanthemum and the potential for its biological control with Verticillium lecanii (Zimm.) Viegas. Ann Appl Biol. 1993; 122(1): 173–187. [Google Scholar]
- 5.De Backer M, Bonants P, Pedley KF, Maes M, Roldan-Ruiz I, Van Bockstaele E, et al. Genetic relationships in an international collection of Puccinia horiana isolates based on newly identified molecular markers and demonstration of recombination. Phytopathology. 2013; 103(11), 1169–1179. doi: 10.1094/PHYTO-01-13-0007-R [DOI] [PubMed] [Google Scholar]
- 6.Hennings P. Einige neue japanische Uredineen. Hedwigia. 1901; 40: 25–26. [Google Scholar]
- 7.Hiratsuka N. Three species of chrysanthemum rust in Japan and its neighboring districts. Sydowia 1957; 2(1): 34–44. [Google Scholar]
- 8.Punithalingam E. Puccinia horiana. CMI descriptions of pathogenic fungi and bacteria. vol. 176 1968. [Google Scholar]
- 9.Bonde MR, Murphy CA, Bauchan GR, Luster DG, Palmer CL, Nester SE, et al. Evidence for systemic infection by Puccinia horiana, causal agent of Chrysanthemum White Rust, in Chrysanthemum. Phytopathology. 2015; 105(1): 91–98. doi: 10.1094/PHYTO-09-13-0266-R [DOI] [PubMed] [Google Scholar]
- 10.Firman ID, Martin PH. White rust of chrysanthemums. Ann Appl Biol. 1968; 62(3): 429–442. [Google Scholar]
- 11.Zandvoort R. Wind dispersal of Puccinia horiana. Eur J Plant Pathol. 1968; 74(4): 124–127. [Google Scholar]
- 12.Dickens JSW. Studies on the chemical control of chrysanthemum white rust caused by Puccinia horiana. Plant Pathol. 1990; 39(3): 434–442. [Google Scholar]
- 13.Akiko K, Kishi K, Yoshioka A. Ocurrence of oxicarboxin-tolerant isolates of Puccinia horiana P. Hennings in Japan. Nippon Shokubutsu Byori Gakkaiho. 1977; 43(2): 145–150. [Google Scholar]
- 14.Cook RTA. First report in England of changes in the susceptibility of Puccinia horiana, the cause of chrysanthemum white rust, to triazole and strobilurin fungicides. Plant Pathol. 2001; 50(6): 792. [Google Scholar]
- 15.Srivastava AK, Défago G, Kern H. Hyperparasitism of Puccinia horiana and other microcyclic rusts. J Phytopathol. 1985; 114(1): 73–78. [Google Scholar]
- 16.Rodríguez-Navarro JA, Zavaleta-Mejía E, Alatorre-Rojas R. Epidemiología y manejo de la roya blanca (Puccinia horiana P. Henn.) del crisantemo (Dendrathema grandiflora Tzvelev). Fitopatología. 1996; 31(2): 122–132. [Google Scholar]
- 17.García-Velasco R, Zavaleta-Mejía E, Rojas-Martínez R. Antagonismo de Cladosporium sp. contra Puccina horiana Henn. causante de la roya blanca del crisantemo (Dendranthema grandiflora Tzcvelev). Rev Mex Fitopatol. 2005; 23 (1): 79–86. [Google Scholar]
- 18.Bensh K, Braun U, Groenewald JZ, Crous PW. The genus Cladosporium. Stud Mycol. 2012; 72(1): 1–401. doi: 10.3114/sim0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moricca S, Ragazzi A. Biological and Integrated means to control rusts diseases In: Ciancio A. & Mukerji KG, editors. Integrated Management of Diseases caused by fungi, phytoplasma and bacteria Vol. 3 Springer Science & Business Media; 2008. pp. 303–332. [Google Scholar]
- 20.Vandermeer J, Perfecto I, Liere H. Evidence for hyperparasitism of coffee rust (Hemileia vastatrix) by the entomogenous fungus, Lecanicillium lecanii, through a complex ecological web. Plant Pathol. 2009; 58(4): 636–641. [Google Scholar]
- 21.Traquair J, Meloche RB, Jarvis WR, Baker KW. Hyperparasitism of Puccinia violae by Cladosporium uredinicola. Can J Bot. 1984; 62(1): 181–184. [Google Scholar]
- 22.Barros ST, Oliveira N, Bastos S, Maia L. Hyperparasitism of Cladosporium uredinicola over Puccinia puta on the host Ipomea fistulosa. Mycologist. 1999; 13(1): 3–24. [Google Scholar]
- 23.Morgan–Jones G, McKemy JM. Studies in the genus Cladosporium sensu lato: I. Concerning Cladosporium uredinicola, occurring on telial columns of Cronartium quercuum and other hosts. Mycotaxon. 1990; 39(1): 185–200. [Google Scholar]
- 24.Sharma JK, Heather WA. Effect of Cladosporium aecidiicola Thum. on the viability of urediniospores of Melampsora medusae Thum. in storage. Eur J Plant Pathol. 1980; 10(6): 360–364. [Google Scholar]
- 25.Assante G, Maffi D, Saracchi M, Farina G, Moricca S, Ragazzi A. Histological studies on the mycoparasitism of Cladosporium tenuissimum on urediniospores of Uromyces appendiculatus. Mycol Research. 2004; 108(2): 170–182. [DOI] [PubMed] [Google Scholar]
- 26.Moricca S, Ragazzi A, Mitchelson KR, Assante G. Antagonism of the two-needle pine stem rust fungi Cronartium flaccidum and Peridermium pini by Cladosporium tenuissimum in vitro and in planta. Phytopathology. 2001; 91(5): 457–468. doi: 10.1094/PHYTO.2001.91.5.457 [DOI] [PubMed] [Google Scholar]
- 27.Moricca S, Ragazzi A, Assante G. Biocontrol of rust fungi by Cladosporium tenuissimum In: Pei MH, & McCracken AR, Editors. Rust diseases of Willow and poplar. CAB International; 2005. p. 213–219. [Google Scholar]
- 28.Tsuneda A, Hiratsuka Y. Mode of parasitism of a mycoparasite, Cladosporium gallicola, on western gall rust Endocronartum harknessii. Can J Plant Pathol. 1979; 1(1):31–36. [Google Scholar]
- 29.Köhl JJ, Molhoek WW, Groenenboom-de Haas BB, Goossen-van de Geijn HH. Selection and orchard testing of antagonists suppressing conidial production by the apple scab pathogen Venturia inaequalis. Eur J Plant Pathol. 2009; 123(4), 401–414. [Google Scholar]
- 30.Zhan G, Tian Y, Wang F, Chen X, Guo J, Jiao M, et al. A Novel Fungal Hyperparasite of Puccinia striiformis f. sp. tritici, the Causal Agent of Wheat Stripe Rust. PLoS One. 2014; 9(11): e111484 doi: 10.1371/journal.pone.0111484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, et al. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales) with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol. 2007; 58(1): 105–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falcon L and Valera A. Extracción de ácidos nucléicos In: Eguiarte L, Souza V, & Aguirre X, Editors. Ecología molecular. Instituto Nacional de Ecología, Mexico: 2007. [Google Scholar]
- 33.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand D, Sninsky JS and White TJ, editors. PCR protocols: a guide to methods and applications. Academic Press, New York: 1990. pp. 315–322. [Google Scholar]
- 34.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999; 91(3): 553–556. [Google Scholar]
- 35.Bensh K, Groenewald JZ, Starink-Willemse M, Andersen B, Sumerell BA, Shin HD, et al. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud Mycol. 2010; 67(1): 1–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012; 9(8): 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012; 29(8): 1969–197. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30(12): 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zalar P, De Hoog GS, Schroers HJ, Crous PW, Groenewald JZ, & Gunde-Cimerman N. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud Mycol. 2007; 58(1): 157–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruzin SE. Plant microtechnique and microscopy. New York: Oxford University Press; 1999. p 315. [Google Scholar]
- 41.O’Keefe G, Davis D. Morphology of Puccinia horiana, causal agent of chrysanthemum white rust, sampled from naturally infected plants. Plant Dis. 2015; http://dx.doi.org/10.1094/ PDIS-02-15-0239-RE. [DOI] [PubMed] [Google Scholar]
- 42.Voigt K, Wöstemeyer J. Phylogeny and origin of 82 zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1 genes. Gene. 2001; 270(1): 113–120. [DOI] [PubMed] [Google Scholar]
- 43.Kiss L. A review of fungal antagonist of powdery mildews and their potential as biocontrol agents. Pest Manag Sci. 2003; 59(4): 475–483. doi: 10.1002/ps.689 [DOI] [PubMed] [Google Scholar]
- 44.Jackson A, Walters D, Marshall G. Antagonistic Interactions between the Foliar Pathogen Botrytis fabae and Isolates of Penicillium brevicompactum and Cladosporium cladosporioides on Faba Beans. Biological Control. 1997; 8(2): 97–106. [Google Scholar]
- 45.Boland GJ, Hunter JE. Influence of Alternaria alternata and Cladosporium cladosporioides on white mold of bean caused by Sclerotinia sclerotiorum. Can J Plant Pathol. 1988; 10(2): 172–177. [Google Scholar]
- 46.Mims CW, Hanlin RT, Richardson EA. Light- and electron-microscopic observations of Cladosporium sp. growing on basidia of Exobasidum camelliae var. gracilis. Can J Bot. 2007; 85(1): 76–82. [Google Scholar]
- 47.Heuchert B, Braun U, Schubert K. Morphotaxonomic revision of fungicolous Cladosporium species (hyphomycetes). Schlechtendalia. 2005; 13(1): 1–78. [Google Scholar]
- 48.Harder DE, Chong J, Rohringer R, Kim WK. Structure and cytochemistry of the walls of urediospores, germ tubes, and appressoria of Puccinia graminis tritici. Can J Bot. 1986; 64(3): 476–485. [Google Scholar]
- 49.Littlefield L, Heath M. Ultrastructure of rust fungi. New York: Academic Press; 1979. p. 277. [Google Scholar]
- 50.Deising H, Heiler S, Rauscher M, Xu H, Mendgen K. Cellular aspects of rust infection structure differentiation: spore adhesion and fungal morphogenesis In: Nicole M & Gianinazzi V, editors. Histology, ultrastructure and molecular cytology of Plant Microorganisms Interactions vol. 7 Springer Science & Business Media; 1996. pp. 135–156. [Google Scholar]
- 51.Freytag S, Mendgen K. Surface carbohydrates and cell wall structure of in vitro-induced uredospore infection structures of Uromyces riciae-fabae before and after treatment with enzymes and alkali. Protoplasma. 1991; 161(1): 94–103. [Google Scholar]
- 52.Omar M, Heather WA. Effect of saprophytic phylloplane fungi on germination and development of Melampsora larici-populina. Mycol Res. 1979; 72(2): 225–231. [Google Scholar]
- 53.Pandey RR, Arora DK, Dubey RC. Antagonistc interactions between fungal pathogens and phylloplane fungi of guava. Mycopathologia. 1993; 124(1): 31–39. [Google Scholar]
- 54.Wang X, Radwan M, Taráwneh A, Gao J, Wedge D, Rosa L, et al. Antifungal activity against plant pathogens of metabolites from the endophytic fungus Cladosporium cladosporioides. J Agric Food Chem. 2013; 61(19): 4551–4555. doi: 10.1021/jf400212y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellswort K, Clark T, Gray C, Johnson J. Isolation and bioassay screening of medicinal plant endophytes from eastern Canada. Can J Microbiol. 2013. 59(11): 761–765. doi: 10.1139/cjm-2013-0639 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) EFα1 partition; (B) ACT partition. (Using the same sequences as in Table 1).
(TIF)
Data Availability Statement
All DNA sequences are available from the GenBank database (accession numbers: KT877404, KT887880, KT721703, KT877405, KT887881, KT721704, KT877407, KT887883, KT887879, KT877406, KT887882, KT887878).