Abstract
In sea urchin development, structures derived from neurogenic territory control the swimming and feeding responses of the pluteus as well as the process of metamorphosis. We have previously isolated an alpha tubulin family member of Paracentrotus lividus (Pl-Tuba1a, formerly known as Pl-Talpha2) that is specifically expressed in the ciliary band and animal pole neurogenic domains of the sea urchin embryo. In order to identify cis-regulatory elements controlling its spatio-temporal expression, we conducted gene transfer experiments, transgene deletions and site specific mutagenesis. Thus, a genomic region of about 2.6 Kb of Pl-Tuba1a, containing four Interspecifically Conserved Regions (ICRs), was identified as responsible for proper gene expression. An enhancer role was ascribed to ICR1 and ICR2, while ICR3 exerted a pivotal role in basal expression, restricting Tuba1a expression to the proper territories of the embryo. Additionally, the mutation of the forkhead box consensus sequence binding site in ICR3 prevented Pl-Tuba1a expression.
Introduction
Cis-regulatory elements, such as promoters, enhancers and silencers, are involved in determining the spatio-temporal patterns of gene expression. Accordingly, the precise identification of these elements is an essential step in understanding the regulatory networks operating at the cellular, tissue or organismal level [1]. It is also known that the cis-regulatory system may be separated into several cis-regulatory modules, whose topological integration establishes a specific pattern of each gene expression and orchestrates the dynamic regulation of gene expression of an embryonic domain in response to environmental and developmental stimuli [2–4]. The Gene Regulatory Networks involved in each specification step of differentiation processes have been extensively studied in echinoderms [5–9]. In particular, the neural differentiation process of sea urchin embryos has been thoroughly analysed [10–15].
All echinoderm larvae possess a nervous system consisting of a ciliary band and associated sensory ganglia (apical, oral and lateral ganglia) that control swimming and feeding [16–21]. Neurons of the larval nervous system of euechinoids first appear as neuroblasts in the thickened ectoderm of the animal plate (anterior neuroectoderm, ANE) at the late blastula–early gastrula stage. Neurons continue to be added throughout larval development, and the fully formed eight-armed pluteus nervous system is organized as a complex array of sensory neurons, interneurons, tracts of axons and ganglia that are closely associated with the larval ciliary band neuroectoderm (CBE) and larval muscles [13].
We have previously identified and characterized a specific alpha tubulin gene (Pl-Tuba1a, formerly named Pl-Talpha2) in the Paracentrotus lividus sea urchin whose expression begins at the hatching blastula stage and is restricted in the major structures that will give rise to the larval nervous system [9, 22–24]. Interestingly, in the same territories is also specifically expressed a beta tubulin gene [25] encoding an isotype containing a carboxy terminal domain that is typical of neural specific tubulin isoforms. Gene transfer experiments showed that a Pl-Tuba1a 5.3 Kb genomic region is involved in the specific temporal and spatial regulation of this gene [26]. Moreover, mechanisms of epigenetic modifications contributing to its expression during embryo development were characterized [27].
Previously, we have identified several putative Interspecific Conserved Regions (ICRs) using computational techniques [26]. In this work, we identify a genomic region of about 2.6 Kb of Pl-Tuba1a, containing four ICRs, as responsible for proper gene expression. In addition, an overall analysis of the ICRs was performed by the gene transfer of properly modified reporter constructs, revealing their role in the regulation of Pl-Tuba1a gene expression.
Materials and Methods
Preparation of reporter constructs
The 5’ deletion constructs were generated by PCR amplification of the full-length clone (Pl-Tuba1a-GFP [26] using appropriate HindIII primer sets (see S1 Table) and subsequent cloning into the HindIII site of pBluescript II SK(+) (pBSK) vector (Stratagene). The GFP reporter constructs maintain the GFP coding sequence in frame with the first three codons of Pl-Tuba1a and are under the control of progressively reduced amounts of Pl-Tuba1a regulatory sequences.
Internal (ICR3 and/or ICR4) deletions were generated by PCR amplification of the -1.8KbGFP construct, excluding each conserved region, using the appropriate primer set and subsequent self-ligation of the two PCR products, permitted by XbaI restriction sites harboured by primers, and cloning into the HindIII site of pBSK vector.
The -1.8(ΔIntron) was obtained by PCR amplifications of the -1.8KbGFP construct, excluding the first intron, using the appropriate primer sets and subsequent self-ligation of the two PCR products, exploiting a KpnI restriction site neighboring the 5’ end of the GFP coding sequence and putting in frame the first three codons of Pl-Tuba1a with GFP ORF.
All the corresponding Luc clones were prepared by replacing the GFP coding sequence via KpnI digestion, with the Luc coding sequence amplified from pXP1 plasmid (ATCC) with a proper 5’ KpnI modified primer set.
All the PCR amplifications were performed using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific), and resultant clones were sequenced to confirm correct insertion and frame maintenance.
The -1.8 Mutant clone was obtained via the QuickChange II Site-Directed Mutagenesis kit, following the manufacturer’s instructions (Agilent Technologies). The -1.8Kb clone was used as a DNA template with the primer set indicated in S1 Table.
Microinjection of constructs and reporter analysis
Sea urchin eggs were injected with 2 pl of a solution containing 5 ng/μl of linearized plasmid (GFP or Luc reporter) together with 5% Texas Red-conjugated dextran, 25 ng/μl carrier DNA (prepared by enzymatic digestion of P. lividus sperm DNA size selected to average length of 5 to 10 Kb), 1M KCl, and 20% glycerol, following the microinjection and embryo culture procedures previously described [23, 28, 29]. Each construct was microinjected at least in triplicate (almost 300 embryos microinjected/experiment) using different batches of sea urchin eggs. As negative controls, pBSK vectors containing GFP or Luc coding sequences were used.
At the desired developmental stage, injected embryos were harvested and, in the case of GFP reporter injected embryos, mounted on glass slides and examined under an epifluorescence Olympus BX50 microscope. Bright-field, GFP fluorescence, and Texas Red fluorescence images were captured with a Nikon digital camera and processed using the Nikon Nis-Elements software. Statistical analysis of expression patterns performed at the pluteus stage (where neurogenic territories are easily identified) showed GFP proper localization in approximately 90% of injected embryos, reflecting the way that exogenous DNA is incorporated in a mosaic fashion.
Luc assay was performed injecting about 300 eggs for every reporter construct. Embryos, immediately after reaching the gastrula stage, were preventively checked for Texas Red staining, counted and recovered in presence of CCLR lysis buffer at the ratio of 1μl to 1 injected embryo. Lysis was than executed as indicated in the Luciferase Assay System from the Promega Corporation. Samples corresponding to 5 μl of each lysate were read for light intensity after the addition of 100 μl of the Luciferase Assay Reagent included in the kit, with a typical delay time of 2 seconds and a read time of 10 seconds on a Promega GloMax-Multi Detection System. Each experiment was repeated in triplicate.
Results
Identification of promoter regulatory elements
In order to identify the minimum regulatory regions necessary and sufficient to drive the proper enhanced spatio/temporal expression of the Pl-Tuba1a gene (GenBank: JF272003), several GFP reporter constructs were generated starting from the recombinant full construct (Pl-Tuba1a-GFP) containing a genomic region from -4.5 Kb to +0.8 Kb. This genomic region contains the transcription start site (TSS), the first exon containing the 5’ UTR and including the ATG, the first intron and two codons of the second exon. Therefore, the construct encodes a fusion protein containing the amino terminal three alpha tubulin amino acids fused to GFP [26]. A series of 5’ shortened constructs was generated and used in gene transfer analysis assays (see S1 Fig in supplementary materials).
Using this approach, we found that the genomic sequences included between -1.8 Kb and +0.8 Kb (the -1.8KbGFP construct) are still able to drive the proper spatio-temporal expression of the Tub-GFP fusion protein. Indeed, GFP expression was detected from the blastula to pluteus stages and was strictly localized in the animal pole and ciliary band domains of the embryos, as previously described for the Pl-Tuba1a-GFP transgene [26]. Fig 1 shows the results of the gene transfer experiments performed with the -1.8Kb GFP construct herein selected as reference for the successive experiments and as template to produce further clones.
Fig 1. Expression of the -1.8KbGFP transgene construct during P. lividus embryo development.
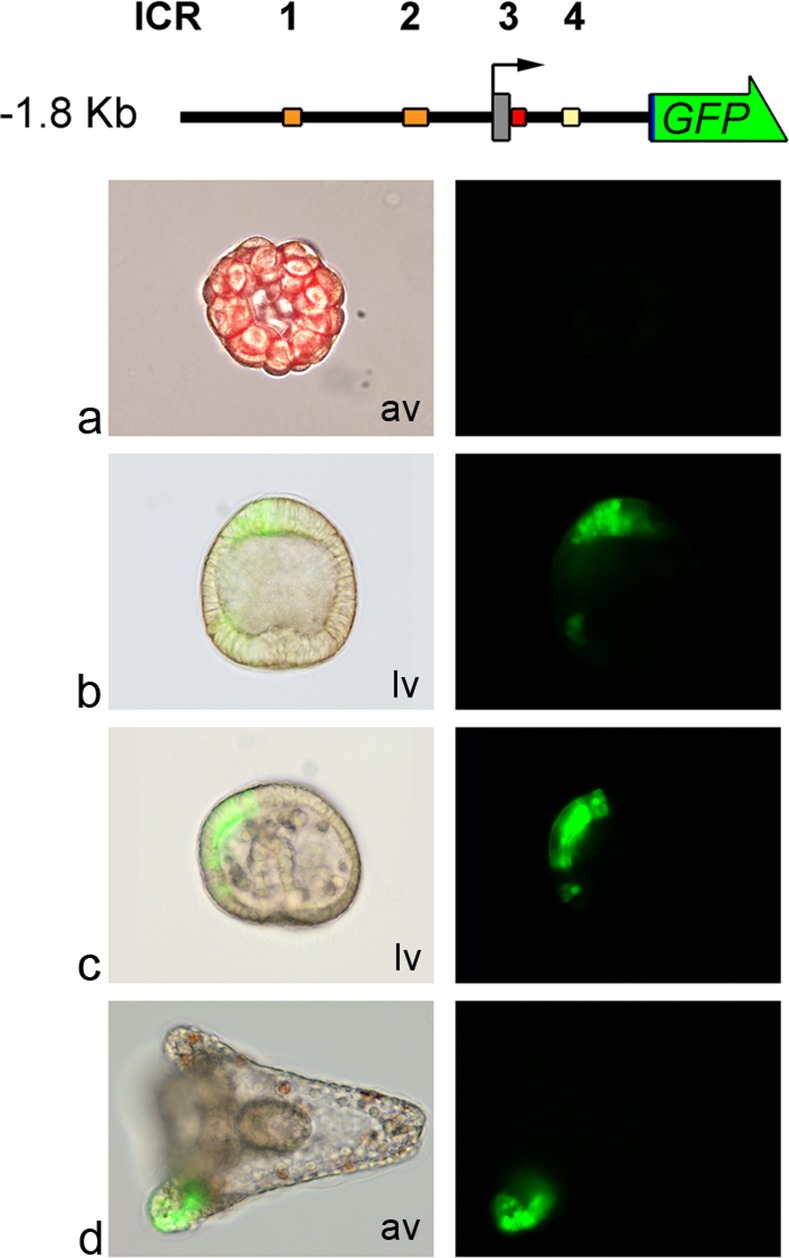
At the top is a schematic structure (drawn to scale) of the Pl-Tuba1a -1.8KbGFP reporter construct. The bent arrow indicates the TSS. A grey box represents the first exon (5’UTR and ATG start codon). Downstream of the first exon there are the first intron and two codons of the second exon. Coloured boxes indicate the four ICRs. For sake of simplicity, only the section of ICR3 inside the intron is shown. The arrowed green box represents the GFP reporter gene cloned in frame with the alpha tubulin codons. Left: triple-merged images (bright-field, GFP fluorescence and Texas Red fluorescence-a) or merged fluorescence and bright-field images (b, c, d). Right: GFP fluorescence images from microinjected embryos. × 20 magnification. (a) 32-cell stage; (b) Blastula stage; (c) Gastrula stage; (d) Pluteus stage. Lv: lateral view; av: animal view.
Interestingly, this region contains four of the six ICRs which were previously identified by an in silico approach. Two of them, ICR1 (approximately 100 bp, between -1200 and -1100 bp) and ICR2 (130 bp, between -510 and -380 bp), are located in the 5’ upstream region; ICR3 (about 240 bp, located between -50 and + 190 bp) contains the TATA box, the transcript leader sequence and a portion of the first intron sequence; ICR4 (located between +400 and +500 bp) is contained in the first intron [26].
Functional analysis of interspecific conserved regions located upstream of the TSS
In order to understand the functional role of each ICR, we performed additional gene transfer experiments; thus, other constructs lacking ICR1 or ICR1-2 were microinjected. These constructs provided similar GFP expression patterns, and results are reported in S1 Fig.
GFP expression was properly detected from the blastula stage (data not shown) to the pluteus stage, and its expression was restricted to the proper territories. Therefore, their removal does not affect spatio/temporal regulation. However, it appears that a progressive reduction of GFP expression occurred after each ICR deletion with respect to the -1.8KbGFP transgene.
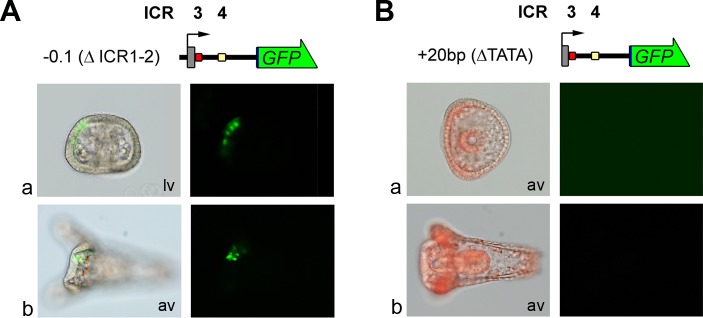
Further deletions, including -0.1(ΔICR1-2)GFP which contains solely the TATA box and the first intron, did not result in any change in spatio/temporal regulation (Fig 2A).
Fig 2. Transgene basal expression and loss of expression by TATA box deletion.
Gene transfer assay results performed using the -0.1(ΔICR1-2)GFP (A) and the +20(ΔTATA)GFP (B) constructs. Structure and conventions are the same as in Fig 1. (a) Gastrula stage; (b) Pluteus stage. (A) Merged fluorescence and bright-field images (left) and GFP fluorescence images (right) from microinjected embryos are shown. (B) Triple-merged bright-field, GFP fluorescence and Texas Red fluorescence images (left) proving embryos are microinjected and GFP fluorescence images (right). × 20 magnification. Lv: lateral view; av: animal view.
Conversely, different results were obtained when the TATA box was removed. Indeed, embryos microinjected with the +20bp(ΔTATA)GFP construct did not show any GFP expression (Fig 2B).
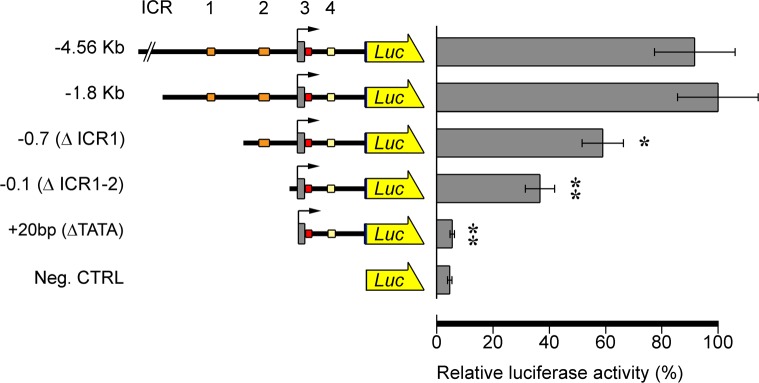
To confirm all of these qualitative results, we performed quantitative targeted gene transfer assays using the luciferase reporter gene. Progressive 5’ deletions, from the recombinant clone Pl-Tuba1a-Luc to -1.8Kb (-1.8KbLuc construct) did not show any statistically significant difference, confirming that regulatory modules are not located in the region between -4.5 and -1.8 Kb (Fig 3). Differently, microinjections performed using the -0.7(ΔICR1)Luc construct showed a significant reduction of luciferase activity with respect to -1.8KbLuc (about 60% residual relative luciferase activity). A very large reduction was measured after microinjecting the -0.1(ΔICR1-2)Luc construct (about 40% residual relative luciferase activity, see Fig 3). Otherwise, gene transfer assays performed by microinjecting the +20bp(ΔTATA)Luc construct showed luciferase activity similar to negative controls (Fig 3).
Fig 3. Enhancer functions of ICR1 and ICR2.
Upstream deletions: luciferase activity progressively decreases after ICR1 and ICR2 sequential deletions. Left: schematic pictures of luciferase reporter constructs. Structure and conventions are the same as in Fig 1, except that the arrowed yellow box represents the Luc reporter gene. Right: luciferase activity measured at gastrula stage (24 hours post fertilization-hpf). The activity of the -1.8KbLuc construct is defined as 100%. Data are expressed as means ± standard deviation (SD) of triplet measurements of at least three independent experiments. Asterisks denote statistical significance: ** P value ≤ 0.0021, * P value = 0.0122.
All these results suggest that ICR1 and 2 may play a role as enhancers of Pl-Tuba1a gene expression, also confirming the functional role of the TATA box. Additionally, they provide evidence of the presence of regulatory modules in the first intron which may act activating the expression in the neurogenic territories.
Functional analysis of interspecific conserved regions in the intron
While our results indicated that the first intron may contain regulatory modules activating and/or enhancing Pl-Tuba1a expression, it is also true that gene regulation is often controlled by quantitative, temporal and/or spatial regulatory modules that may be variously organized and potentially overlapping. Thus, we continued to use the double GFP/Luc approach.
Initially, in order to study the functional role of the intron, we performed gene transfer experiments using defective constructs in the whole first intron (from +96 to +898), which is localized after the tubulin start codon. Although this -1.8(ΔIntron)GFP construct encodes the tub-GFP fusion protein without splicing, microinjected embryos did not show any reporter expression during development, as shown in Figs 4 and 5. This result was confirmed by corresponding -1.8(ΔIntron)Luc construct microinjections (Fig 6). Indeed, intron removal dramatically affected luciferase activity, with results similar to negative controls.
Fig 4. Transgene loss of expression by intron deletion.
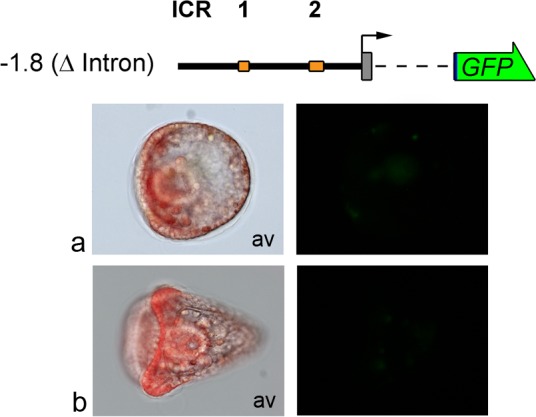
Embryos were microinjected with the -1.8(ΔIntron)GFP construct. Left: triple-merged images (bright-field, GFP fluorescence and Texas Red fluorescence) proving embryos are microinjected. Right: GFP fluorescence images. Structure and conventions are the same as in Fig 1. (a) Gastrula stage; (b) Pluteus stage. Av: animal view.
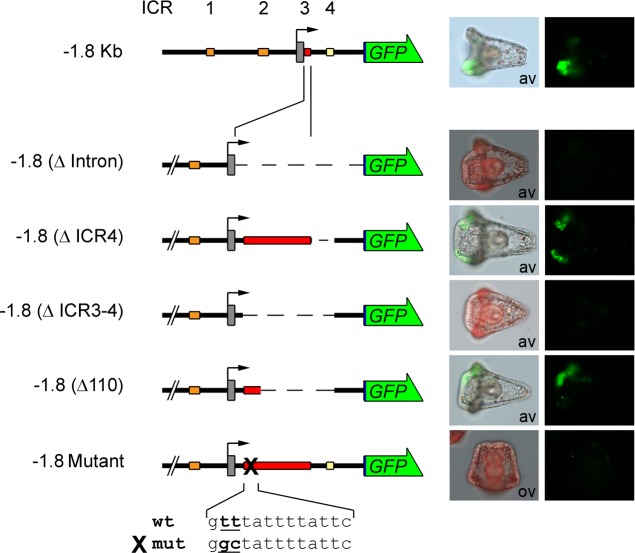
Fig 5. ICR3 and ICR4 function: spatial expression.
In these construct depictions, ICR3 was enlarged to show deletion details (not to scale). Left: schematic pictures of GFP reporter constructs. Structure and conventions are the same as in Fig 1. Right: merged fluorescence and bright-field images or triple-merged images and fluorescence images from microinjected pluteus stage embryos are shown to observe GFP localization. × 20 magnification. Av: animal view; ov: oral view.
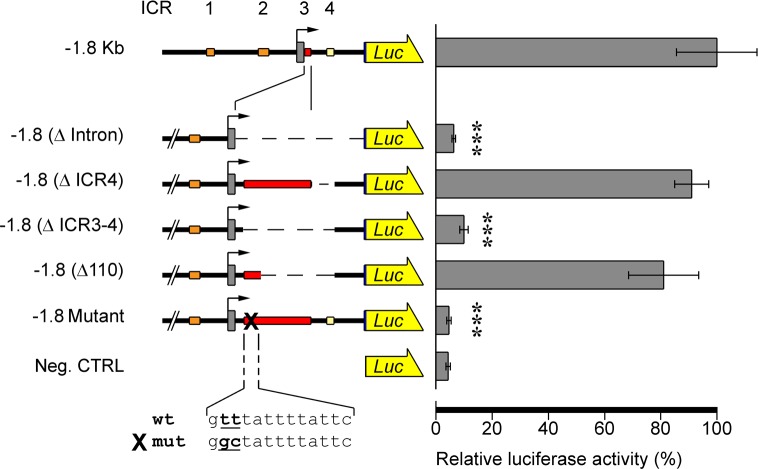
Fig 6. ICR3 and ICR4 function: quantitative analysis.
In these construct depictions, ICR3 was enlarged to show deletion details (not to scale). Left: schematic pictures of luciferase reporter constructs. Structure and conventions are the same as in Fig 2. Right: luciferase activity measured at gastrula stage (24 hpf). The activity of the -1.8KbLuc construct is defined as 100%. Data are expressed as means ± SD of triplet measurements of at least three independent experiments. Asterisks denote statistical significance: *** P value < 0.0005.
Interestingly, the intron contains ICR3 and ICR4; therefore, to validate their involvement in Pl-Tuba1a regulation, we dissected intron sequences by microinjecting several GFP constructs lacking one or both ICRs. Obviously, because the intron contains important elements for transcript maturation whose mishandling could impair the proper functionality of the process, we took care to maintain all of the splice sites (5′ donor, 3′ acceptor and branch point) in the planning of the constructs.
Embryos microinjected with the -1.8(ΔICR4)GFP (deletion from +206 to +483) showed reporter expression similar to -1.8KbGFP, used as a positive control. Indeed, fluorescence appeared from the blastula stage (data not shown) to the pluteus stage, when it was localized in the ciliary band and the ganglia (Fig 5) as the endogenous transcript. Similarly, luciferase activity from embryos expressing -1.8(ΔICR4)Luc was comparable to the activity measured when the -1.8KbLuc construct was used (Fig 6). Thus, it is reasonable to suppose that the critical regulatory sequences are located in ICR3.
ICR3 contains the TATA box (from -29 bp to -22 bp), the first exon (from +1 bp to +95 bp) and 95 bp of the intron sequence. We have already demonstrated the pivotal role of the TATA box; additionally, an in silico analysis for transcription factor (TF) binding sites suggested the presence of putative consensus sequences located between +100 and +190 bp. Therefore, we extended intron deletion to +100 bp (deletion from +100 to +483), obtaining the -1.8(ΔICR3-4)GFP/Luc constructs.
The -1.8(ΔICR3-4)GFP injected embryos failed to express GFP in a manner that resembled whole intron deletion (Fig 5). The same results were obtained from the quantitative analysis of the corresponding Luc assays (Fig 6).
These results, then, suggest a transcriptional regulatory role for the region from +100 bp to +206 (ICR3 core).
Detailed analysis of the ICR3 core and site-directed mutagenesis
In order to analyze the ICR3 core, new deletion constructs were built. In particular, microinjected constructs named -1.8(Δ110)GFP/Luc (deletion from +110 to +483), containing only 10 bp in addition to the non-functional -1.8(ΔICR3-4) construct, again showed the ability to drive the expression of the reporter genes in the apical organ and in the ciliary band (see Figs 5 and 6). Thus, we looked for a putative binding site of TFs located in the sequence near +100 bp. Both the Ncx and forkhead box (Fox) classes of TFs (best match of FoxD3 matrix is shown in Fig 7) could bind to this region. After a detailed match analysis (i.e. position, score), we decided to mutagenize two thymine residues in the Fox binding site. Therefore, by site-specific mutagenesis, we obtained a mutant clone (-1.8 Mutant) which differed from the wild-type by two base changes at positions 101–102 (T101 to G and T102 to C). Embryos microinjected with the mutant construct showed neither GFP expression nor luciferase activity (see Figs 5 and 6). Thus, these results suggest that this binding site is necessary to drive the proper expression of the Pl-Tuba1a gene.
Fig 7. Transcription factor binding sites in the sequence necessary for proper Pl-Tuba1a expression.
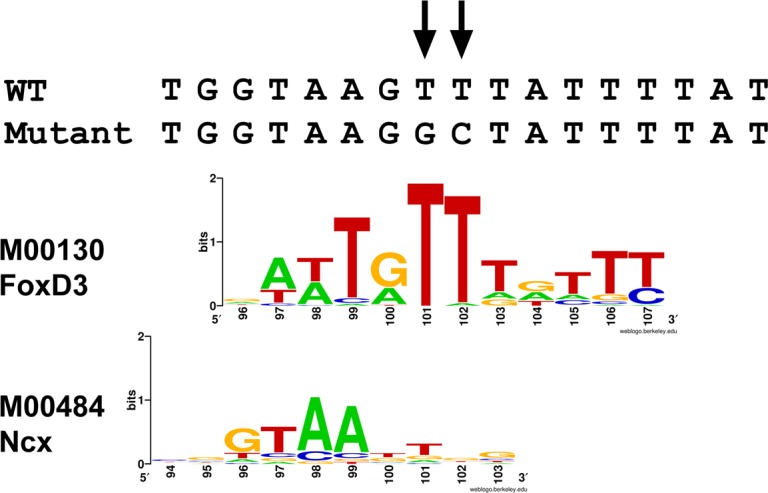
Top: wild-type and mutant construct sequences from +94 to +110 bp. Arrows indicate specific nucleotide changes generated by site-directed mutagenesis (T101 to G and T102 to C). Center and bottom: FoxD3 matrix logo (M00130) and Ncx (M00484) matrix logo aligned to sequences. Logos were generated using WebLogo [30].
Conclusions
The sea urchin embryo is an excellent system for identifying the regulatory activities that coordinate neurogenesis events. It has also been shown that the gene products essential for neuroectoderm patterning in the deuterostome lineage, which includes the sea urchin, similarly act in other organisms and are also necessary for patterning the nervous system in chordates [31–33].
At the beginning of sea urchin embryogenesis, two neurogenic regions are specified by separate signals, including Wnt and TGFβ: the ANE and the more posterior CBE [12, 34]. ANE contains serotonergic and non-serotonergic neurons, as well as some cells that produce long, immotile cilia which might have a sensory function, whereas the CBE is predominantly composed of specialized ciliated cells and functions as a swimming and feeding organ. Interestingly, the apical organ was recently considered the central integrating component of the nervous system and the ciliary band a peripheral component that responds to stimuli and controls ciliary activity [9]. The Pl-Tuba1a gene, encoding an alpha tubulin isotype that might be used for primary cilia axoneme and for axon microtubules, is expressed in both the ANE and CBE regions [22].
In this study, we show that a genomic region of about 2.6 Kb of Pl-Tuba1a can recreate the spatio-temporal pattern of endogenous Pl-Tuba1a gene expression. It is well known that the analysis of ICRs is a useful tool for investigating cis-regulatory elements [35]. Thus, in order to look for cis-regulatory sequences in the Pl-Tuba1a gene, we have previously compared P. lividus and Strongylocentrotus purpuratus tuba1a orthologues. Using this strategy, we found six ICRs, and here we show that three of them have a regulatory role [26].
In particular, the data presented in this paper show that proper Pl-Tuba1a gene expression is enhanced by two upstream regions (ICR1 and 2) containing several TF binding sites, including a Fox consensus sequence.
The removal of ICR1 and 2 causes a decrease in Pl-Tuba1a expression level, suggesting that these modules may act by boosting Pl-Tuba1a gene expression in the ANE and CBE regions of the sea urchin embryo. Moreover, Pl-Tuba1a expression depends on a very short sequence spanning the transcription start site that contains a TATA box and a binding site for some members of the Fox transcriptional regulator family (FoxD, Q, J, etc.) whose consensus binding sequences are extremely similar [36]. Fox TFs bind DNA through a winged helix domain and play a pivotal role in differentiation and specification events, including the maturation process of neurons [37].
Fox TFs have also been extensively studied in other systems (mouse, frog, zebrafish, fruit fly). For instance, it has been demonstrated that FoxJ1 is required to regulate a cohort of ciliary genes to make motile cilia [38, 39], and that FoxD3 is required for neural crest specification [40, 41]. FoxA1 participates in stimulating the neuronal differentiation of pluripotent stem cells and activates the neural betaIII tubulin gene [42]; moreover, FoxA1 and FoxA2 are crucial for the specification, differentiation and maintenance of dopamine neurons during mouse embryonic development [43].
Interestingly, FoxD3 and FoxA1 are believed to scan chromatin for enhancers with forkhead motifs and to trigger their transcriptional competency through initial chromatin decompaction. According to this specific role, they have been designated “pioneer” TFs [36, 44]. Notably, FoxD3 pioneer factor occupancy can modulate the local epigenetic state of chromatin, serving as a placeholder until the later appearance of FoxA1 during mouse gastrulation [45].
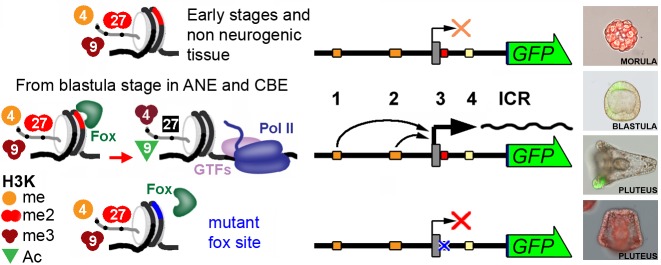
It is noteworthy that a putative Fox binding site is located in ICR3 of Pl-Tuba1a and that its removal (by deletion or mutagenesis) impairs expression, suggesting a fundamental role for a Fox family member for Pl-Tuba1a activation during neural structure development.
In the purple sea urchin (S. purpuratus), 22 fox genes have been identified, and it has been shown that foxD, foxJ1 and foxQ2 are expressed in all or in a subset of cells derived from what is initially the animal pole of the embryo, and in the ciliary band by the end of gastrulation [43, 44]. In particular, FoxQ2, that at the gastrula stage is an animal pole marker, plays an essential role in the formation of the apical tuft cilia and is required for the development of serotonergic neurons [45–51]. Moreover, foxG (also known as Brain factor1) is expressed in the periphery of the ventral ectoderm at mesenchyme blastula, then strictly in the ciliary band [52].The finding that FoxD3 performs a priming activity is in good agreement with the previously demonstrated existence of developmentally dependent changes of the Pl-Tuba1a gene epitype. We have indeed shown that the Pl-Tuba1a gene begins to be expressed only when specific histone modifications (H3K9Ac, H3K4me3) induce an accessible chromatin conformation of the core promoter [27].
Finally, it is remarkable that the expression profile of Pl-Tuba1a seems to overlap with that of foxJ1/Q2 and foxG, suggesting a possible sequential enhancement/priming role of these factors in the sequential Pl-tuba1a expression during the differentiation of the animal plate and ciliary band.
Therefore, it could be hypothesized that the binding site in ICR3 likely represents a holder platform for a Fox pioneer factor inducing both early specific neurogenic activation and later expression maintenance. Accordingly, a model in which FoxQ2 initiates an anterior patterning centre that implements correct size and positions of ANE structures was recently proposed [53]. Additionally, after chromatin remodeling and increased accessibility, Fox member(s) bound to ICR2 might enhance Pl-Tuba1a gene expression (Fig 8).
Fig 8. A summary of the transcription regulation of the Pl-Tuba1a gene.
Chromatin modifications occurring on the Pl-Tuba1a promoter, according to [27] are shown. Transgenic wild type and mutagenized constructs and their expression during embryo development are also shown. See text for details.
Supporting Information
Underlined sequences indicates restriction enzyme recognition sites.
(DOCX)
At left is a schematic structure (drawn to scale) of the Pl-Tuba1a-GFP reporter constructs. The bent arrow indicates the transcription start site. A grey box represents the first exon (5’UTR and ATG start codon). Downstream of the first exon there are the first intron and two codons of the second exon. Coloured boxes indicate the four ICRs. For sake of simplicity, only the section/segment of ICR3 inside the intron is shown. The arrowed green box represents the GFP reporter gene cloned in frame with the alpha tubulin codons. At right: (left column) merged fluorescence and bright-field images or triple-merged images (bright-field, GFP fluorescence and Texas Red fluorescence, last construct); (right column) GFP fluorescence images from microinjected embryos (animal views). × 20 magnification.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Università degli Studi di Palermo FFR - EX60% [grant numbers 4201/2007 to FG and 3098/2012 to MAR]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C-H, et al. A Genomic Regulatory Network for Development. Science. 2002. March 1;295(5560):1669–78. 10.1126/science.1069883 [DOI] [PubMed] [Google Scholar]
- 2.Ochoa-Espinosa A, Small S. Developmental mechanisms and cis-regulatory codes. Current Opinion in Genetics & Development. 2006. April;16(2):165–70. [DOI] [PubMed] [Google Scholar]
- 3.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. PNAS. 2008. December 23;105(51):20063–6. 10.1073/pnas.0806007105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yáñez-Cuna JO, Kvon EZ, Stark A. Deciphering the transcriptional cis-regulatory code. Trends Genet. 2013. January;29(1):11–22. 10.1016/j.tig.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 5.Li E, Materna SC, Davidson EH. New regulatory circuit controlling spatial and temporal gene expression in the sea urchin embryo oral ectoderm GRN. Dev Biol. 2013. October 1;382(1):268–79. 10.1016/j.ydbio.2013.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Tabou de-Leon S, Su Y-H, Lin K-T, Li E, Davidson EH. Gene regulatory control in the sea urchin aboral ectoderm: spatial initiation, signaling inputs, and cell fate lockdown. Dev Biol. 2013. February 1;374(1):245–54. 10.1016/j.ydbio.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barsi JC, Li E, Davidson EH. Geometric control of ciliated band regulatory states in the sea urchin embryo. Development. 2015. March 1;142(5):953–61. 10.1242/dev.117986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yankura KA, Koechlein CS, Cryan AF, Cheatle A, Hinman VF. Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. PNAS. 2013. May 21;110(21):8591–6. 10.1073/pnas.1220903110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner S, Zysk I, Byrne G, Kramer M, Moller D, Taylor V, et al. Neurogenesis in sea urchin embryos and the diversity of deuterostome neurogenic mechanisms. Development. 2016. January 15;143(2):286–97. 10.1242/dev.124503 [DOI] [PubMed] [Google Scholar]
- 10.Israel JW, Martik ML, Byrne M, Raff EC, Raff RA, McClay DR, et al. Comparative Developmental Transcriptomics Reveals Rewiring of a Highly Conserved Gene Regulatory Network during a Major Life History Switch in the Sea Urchin Genus Heliocidaris. PLoS Biol. 2016. March;14(3):e1002391 10.1371/journal.pbio.1002391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakajima Y, Kaneko H, Murray G, Burke RD. Divergent patterns of neural development in larval echinoids and asteroids. Evol Dev. 2004. April;6(2):95–104. [DOI] [PubMed] [Google Scholar]
- 12.Yaguchi S, Yaguchi J, Burke RD. Specification of ectoderm restricts the size of the animal plate and patterns neurogenesis in sea urchin embryos. Development. 2006. June;133(12):2337–46. 10.1242/dev.02396 [DOI] [PubMed] [Google Scholar]
- 13.Angerer LM, Yaguchi S, Angerer RC, Burke RD. The evolution of nervous system patterning: insights from sea urchin development. Development. 2011. September;138(17):3613–23. 10.1242/dev.058172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapraz F, Haillot E, Lepage T. A deuterostome origin of the Spemann organiser suggested by Nodal and ADMPs functions in Echinoderms. Nat Commun. 2015;6:8434 10.1038/ncomms9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Z, Angerer LM, Angerer RC. Neurogenic gene regulatory pathways in the sea urchin embryo. Development. 2016. January 15;143(2):298–305. 10.1242/dev.125989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop CD, MacNeil KEA, Patel D, Taylor VJ, Burke RD. Neural development in Eucidaris tribuloides and the evolutionary history of the echinoid larval nervous system. Dev Biol. 2013. May 1;377(1):236–44. 10.1016/j.ydbio.2013.03.006 [DOI] [PubMed] [Google Scholar]
- 17.Byrne M, Nakajima Y, Chee FC, Burke RD. Apical organs in echinoderm larvae: insights into larval evolution in the Ambulacraria. Evol Dev. 2007. October;9(5):432–45. 10.1111/j.1525-142X.2007.00189.x [DOI] [PubMed] [Google Scholar]
- 18.Hirokawa T, Komatsu M, Nakajima Y. Development of the nervous system in the brittle star Amphipholis kochii. Dev Genes Evol. 2008. January;218(1):15–21. 10.1007/s00427-007-0196-6 [DOI] [PubMed] [Google Scholar]
- 19.Murabe N, Hatoyama H, Hase S, Komatsu M, Burke RD, Kaneko H, et al. Neural architecture of the brachiolaria larva of the starfish, Asterina pectinifera. J Comp Neurol. 2008. July 20;509(3):271–82. 10.1002/cne.21742 [DOI] [PubMed] [Google Scholar]
- 20.Nakano H, Nakajima Y, Amemiya S. Nervous system development of two crinoid species, the sea lily Metacrinus rotundus and the feather star Oxycomanthus japonicus. Dev Genes Evol. 2009. December;219(11–12):565–76. 10.1007/s00427-010-0317-5 [DOI] [PubMed] [Google Scholar]
- 21.Nakano H, Murabe N, Amemiya S, Nakajima Y. Nervous system development of the sea cucumber Stichopus japonicus. Dev Biol. 2006. April 1;292(1):205–12. 10.1016/j.ydbio.2005.12.038 [DOI] [PubMed] [Google Scholar]
- 22.Gianguzza F, Casano C, Ragusa M. Alpha-tubulin marker gene of neural territory of sea urchin embryos detected by whole-mount in situ hybridization. Int J Dev Biol. 1995. June;39(3):477–83. [PubMed] [Google Scholar]
- 23.Costa S, Ragusa MA, Drago G, Casano C, Alaimo G, Guida N, et al. Sea urchin neural alpha2 tubulin gene: isolation and promoter analysis. Biochem Biophys Res Commun. 2004. April 2;316(2):446–53. 10.1016/j.bbrc.2004.02.070 [DOI] [PubMed] [Google Scholar]
- 24.Wei Z, Angerer LM, Angerer RC. Neurogenic gene regulatory pathways in the sea urchin embryo. Development. 2016. January 15;143(2):298–305. 10.1242/dev.125989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casano C, Ragusa M, Cutrera M, Costa S, Gianguzza F. Spatial expression of alpha and beta tubulin genes in the late embryogenesis of the sea urchin Paracentrotus lividus. Int J Dev Biol. 1996. October;40(5):1033–41. [PubMed] [Google Scholar]
- 26.Ragusa MA, Longo V, Emanuele M, Costa S, Gianguzza F. In silico characterization of the neural alpha tubulin gene promoter of the sea urchin embryo Paracentrotus lividus by phylogenetic footprinting. Mol Biol Rep. 2012. March;39(3):2633–44. 10.1007/s11033-011-1016-7 [DOI] [PubMed] [Google Scholar]
- 27.Emanuele M, Costa S, Ragusa MA, Gianguzza F. Chromatin dynamics of the developmentally regulated P. lividus neural alpha tubulin gene. Int J Dev Biol. 2011;55(6):591–6. 10.1387/ijdb.103264me [DOI] [PubMed] [Google Scholar]
- 28.McMahon AP, Flytzanis CN, Hough-Evans BR, Katula KS, Britten RJ, Davidson EH. Introduction of cloned DNA into sea urchin egg cytoplasm: replication and persistence during embryogenesis. Dev Biol. 1985. April;108(2):420–30. [DOI] [PubMed] [Google Scholar]
- 29.Arnone MI, Dmochowski IJ, Gache C. Using reporter genes to study cis-regulatory elements. Methods Cell Biol. 2004;74:621–52. [DOI] [PubMed] [Google Scholar]
- 30.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004. June;14(6):1188–90. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burke RD, Angerer LM, Elphick MR, Humphrey GW, Yaguchi S, Kiyama T, et al. A genomic view of the sea urchin nervous system. Dev Biol. 2006. December 1;300(1):434–60. 10.1016/j.ydbio.2006.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke RD, Moller DJ, Krupke OA, Taylor VJ. Sea urchin neural development and the metazoan paradigm of neurogenesis. Genesis. 2014. March;52(3):208–21. [DOI] [PubMed] [Google Scholar]
- 33.Range R. Specification and positioning of the anterior neuroectoderm in deuterostome embryos. Genesis. 2014. March;52(3):222–34. 10.1002/dvg.22759 [DOI] [PubMed] [Google Scholar]
- 34.Yaguchi S, Yaguchi J, Angerer RC, Angerer LM, Burke RD. TGFβ signaling positions the ciliary band and patterns neurons in the sea urchin embryo. Dev Biol. 2010. November 1;347(1):71–81. 10.1016/j.ydbio.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper GM, Sidow A. Genomic regulatory regions: insights from comparative sequence analysis. Curr Opin Genet Dev. 2003. December;13(6):604–10. [DOI] [PubMed] [Google Scholar]
- 36.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011. November 1;25(21):2227–41. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stott SRW, Metzakopian E, Lin W, Kaestner KH, Hen R, Ang S-L. Foxa1 and foxa2 are required for the maintenance of dopaminergic properties in ventral midbrain neurons at late embryonic stages. J Neurosci. 2013. May 1;33(18):8022–34. 10.1523/JNEUROSCI.4774-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas J, Morlé L, Soulavie F, Laurençon A, Sagnol S, Durand B. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol Cell. 2010. September;102(9):499–513. 10.1042/BC20100035 [DOI] [PubMed] [Google Scholar]
- 39.Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014. April;141(7):1427–41. 10.1242/dev.074666 [DOI] [PubMed] [Google Scholar]
- 40.Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001. April;128(8):1467–79. [DOI] [PubMed] [Google Scholar]
- 41.Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, et al. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006. April 1;292(1):174–88. 10.1016/j.ydbio.2005.12.035 [DOI] [PubMed] [Google Scholar]
- 42.Tan Y, Xie Z, Ding M, Wang Z, Yu Q, Meng L, et al. Increased levels of FoxA1 transcription factor in pluripotent P19 embryonal carcinoma cells stimulate neural differentiation. Stem Cells Dev. 2010. September;19(9):1365–74. 10.1089/scd.2009.0386 [DOI] [PubMed] [Google Scholar]
- 43.Domanskyi A, Alter H, Vogt MA, Gass P, Vinnikov IA. Transcription factors Foxa1 and Foxa2 are required for adult dopamine neurons maintenance. Front Cell Neurosci. 2014;8:275 10.3389/fncel.2014.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelms BL, Labosky PA. Transcriptional Control of Neural Crest Development. Colloquium Series on Developmental Biology. 2010. January 1;1(1):1–227. [PubMed] [Google Scholar]
- 45.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009. December 15;23(24):2824–38. 10.1101/gad.1861209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol. 2006. December 1;300(1):49–62. 10.1016/j.ydbio.2006.09.031 [DOI] [PubMed] [Google Scholar]
- 47.Poustka AJ, Kühn A, Groth D, Weise V, Yaguchi S, Burke RD, et al. A global view of gene expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol. 2007;8(5):R85 10.1186/gb-2007-8-5-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn EF, Moy VN, Angerer LM, Angerer RC, Morris RL, Peterson KJ. Molecular paleoecology: using gene regulatory analysis to address the origins of complex life cycles in the late Precambrian. Evol Dev. 2007. February;9(1):10–24. 10.1111/j.1525-142X.2006.00134.x [DOI] [PubMed] [Google Scholar]
- 49.Yaguchi S, Yaguchi J, Angerer RC, Angerer LM. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell. 2008. January;14(1):97–107. 10.1016/j.devcel.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 50.Range RC, Angerer RC, Angerer LM. Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol. 2013;11(1):e1001467 10.1371/journal.pbio.1001467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yaguchi J, Takeda N, Inaba K, Yaguchi S. Cooperative Wnt-Nodal Signals Regulate the Patterning of Anterior Neuroectoderm. PLoS Genet. 2016. April;12(4):e1006001 10.1371/journal.pgen.1006001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saudemont A, Haillot E, Mekpoh F, Bessodes N, Quirin M, Lapraz F, et al. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 2010;6(12):e1001259 10.1371/journal.pgen.1001259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Range RC, Wei Z. An anterior signaling center patterns and sizes the anterior neuroectoderm of the sea urchin embryo. Development. 2016. May 1;143(9):1523–33. 10.1242/dev.128165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Underlined sequences indicates restriction enzyme recognition sites.
(DOCX)
At left is a schematic structure (drawn to scale) of the Pl-Tuba1a-GFP reporter constructs. The bent arrow indicates the transcription start site. A grey box represents the first exon (5’UTR and ATG start codon). Downstream of the first exon there are the first intron and two codons of the second exon. Coloured boxes indicate the four ICRs. For sake of simplicity, only the section/segment of ICR3 inside the intron is shown. The arrowed green box represents the GFP reporter gene cloned in frame with the alpha tubulin codons. At right: (left column) merged fluorescence and bright-field images or triple-merged images (bright-field, GFP fluorescence and Texas Red fluorescence, last construct); (right column) GFP fluorescence images from microinjected embryos (animal views). × 20 magnification.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.