Abstract
This paper elucidates the teixobactin pharmacophore by comparing the arginine analogue of teixobactin Arg10-teixobactin to seven homologues with varying structure and stereochemistry. The roles of the guanidinium group at position 10, the stereochemistry of the macrolactone ring, and the “tail” comprising residues 1–5 are investigated. The guanidinium group is not necessary for activity; Lys10-teixobactin is more active than Arg10-teixobactin against gram-positive bacteria in minimum inhibitory concentration (MIC) assays. The relative stereochemistry of the macrolactone ring is important; diastereomer l-Thr8,Arg10-teixobactin is inactive, and diastereomer d-allo-Ile11,Arg10-teixobactin is less active. The macrolactone ring is critical; seco-Arg10-teixobactin is inactive. The absolute stereochemistry is not important; the enantiomer ent-Arg10-teixobactin is comparable in activity. The hydrophobic N-terminal tail is important; truncation of residues 1–5 results in loss of activity, and replacement of residues 1–5 with a dodecanoyl group partially restores activity. These findings pave the way for developing simpler homologues of teixobactin with enhanced pharmacological properties.
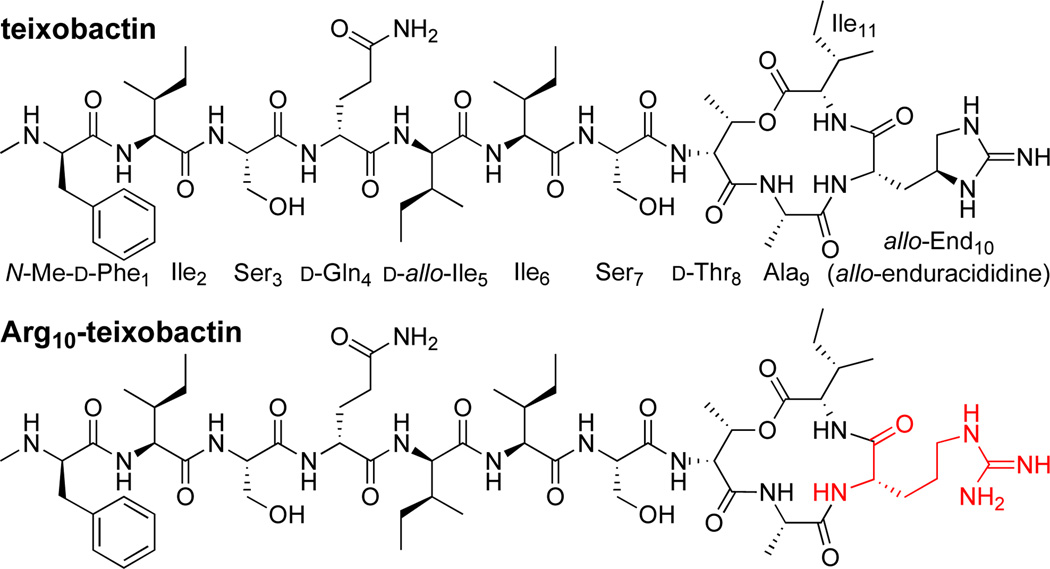
At the beginning of 2015, a new antibiotic, teixobactin, was reported in Nature,1 with great attention in the scientific press2,3,4,5 and the popular press.6 Teixobactin is a non-ribosomal undecapeptide containing a macrocyclic depsipeptide group (Figure 1). It contains four d-amino acids and seven l-amino acids, and the C-terminal Ile11 is cyclized onto the side chain of d-Thr8 to form a 13-membered lactone. Residue 10 of teixobactin is the non-proteinogenic amino acid, l-allo-enduracididine (allo-End10), which is a cyclic analogue of arginine. Teixobactin acts against gram-positive bacteria by binding to the prenyl-pyrophosphate-GlcNAc region of lipid II.1 This region is highly conserved in bacteria and cannot easily mutate to impart drug-resistance.7,8 It is thus an attractive antibiotic target.
Figure 1.
Structures of teixobactin and Arg10-teixobactin.
Recently, Jad et al. and Parmar et al. reported syntheses of the arginine analogue of teixobactin Arg10-teixobactin.9,10 Both syntheses involve solid-phase peptide synthesis (SPPS) of a branched precursor on 2-chlorotrityl resin, followed by solution-phase macrolactamization to form the Ala9–Arg10 amide bond. The former synthesis requires both Fmoc and Alloc groups as orthogonal α-amino protecting groups; the latter requires Fmoc, Alloc, and trityl groups. Both syntheses introduce d-Thr8 without protecting the alcohol group and O-acylate it before completing the N-terminal tail. Both sets of authors reported that Arg10-teixobactin is about an order of magnitude less active against gram-positive bacteria than teixobactin in minimum inhibitory concentration (MIC) assays.11,12,13
In the current study, we set out to elucidate the teixobactin pharmacophore by synthesizing and evaluating a series of teixobactin homologues. We examine the roles of the guanidinium group at position 10, the stereochemistry of the macrolactone ring, and the “tail” comprising residues 1–5. We also report a simpler synthesis of teixobactin analogues and a simpler homologue, which we term lipobactin 1.
We synthesized Arg10-teixobactin and other homologues by SPPS on 2-chlorotrityl resin, followed by solution-phase macrolactamization to form the Arg10–Ile11 amide bond (Scheme 1).14,15,16,17 We used only Fmoc protecting groups to construct all of the amide bonds and carried d-Thr8 through the entire synthesis without side chain protection. All homologues were prepared and studied as the trifluoroacetic acid (TFA) salts.
Scheme 1.
Synthesis of Arg10-teixobactin.
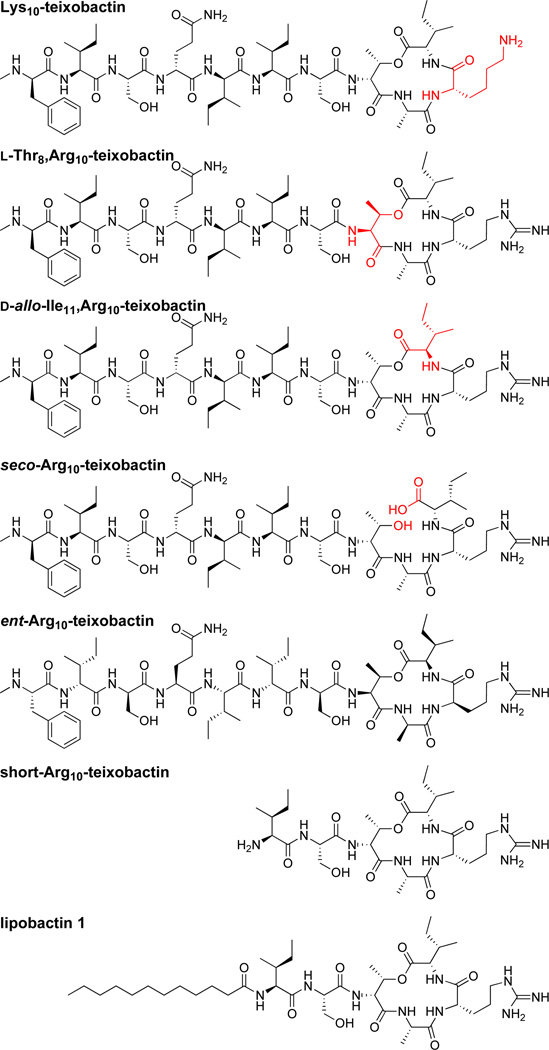
We began the synthesis by attaching Fmoc-Arg(Pbf)-OH to 2-chlorotrityl resin. Residues 9 through 1 were then introduced by standard Fmoc-based SPPS using HCTU as the coupling reagent. d-Thr8 was introduced without a protecting group at the hydroxy position. No O-acylation of d-Thr8 was observed in the subsequent rounds of SPPS. d-Thr8 was then O-acylated with Fmoc-Ile-OH using DIC and DMAP.18,19,20 Fmoc-deprotection, followed by cleavage from the resin with 20% hexafluoroisopropanol (HFIP) in CH2Cl2 afforded the acyclic protected precursor. Macrolactamization with HBTU and HOBt, followed by global deprotection with trifluoroacetic acid (TFA) and RP-HPLC purification afforded Arg10-teixobactin. We also prepared a series of homologues using similar procedures (Figure 2).
Figure 2.
Structures of teixobactin homologues.
We investigated the antibiotic activity of Arg10-teixobactin and homologues in MIC assays against four types of gram-positive bacteria. We used the antibiotic vancomycin as a positive control and the gram-negative bacterium E. coli as a negative control. We selected non-pathogenic strains of bacteria to facilitate the safe and rapid screening of Arg10-teixobactin and other homologues in a biosafety level 1 (BSL-1) environment.
To explore the role of a guanidinium group in residue 10, we compared the MIC of Arg10-teixobactin to Lys10-teixobactin. The arginine residue serves as a surrogate for allo-enduracididine, which is not commercially available and has only been prepared by cumbersome multistep syntheses.21,22,23,24,25 Arg10-teixobactin gave MIC values of 1–4 µg/mL against the four gram-positive bacteria studied (Table 1). Although side-by-side comparison to an authentic sample of teixobactin was not possible, comparison to the original published values in related bacteria suggests that Arg10-teixobactin is about an order of magnitude less active (Table 1). Surprisingly, Lys10-teixobactin gave MIC values 2–4 times lower than Arg10-teixobactin. While the MIC values for Lys10-teixobactin are slightly higher than those reported for teixobactin, they are comparable to those of vancomycin (Table 1). This interesting finding indicates that the guanidinium group at position 10 is not necessary for activity and lays the foundation for the future discovery of homologues that lack allo-enduracididine and are even more potent.
Table 1.
MIC of teixobactin homologues in µg/mL.
|
Staphylococcus epidermidis ATCC 14990 |
Streptococcus salivarius ATCC 13419 |
Enterococcus durans ATCC 6056 |
Bacillus subtilis ATCC 6051 |
Escherichia coli ATCC 10798 |
|
|---|---|---|---|---|---|
| Arg10-teixobactin | 1 | 1 | 4 | 2 | >32 |
| Lys10-teixobactin | 0.25 | 0.5 | 1 | 0.5 | >32 |
| l-Thr8,Arg10-teixobactin | >32 | >32 | >32 | >32 | >32 |
| d-allo-Ile11,Arg10-teixobactin | 2 | 2 | 8 | 4 | >32 |
| seco-Arg10-teixobactin | >32 | >32 | >32 | >32 | >32 |
| ent-Arg10-teixobactin | 2 | 1 | 4 | 2 | >32 |
| short-Arg10-teixobactin | >32 | >32 | >32 | >32 | >32 |
| lipobactin 1 | 4 | 4 | 8 | 4 | >32 |
| vancomycin | 0.5 | 0.5 | 0.5 | 1 | >32 |
| teixobactin1 | 0.08–0.3 | 0.02–0.15 | 0.3–0.6 | 0.02–0.6 | 25 |
| various Staphylococcus1 |
various Streptococcus1 |
various Enterococcus1 |
various Bacillus1 |
E. coli1 | |
To investigate the role of the macrolactone ring stereochemistry, we compared the diastereomer l-Thr8,Arg10-teixobactin and d-allo-Ile11,Arg10-teixobactin to Arg10-teixobactin. The former proved inactive (MIC > 32 µg/mL) against the gram positive bacteria, while the latter proved half as active (Table 1). Collectively, these results suggest that the ring stereochemistry and the conformation are important in teixobactin activity. Seco-Arg10-teixobactin also proved inactive (MIC > 32 µg/mL), further supporting the importance of the cyclic depsipeptide structure (Table 1).
To further investigate the role of the macrolactone ring stereochemistry, we compared ent-Arg10-teixobactin to Arg10-teixobactin. Ent-Arg10-teixobactin exhibits comparable activity to Arg10-teixobactin. This exciting finding supports a model in which the amide NH groups on macrolactone ring bind to the achiral pyrophosphate group of lipid II through hydrogen-bonding interactions. This mode of binding has previously been reported in the NMR structure of the complex of nisin with lipid II (PDB 1WCO)26 and appears to occur for teixobactin as well.
To investigate the role of the N-terminal tail, we truncated residues 1–5. The resulting short-Arg10-teixobactin also proved inactive (MIC > 32 µg/mL). To investigate the possibility that the hydrophobic residues N-Me-d-Phe, Ile, and d-allo-Ile at positions 1, 2, and 5 help to anchor teixobactin into the plasma membrane, we replaced residues 1–5 with a dodecanoyl group.27,28 The resulting homologue, lipobactin 1, proved only 2–4 times less active than Arg10-teixobactin (Table 1). This finding confirms the importance of the hydrophobicity of the N-terminal tail and paves the way for further developing simpler homologues of teixobactin with enhanced pharmacological properties.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) for funding (grant 1R21AI121548-01). The authors thank M. McClelland and L. Mota-Bravo for helpful advice on MIC assays.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:.
Procedures for the synthesis of Arg10-teixobactin and homologues and minimum inhibitory concentration (MIC) assays; spectral and other characterization data.
The authors declare no competing financial interest.
REFERENCES AND NOTES
- 1.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Editorial. Listen up, (2015) Nature. 517:121. doi: 10.1038/517121b. [DOI] [PubMed] [Google Scholar]
- 3.Ledford H. Promising antibiotic discovered in microbial ‘dark matter’. [accessed January 7, 2015];Nature News. 2015 Jan 7; [Online], http://www.nature.com/news/promising-antibiotic-discovered-in-microbial-dark-matter-1.16675. [Google Scholar]
- 4.Wright G. Antibiotics: An irresistible newcomer. Nature. 2015;517:442–444. doi: 10.1038/nature14193. [DOI] [PubMed] [Google Scholar]
- 5.Kahrstrom CT. Antibacterial drugs: a new drug for resistant bugs. Nature Rev. Drug Discov. 2015;14:94. doi: 10.1038/nrd4544. [DOI] [PubMed] [Google Scholar]
- 6.Grady D. New Antibiotic Stirs Hope Against Resistant Bacteria. [accessed January 7, 2015];The New York Times. 2015 Jan 7; [Online], http://www.nytimes.com/2015/01/08/health/from-a-pile-of-dirt-hope-for-a-powerful-new-antibiotic.html?_r=0. [Google Scholar]
- 7.Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 8.de Kruijff B, van Dam V, Breukink E. Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fatty Acids. 2008;79:117–121. doi: 10.1016/j.plefa.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Jad YE, Acosta GA, Naicker T, Ramtahal M, El-Faham A, Govender T, Kruger HG, De la Torre BG, Albericio F. Synthesis and biological evaluation of a teixobactin analogue. Org. Lett. 2015;17:6182–6185. doi: 10.1021/acs.orglett.5b03176. [DOI] [PubMed] [Google Scholar]
- 10.Parmar A, Iyer A, Vincent CS, Van Lysebetten D, Prior SH, Madder A, Taylor EJ, Singh I. Efficient total syntheses and biological activities of two teixobactin analogues. Chem. Commun. 2016;52:6060–6063. doi: 10.1039/c5cc10249a. [DOI] [PubMed] [Google Scholar]
- 11.The MIC values reported in Table 1 of reference 9 appear to be in error by a factor of 103, being reported as nM rather than as µM or as µg/mL
- 12.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 13.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard―Ninth Edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. CLSI document M07-A9. [Google Scholar]
- 14.Davies JS. The cyclization of peptides and depsipeptides. J. Pept. Sci. 2003;9:471–501. doi: 10.1002/psc.491. [DOI] [PubMed] [Google Scholar]
- 15.Sarabia F, Chammaa S, Ruiz AS, Ortiz LM, Herrera FJ. Chemistry and biology of cyclic depsipeptides of medicinal and biological interest. Curr. Med. Chem. 2004;11:1309–1332. doi: 10.2174/0929867043365224. [DOI] [PubMed] [Google Scholar]
- 16.Lambert JN, Mitchell JP, Roberts KD. The synthesis of cyclic peptides. J. Chem. Soc., Perkin Trans. 2001;1:471–484. [Google Scholar]
- 17.White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat. Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 18.Neises B, Steglich W. Simple method for the esterification of carboxylic acids. Angew. Chem., Int. Ed. Engl. 1978;17:522. [Google Scholar]
- 19.Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, Konig C, Ammerman NC, Barrio MB, Borchers K, Bordon-Pallier F, Bronstrup M, Coutemanche G, Gerlitz M, Geslin M, Hammann P, Heinz DW, Hoffmann H, Klieber S, Kohlmann M, Kurz M, Lair C, Matter H, Nuermberger E, Tyagi S, Fraisse L, Grosset JH, Lagrange S, Muller R. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science. 2015;348:1106–1112. doi: 10.1126/science.aaa4690. [DOI] [PubMed] [Google Scholar]
- 20.Esterification with DIC and DMAP is known to epimerize amino acids. 1H NMR analysis of the unpurified Arg10-teixobactin, and comparison to an authentic sample of d-allo-Ile11,Arg10-teixobactin, showed a 2:1 ratio of Arg10-teixobactin and the epimeric d-allo-Ile11,Arg10-teixobactin (Figure S1). HPLC purification of the crude product afforded Arg10-teixobactin in approximately 95% diasteromeric purity
- 21.Tsuji S, Kusumoto S, Shiba T. Synthesis of enduracididine, a component amino acid of antibiotic enduracidin. Chem. Lett. 1975;12:1281–1284. [Google Scholar]
- 22.Sanière L, Leman L, Bourguignon J, Dauban P, Dodd RH. Iminoiodane mediated aziridination of α-allylglycine: Access to a novel rigid arginine derivative and to the natural amino acid enduracididine. Tetrahedron. 2004;60:5889–5897. [Google Scholar]
- 23.Möschwitzer VD, Kariuki BM, Redman JE. Asymmetric synthesis of aminopyrimidine and cyclic guanidine amino acids. Tetrahedron Lett. 2013;54:4526–4528. [Google Scholar]
- 24.Olson DE, Su JY, Robert DA, Du Bois J. Vicinal diamination of alkenes under Rh-catalysis. J. Am. Chem. Soc. 2014;136:13506–13509. doi: 10.1021/ja506532h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig W, Chen J, Richardson D, Thorpe R, Yuan Y. A highly stereoselective and scalable synthesis of l-allo-enduracididine. Org. Lett. 2015;17:4620–4623. doi: 10.1021/acs.orglett.5b02362. [DOI] [PubMed] [Google Scholar]
- 26.Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 2004;11:963–937. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 27.Straus SK, Hancock REW. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta, Biomembr. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Steenbergen JN, Alder J, Thorne GM, Tally FP. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J. Antimicrob. Chemother. 2005;55:283–288. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]