Abstract
Background
APOBEC3B was recently identified as a gain-of-function enzymatic source of mutagenesis, which may offer novel therapeutic options with molecules that specifically target this enzyme. In primary breast cancer, APOBEC3B mRNA is deregulated in a substantial proportion of cases and its expression is associated with poor prognosis. However, its expression in breast cancer metastases, which are the main causes of breast cancer-related death, remained to be elucidated.
Patients and methods
RNA was isolated from 55 primary breast cancers and paired metastases, including regional lymph node (N = 20) and distant metastases (N = 35). APOBEC3B mRNA levels were measured by RT-qPCR. Expression levels of the primary tumors and corresponding metastases were compared, including subgroup analysis by estrogen receptor (ER/ESR1) status.
Results
Overall, APOBEC3B mRNA levels of distant metastases were significantly higher as compared to the corresponding primary breast tumor (P = 0.0015), an effect that was not seen for loco-regional lymph node metastases (P = 0.23). Subgroup analysis by ER-status showed that increased APOBEC3B levels in distant metastases were restricted to metastases arising from ER-positive primary breast cancers (P = 0.002). However, regarding ER-negative primary tumors, only loco-regional lymph node metastases showed increased APOBEC3B expression when compared to the corresponding primary tumor (P = 0.028).
Conclusion
APOBEC3B mRNA levels are significantly higher in breast cancer metastases as compared to the corresponding ER-positive primary tumors. This suggests a potential role for APOBEC3B in luminal breast cancer progression, and consequently, a promising role for anti-APOBEC3B therapies in advanced stages of this frequent form of breast cancer.
Highlights
APOBEC3B is a gain-of-function enzymatic source of mutagenesis.
Levels are higher in breast metastases as compared to corresponding primary tumors.
This implies a novel role for APOBEC3B during breast cancer progression.
This makes APOBEC3B a promising target for anti-APOBEC3B therapies.
Especially in advanced stages of breast cancer.
Introduction
Breast cancer is the fifth cause of overall cancer related death [1] and this mortality is largely caused by progression of metastatic disease [2]. Therefore, one of the most important challenges in breast cancer research includes the genetic changes and molecular mechanisms by which cancer cells acquire their metastatic ability. The generally accepted hypothesis is that metastases are caused by multiple intricate steps that arise in the primary tumor site [3]. Nevertheless, discordances between primary tumors and corresponding metastases are often encountered [4]. However, therapies applied for disseminated disease are mainly based on primary tumor characteristics only. The study of molecular differences between matched primary tumors and metastatic lesions may improve our understanding of disease progression and has the potential to reveal novel, potentially targetable drivers of metastatic progression.
Apolipoprotein B mRNA Editing Enzyme, Catalytic Polypeptide-Like 3B (APOBEC3B) is a member of the AID/APOBEC family of deaminases, which is recognized for its ability to deaminate genomic DNA cytosines. APOBEC enzymes normally function in the innate immune system and in the protection against viral pathogens, but these enzymes can also generate C→T mutations in the host genome [5]. Recently, several studies showed that APOBEC3B is a common enzymatic mutagenic factor affecting the evolution of different cancer types, including breast cancer [5–19].
In breast cancer, APOBEC3B mRNA is substantially upregulated in one third of cases and its expression is associated with mutational load, including certain driver mutations in PIK3CA and TP53 [18,20]. Besides, multiple studies have postulated that APOBEC3B influences the development of metastases and drug resistance, especially in estrogen receptor alpha (ERα)-positive breast cancer [5,21,22]. In line with this, we previously reported an association between high APOBEC3B mRNA expression and poor outcome in a large cohort of patients with ERα-positive breast cancer [23].
Since APOBEC3B is a gain-of-function mutagenic enzyme, it may be treatable with small molecules [5,24], which could have an important role in the management of metastatic disease. However, the expression of APOBEC3B in breast cancer metastases remained to be elucidated.
In this study, we therefore quantified APOBEC3B mRNA in primary breast cancers and paired metastases to gain more insight into the levels of expression during breast cancer progression.
Materials and methods
Clinical pathological data
In this study we adhered to the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands (http://www.fmwv.nl) and the study making secondary use of human materials has been approved by our ‘Medische Ethnische Toetsing Commissie’ (METC; MEC 02.953). The use of anonymous or coded left over material for scientific purposes is part of the standard treatment agreement with patients and therefore informed consent was not required according to Dutch law [25]. We selected 73 formalin-fixed paraffin-embedded (FFPE) primary breast cancers and corresponding metastases from the pathology archives of the University Medical Center Utrecht and Erasmus Medical Center Rotterdam. Each specimen was reviewed by a pathologist to determine the percentage of invasive tumor cells. Inclusion criteria were: availability of clinical and pathological data, the presence of enough tumor tissue with the possibility to macro-dissect an area containing at least 50% tumor cells and good RNA quality and quantity to reliably determine expression levels by RT-qPCR (see also below). After applying these inclusion criteria, 55 paired primary tumors and metastases from different sites remained, including those from regional lymph nodes (N = 20), brain (N = 14), liver (N = 6), ovary (N = 4), lung (N = 4), bone (N = 4) and gastrointestinal tract (N = 3). Clinicopathological characteristics included age, primary tumor size, histological subtype, Bloom & Richardson score, ERα and human epidermal growth factor receptor 2 (HER2) expression and regional lymph node status. Furthermore, overall survival (death due to any cause) was reported. Detailed clinical information of this cohort is summarized in Table 1.
Table 1. Association of APOBEC3B mRNA expression with clinicopathological characteristics of the primary tumor.
| APOBEC3B mRNA log2 | AVG epithelial mRNA log2 | PTPRC (CD45) mRNA log2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Clinical characteristics | No of patients* | Percentage of patients | Median | IQR | Median | IQR | Median | IQR≠ |
| All patients in this cohort | 55 | 100% | -6.14 | -5.28 | -3.11 | -1.25 | -2.98 | -2.31 |
| Age at surgery (years) | ||||||||
| ≤ 40 | 10 | 18% | -6.69 | -4.62 | -2.74 | -1.40 | -2.31 | -2.89 |
| 41–55 | 21 | 38% | -5.80 | -5.95 | -3.23 | -1.46 | -3.14 | -1.41 |
| 56–70 | 20 | 36% | -6.64 | -4.16 | -3.03 | -1.25 | -3.48 | -7.68 |
| > 70 | 5 | 9% | -6.04 | -2.08 | -2.93 | -0.54 | -2.40 | -1.09 |
| P≠ | 0.76 | 0.52 | 1.00 | |||||
| Tumor size | ||||||||
| ≤ 2 cm | 17 | 31% | -6.14 | -6.31 | -3.03 | -0.94 | -2.72 | -2.28 |
| 2 ≤ 5 cm | 27 | 49% | -5.92 | -2.25 | -2.93 | -1.40 | -3.62 | -7.65 |
| > 5 cm | 7 | 13% | -6.91 | -4.07 | -3.17 | -1.19 | -3.27 | -1.69 |
| P≠ | 0.70 | 0.95 | 0.88 | |||||
| Histopathological subtypes† | ||||||||
| Ductal | 43 | 78% | -5.80 | -4.65 | -2.93 | -1.41 | -3.27 | -1.76 |
| Lobular | 8 | 15% | -8.50 | -3.92 | -3.14 | -0.85 | -2.31 | -4.90 |
| Other | 4 | 7% | -6.43 | -3.19 | -3.37 | -0.58 | -1.23 | -2.24 |
| P$ | 0.07 | 0.41 | 0.31 | |||||
| Bloom & Richardson grade | ||||||||
| I + II | 10 | 18% | -6.39 | -4.78 | -3.11 | -1.51 | -6.94 | -7.56 |
| III | 38 | 69% | -5.78 | -4.65 | -3.15 | -1.33 | -2.92 | -2.37 |
| P$ | 0.43 | 0.52 | 0.08 | |||||
| ESR1 status | ||||||||
| Negative | 22 | 40% | -5.64 | -3.79 | -2.91 | -1.24 | -3.06 | -2.00 |
| Positive | 33 | 60% | -6.40 | -4.69 | -3.17 | -1.35 | -2.98 | -2.81 |
| P$ | 0.15 | 0.39 | 0.88 | |||||
| ERBB2 status | ||||||||
| Negative | 43 | 78% | -5.80 | -5.01 | -3.19 | -1.06 | -2.97 | -2.90 |
| Positive/amplified | 12 | 22% | -7.35 | -3.70 | -2.77 | -1.59 | -3.31 | -1.67 |
| P$ | 0.04 | 0.11 | 0.96 | |||||
| Regional lymph node status | ||||||||
| Negative | 16 | 29% | -5.64 | -4.40 | -3.17 | -1.14 | -2.84 | -2.63 |
| Positive | 33 | 60% | -6.40 | -4.62 | -3.00 | -1.17 | -2.98 | -1.84 |
| P$ | 0.23 | 0.74 | 0.90 | |||||
| Time between primary tumor and studied metastasis | ||||||||
| ≤ 24 months | 33 | 60% | -5.92 | -5.45 | -2.88 | -1.16 | -2.85 | -2.09 |
| > 24 months | 22 | 40% | -6.43 | -4.09 | -3.24 | -0.93 | -3.20 | -2.28 |
| P≠ | 0.97 | 0.42 | 0.33 | |||||
| Overall survival status | ||||||||
| Alive | 22 | 40% | -5.92 | -2.52 | -3.11 | -1.48 | -3.67 | -8.34 |
| Deceased | 33 | 60% | -6.65 | -5.03 | -3.11 | -0.95 | -2.81 | -1.68 |
| P$ | 0.21 | 0.62 | 0.20 | |||||
AVG epithelial; average mRNA level of KRT19 and EPCAM. IQR; interquartile range.
* Due to missing values numbers do not always add up to 55.
≠ Spearman correlation significance (2-tailed).
$ Mann-Whitney Test significance (2-tailed).
† mRNA expression of ductal and lobular breast cancer was compared.
RNA isolation and quantitative reverse transcriptase polymerase chain reaction (RT-qPCR)
Ten 10 μm slides were cut from the primary tumors and paired metastases. The first and last sections (5 μm) were stained with hematoxylin and eosin to guide macro-dissection of the tumor cells for RNA extraction. Total RNA was isolated from the macro-dissected sections with the AllPrep DNA/RNA FFPE Kit (Qiagen) and resulting nucleic acid concentrations were measured with a Nanodrop 2000 system (ThermoFisher Scientific). cDNA was generated for 30 min at 48°C with RevertAid H minus (ThermoFisher Scientific) and gene-specific pre-amplified with Taqman PreAmp Master mix (ThermoFisher Scientific) for 15 cycles, followed by Taqman probe—based real time PCR according to the manufacturer’s instructions in a MX3000P Real-Time PCR System (Agilent). The following gene expression assays were evaluated (all from ThermoFisher Scientific): APOBEC3B, Hs00358981_m1; EPCAM, Hs00158980_m1; ESR1, Hs00174860_m1; ERBB2, Hs01001580_m1, KRT19, Hs01051611_gH; PTPRC, Hs00236304_m1. mRNA levels were quantified relative to the average expression of GUSB, Hs9999908_m1 and HMBS, Hs00609297_m1 using the delta Cq (dCq = 2^(average Cq reference genes—Cq target gene)) method.
Quality and quantity control measurements for reliable quantitative reverse transcriptase polymerase chain reaction (RT-qPCR)
For reliable RT-qPCR measurements, only samples that resulted in amplifiable products within 25 cycles for the used reference gene set at an input of 50 ng total RNA (92.9% of the samples) were considered to be of good quality to reliably determine RT-qPCR levels. Furthermore, a serially diluted FFPE breast tumor sample was included in each experiment to evaluate the linear amplification and efficiencies for all genes included in the panel and absence of amplification in the absence of reverse transcriptase. All gene transcripts were 100% efficient amplified (range 94%-102%) and were negative in the absence of reverse transcriptase.
Estrogen receptor (ER/ESR1) and HER2 (HER2/ERBB2) status
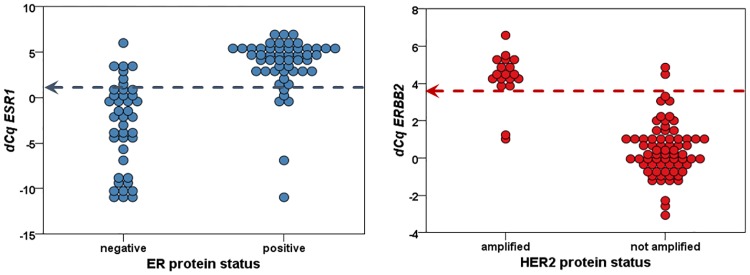
Because data regarding ER and HER2 expression on protein level of our data set was incomplete, ESR1 and ERBB2 mRNA expression was used to determine ESR1 and ERBB2 mRNA status (using a cut-off dCq for ESR1>1 and ERBB2>3.5 by optimal binning for n = 92 and n = 87 overlapping samples, respectively (Fig 1)). Because ER and HER2 are determined on protein level in daily clinical practice (using a scoring system according to national guidelines [26,27]), we investigated whether the ESR1 and ERBB2 mRNA status accurately reflected the ER and HER2 protein status as reported in the pathology reports in samples with known receptor protein status (Fig 1). These cut-offs resulted in a sensitivity of 0.88 and specificity of 0.85 for ESR1 and in a sensitivity of 0.89 and specificity of 0.97 for ERBB2.
Fig 1. Correlation of ER and HER2 protein status with ESR1 and ERBB2 mRNA levels.
Arrows indicate used cut-off value.
Statistics
SPSS version 23 was used for all statistical analyses. The Kolmogorov-Smirnov and Shapiro-Wilk tests were used to test for normality of the distributions. To compare mean values between two or more groups, the Mann-Whitney U Test or Kruskal-Wallis test was used, followed by a test for trend if appropriate. To compare values measured in primary cancers and paired metastases, the paired Wilcoxon Signed Ranks test was used. To correlate linear variables, the Spearman Rank Correlation test was used. P-values ≤ 0.05 were considered statistically significant.
Results
APOBEC3B mRNA expression in primary breast cancer
Since the Kolmogorov-Smirnov and Shapiro-Wilk tests showed that our data were not always normally distributed, we tested all our data non-parametrically. First, we correlated the levels of APOBEC3B mRNA measured in the primary tumors with traditional clinicopathological characteristics (Table 1). Besides a higher expression of APOBEC3B mRNA in ERBB2 negative tumors when compared to ERBB2 positive tumors (Spearman’s Rho = -0.46, P = 0.002), APOBEC3B mRNA levels were not correlated to any of the studied parameters.
To ensure that different levels of tumor cells or inflammatory cells in the primary tumors and their matched metastasis did not bias our data, we quantified the mRNA levels of KRT19 and EPCAM (as a measure for epithelial content) and PTPRC (the gene for the common leukocyte antigen CD45, as a measure for the presence of lymphocytes). Although a weak positive correlation was observed between APOBEC3B mRNA levels and epithelial content in the complete cohort (Spearman’s Rho = 0.21, P = 0.031, N = 110), no significant correlations were observed between the mRNA levels of APOBEC3B and epithelial or infiltrate content when analyzed separately for the primary tumors and the metastases (Spearman correlation significance P > 0.05). In addition, epithelial and infiltrate content did not differ significantly between the primary tumors and matched metastases (paired Wilcoxon Signed Rank test P > 0.05).
APOBEC3B mRNA expression in primary breast cancer and paired metastases
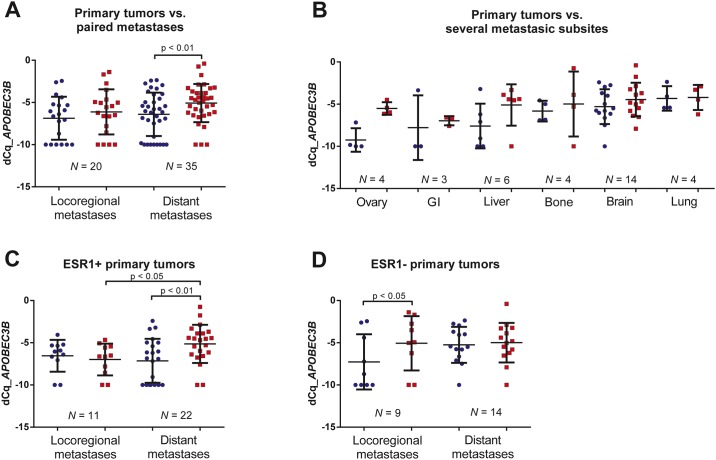
Next, we correlated APOBEC3B mRNA expression in primary tumors and their matched metastases. This analysis revealed that APOBEC3B mRNA levels were significantly higher in the matched distant metastases as compared to the primary tumors (paired Wilcoxon Signed Rank test P = 0.0015, Fig 2A and Table 2). In contrast, no difference was perceived between primary tumors and matched loco-regional lymph node metastases (paired Wilcoxon Signed Rank test P = 0.23)), while levels remained significantly elevated for the cohort with distant metastases (paired Wilcoxon Signed Rank test P = 0.02); (Table 2). Also, APOBEC3B mRNA levels measured in distant metastases (N = 35) showed a trend towards higher expression when compared to regional lymph node metastases (N = 20) of unmatched cases (Mann-Whitney U test P = 0.08, Fig 2A). No such trend was observed in primary tumors that disseminated either to loco-regional or distant locations (Mann-Whitney U test P = 0.42, Fig 2A).
Fig 2. APOBEC3B mRNA expression differences between primary breast tumors and paired metastases.
(A) APOBEC3B mRNA expression in primary breast tumors versus paired loco-regional and distant metastases. (B) APOBEC3B mRNA expression in primary breast tumors versus paired metastases, subdivided per location of metastasis (ovary (N = 4); liver (N = 6); bone (N = 4); brain (N = 14); lung (N = 5) and gastro-intestinal tract (N = 3). (C) APOBEC3B mRNA expression in ESR1-positive primary breast tumors versus paired distant and loco-regional metastases. (D) APOBEC3B mRNA expression in ESR1-negative primary breast tumors versus paired distant and loco-regional metastases. P-values obtained by paired Wilcoxon Signed Ranks test (2-tailed).
Table 2. Association of APOBEC3B mRNA in matched primary tumors and their metastasis according metastatic site.
| APOBEC3B mRNA log2 | AVG epithelial mRNA log2 | PTPRC (CD45) mRNA log2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Clinical parameter | No of patients* | Percentage of patients | Mean | SD | Mean | SD | Mean | SD |
| Tissue origin | ||||||||
| Primary tumor | 55 | 100% | -6.51 | 2.53 | -3.04 | 1.07 | -3.98 | 3.23 |
| Paired Metastasis | 55 | 100% | -5.36 | 2.39 | -2.85 | 1.08 | -4.48 | 2.84 |
| Pǂ | 0.0015 | 0.20 | 0.26 | |||||
| According metastatic site | ||||||||
| Loco-regional lymph node | ||||||||
| Primary tumor | 20 | 36% | -6.87 | 2.55 | -2.82 | 0.91 | -3.88 | 2.89 |
| Paired Metastasis | 20 | 36% | -6.12 | 2.68 | -2.63 | 0.89 | -3.42 | 3.03 |
| Pǂ | 0.23 | 0.31 | 0.49 | |||||
| Distant metastases | ||||||||
| Primary tumor | 35 | 64% | -6.30 | 2.54 | -3.16 | 1.15 | -4.30 | 3.34 |
| Paired Metastasis | 35 | 64% | -4.93 | 2.13 | -2.98 | 1.16 | -4.82 | 2.80 |
| Pǂ | 0.002 | 0.38 | 0.39 | |||||
| Distant metastasis specified | ||||||||
| Ovary | ||||||||
| Primary tumor | 4 | 7% | -9.25 | 1.39 | -3.31 | 1.25 | -5.53 | 5.25 |
| Paired Metastasis | 4 | 7% | -5.51 | 0.74 | -3.03 | 1.20 | -3.99 | 0.93 |
| Pǂ | 0.07 | 0.72 | 0.47 | |||||
| GI tract | ||||||||
| Primary tumor | 3 | 5% | -7.79 | 3.84 | -3.32 | 0.20 | -1.83 | 1.10 |
| Paired Metastasis | 3 | 5% | -6.96 | 0.54 | -3.93 | 1.88 | -4.41 | 4.88 |
| Pǂ | 0.59 | 0.59 | 0.11 | |||||
| Liver | ||||||||
| Primary tumor | 6 | 11% | -7.59 | 2.65 | -3.16 | 1.57 | -2.66 | 1.35 |
| Paired Metastasis | 6 | 11% | -5.10 | 2.45 | -2.20 | 1.17 | -4.76 | 2.71 |
| Pǂ | 0.08 | 0.17 | 0.028 | |||||
| Bone | ||||||||
| Primary tumor | 4 | 7% | -5.82 | 1.23 | -3.01 | 1.28 | -5.04 | 3.31 |
| Paired Metastasis | 4 | 7% | -4.98 | 3.85 | -2.63 | 0.96 | -4.11 | 3.97 |
| Pǂ | 0.47 | 0.27 | 0.47 | |||||
| Brain | ||||||||
| Primary tumor | 14 | 25% | -5.29 | 2.06 | -3.10 | 1.03 | -5.53 | 3.58 |
| Paired Metastasis | 14 | 25% | -4.46 | 2.00 | -2.95 | 0.81 | -5.67 | 2.82 |
| Pǂ | 0.20 | 0.68 | 0.98 | |||||
| Lung | ||||||||
| Primary tumor | 4 | 7% | -4.32 | 1.45 | -3.29 | 1.69 | -2.35 | 1.59 |
| Paired Metastasis | 4 | 7% | -4.21 | 1.49 | -3.82 | 1.47 | -3.76 | 1.66 |
| Pǂ | 0.72 | 0.47 | 0.14 | |||||
SD; standard deviation. AVG epithelial; average mRNA level of KRT19 and EPCAM.
* Due to missing values, numbers don’t always add up to 55.
ǂ Paired Wilcoxon Signed Ranks Test significance (2-tailed).
Subgroup analysis by distant metastatic site showed increased APOBEC3B expression for all locations, particularly for liver and ovary, although no significance was reached for any of the relatively small subgroups (paired Wilcoxon Signed Ranks test P > 0.05, Fig 2B and Table 2). We also compared mRNA levels measured in primary tumors according to distant metastatic site. These analyses showed that APOBEC3B mRNA levels were lowest in primary tumors that metastasized to the ovaries and gastro-intestinal sites and highest in primary tumors that metastasized to lung, brain or bone (Kruskal-Wallis test P = 0.030), Fig 2B).
APOBEC3B mRNA expression according to ESR1 and ERBB2 status of the primary tumor
We observed no association between APOBEC3B mRNA levels and ESR1-status (Table 1). Several previous studies, however, showed higher APOBEC3B mRNA expression in ERα-negative tumors compared to ERα-positive tumors [23,28,29]. Notably, in these studies, high APOBEC3B expression levels were only associated with poor prognosis for ERα-positive primary breast tumors. We therefore categorized our primary cohort into ESR1-positive and ESR1-negative primary tumors (Fig 2C and 2D). For the ESR1-positive primary tumors, a significantly higher expression was seen in paired distant metastases, but not in loco-regional metastases (Fig 2C; paired Wilcoxon Signed Ranks Test P = 0.002 and 0.53, respectively). In contrast, for the ESR1-negative primary tumors a significantly higher expression was seen in loco-regional metastases, but not in distant metastases (Fig 2D; paired Wilcoxon Signed Ranks Test P = 0.028 and 0.81, respectively). Receptor conversion from an ESR1-positive primary tumor to an ESR1-negative metastasis could not explain this finding (Table 3).
Table 3. ESR1 conversions from primary tumor to metastasis specified by site of metastasis.
| ESR1 conversion primary to metastasis | Metastasis type | N | dCq APOBEC3B | dCq APOBEC3B | P-value* | P-value* |
|---|---|---|---|---|---|---|
| Primary (Average) | Metastasis (Average) | |||||
| Not converted | loco-regional lymph node | 15 | -6.4 | -5.97 | 0.61 | 0.032 |
| distant metastasis | 27 | -5.86 | -4.87 | 0.015 | ||
| ESR1- primary to ESR1+metastasis | loco-regional lymph node | 5 | -8.26 | -6.57 | 0.11 | 0.041 |
| ESR1+ primary to ESR1- metastasis | distant metastasis | 8 | -7.8 | -5.16 | 0.07 |
* Wilcoxon Signed Ranks Test
No such difference was seen after categorizing our patients according to ERBB2-status. Irrespective of ERBB2-status, APOBEC3B levels were only higher in the distant metastases and not in the loco-regional lymph nodes when compared to the paired primary tumor (paired Wilcoxon Signed Ranks Test P < 0.05 and > 0.05, respectively).
Discussion
APOBEC3B is thought to affect the evolution of breast cancer by somatically mutagenizing the cancer genome, which could potentially be abrogated by therapeutic intervention [5]. Previous studies investigated APOBEC3B mRNA expression in primary breast tumors and paired normal tissue. These studies reported upregulation in primary tumors compared to normal tissue, especially in ERα-negative cases [17,28]. However, metastatic disease remains the major cause of breast cancer related mortality and several studies reported discordances of (epi) genetic and immunohistochemical markers between primary tumors and matched metastases [4,30–34]. To the best of our knowledge, no data is available regarding APOBEC3B expression in breast cancer metastases. Therefore, we set out to evaluate APOBEC3B mRNA expression in primary breast cancer and matched metastases. Importantly, we encountered a significant increase in APOBEC3B mRNA levels in the metastases compared to their corresponding primary tumor. Furthermore, distant metastases showed higher expression than loco-regional lymph node metastases. This implies a role for APOBEC3B not only at the stage of the primary tumor but also, and according to our data even more dominantly, during tumor evolution of metastatic breast cancer.
Previous studies reported an association between APOBEC3B expression and aggressive characteristics of the primary breast cancer, including high histologic grade, genomic grade, advanced stage, negative ERα status and HER2/ERBB2 amplification [21,23,28,29,35]. In this current, more concise study with a special focus on breast cancer metastases, we only observed a negative association between APOBEC3B and ERBB2 mRNA levels in both the primary tumor (Table 1) and the metastases (data not shown). However, our sample size was relatively small with a relatively high number of cases with loco-regional (36%) and brain metastases (25%), which could have biased our results.
Overall, we did not find a correlation between APOBEC3B and ESR1-status of the primary tumor, while previous studies reported higher APOBEC3B mRNA expression in ERα-negative tumors compared to ERα-positive tumors ([23,28,29]. Interestingly, for our ESR1-negative primary cases, a significantly higher expression of APOBEC3B was seen in paired loco-regional metastases only and not in paired distant metastases. For our ESR1-positive primary cases on the other hand, a significantly higher expression was seen in paired distant metastases and not in the loco-regional lymph nodes. This is especially noteworthy in view of our previous finding, that high levels of APOBEC3B were only associated with poor prognosis in ESR1-positive primary breast cancers, and not in ESR1-negative cases [23].
Irrespective of ESR1-status, we showed that APOBEC3B expression was increased in distant metastases compared to the corresponding primary tumor, with highest expression in liver, lung, brain and bone metastases. APOBEC3B thus seems not only needed for breast cancer progression, but also for maintenance of the metastasis in distant environments. Since APOBEC3B is upregulated in numerous cancer types we wondered if these findings could be explained by the micro-environment of the distant site. In an article of Burns et al. [7], APOBEC3B expression levels determined by RNA-seq showed a lower expression in normal brain and ovarian tissue relative to normal breast tissue. Furthermore, brain tumors (low-grade glioma, glioblastoma multiforme) and ovarian tumors (serous cystadenocarcinoma) also had lower APOBEC3B expression levels than breast carcinoma. This might imply that the higher APOBEC3B mRNA levels we found in breast cancer brain and ovarian metastases are independent of the micro-environment at these locations. Furthermore, since the pattern of APOBEC3B expression in primary tumors is retained and even increased in paired metastases, and shows a trend toward a possible metastatic location-specific pattern, one could envision that the primary tumor is already ‘primed’ for an eventual site of dissemination. This should however be validated in a larger cohort and also at the protein level. To this end, we tried 2 commercially available APOBEC3B antibodies (PAB2474 from Abnova and Anti-APOBEC3B antibody—N-terminal ab191695 from Abcam). However, despite various efforts, we had to conclude that detecting APOBEC3B in breast cancer by immunohistochemistry with these currently commercially available antibodies should be considered unreliable due to non-specific staining. Hopefully, now that APOBEC3B is gaining increased interest, more specific antibodies will become available soon to confirm our findings at the protein level.
In theory, tumor heterogeneity could explain some of the observed differences between primary tumors and paired metastases. However, the reported distinct APOBEC3B mRNA expression levels of the primary tumor that were largely retained or increased in the paired metastases could not solely be explained by heterogeneity. In daily practice, the majority of metastases are not resected or biopsied. This likely resulted in a selection bias, since we only included primary tumors with available material of the paired metastasis. Another weakness of our study is the relatively small number of patients with distant metastases, which limited the reliability of subgroup analysis according to metastatic site.
In conclusion, our findings add to the knowledge that APOBEC3B contributes to breast cancer progression and has now extended this to metastatic disease. Since APOBEC3B expression is at least retained and often even increased in distant metastases, our data suggest that it might also be an effective interventional candidate for disseminated breast cancer.
Abbreviations
- APOBEC3B
Apolipoprotein B mRNA Editing Enzyme, Catalytic Polypeptide-Like 3B
- AVG
average
- Cq
quantitative threshold cycle
- dCq
delta quantitative threshold cycle
- ER/ESR1
estrogen receptor
- ERα
estrogen receptor alpha
- FFPE
formalin-fixed paraffin-embedded
- HER2/ERBB2
human epidermal growth factor receptor 2
- NWO
Netherlands Organization of Scientific Research
- PTPRC
gene for the common leukocyte antigen CD45
- RT-qPCR
quantitative reverse transcriptase polymerase chain reaction
Data Availability
All relevant data are within the paper. There are no ethical or legal restrictions to any of the data used for this study.
Funding Statement
This study was supported by Cancer Genomics Netherlands (AMS and JWMM) and Netherlands Organization of Scientific Research (NWO) (JWMM). The supporters had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359–386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massague J (2006) Cancer metastasis: building a framework. Cell 127: 679–695. 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 3.Fidler IJ (2003) The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer 3: 453–458. 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- 4.Kroigard AB, Larsen MJ, Thomassen M, Kruse TA (2016) Molecular Concordance Between Primary Breast Cancer and Matched Metastases. Breast J 22: 420–430. 10.1111/tbj.12596 [DOI] [PubMed] [Google Scholar]
- 5.Harris RS (2015) Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res 17: 8 10.1186/s13058-014-0498-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, et al. (2013) APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494: 366–370. 10.1038/nature11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns MB, Temiz NA, Harris RS (2013) Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45: 977–983. 10.1038/ng.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuong KJ, Loeb LA (2013) APOBEC3B mutagenesis in cancer. Nat Genet 45: 964–965. 10.1038/ng.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, et al. (2013) APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res 73: 7222–7231. 10.1158/0008-5472.CAN-13-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nowarski R, Kotler M (2013) APOBEC3 cytidine deaminases in double-strand DNA break repair and cancer promotion. Cancer Res 73: 3494–3498. 10.1158/0008-5472.CAN-13-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, et al. (2013) An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976. 10.1038/ng.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xuan D, Li G, Cai Q, Deming-Halverson S, Shrubsole MJ, et al. (2013) APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis 34: 2240–2243. 10.1093/carcin/bgt185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gwak M, Choi YJ, Yoo NJ, Lee S (2014) Expression of DNA cytosine deaminase APOBEC3 proteins, a potential source for producing mutations, in gastric, colorectal and prostate cancers. Tumori 100: 112e–117e. [DOI] [PubMed] [Google Scholar]
- 14.Jin Z, Han YX, Han XR (2014) The role of APOBEC3B in chondrosarcoma. Oncol Rep 32: 1867–1872. 10.3892/or.2014.3437 [DOI] [PubMed] [Google Scholar]
- 15.Burns MB, Leonard B, Harris RS (2015) APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed J 38: 102–110. 10.4103/2319-4170.148904 [DOI] [PubMed] [Google Scholar]
- 16.Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, et al. (2015) An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 47: 1067–1072. 10.1038/ng.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Wei W, Jin HC, Ying RC, Zhu AK, et al. (2015) The roles of APOBEC3B in gastric cancer. Int J Clin Exp Pathol 8: 5089–5096. [PMC free article] [PubMed] [Google Scholar]
- 18.Kosumi K, Baba Y, Ishimoto T, Harada K, Nakamura K, et al. (2016) APOBEC3B is an enzymatic source of molecular alterations in esophageal squamous cell carcinoma. Med Oncol 33: 26 10.1007/s12032-016-0739-7 [DOI] [PubMed] [Google Scholar]
- 19.Morganella S, Alexandrov LB, Glodzik D, Zou X, Davies H, et al. (2016) The topography of mutational processes in breast cancer genomes. Nat Commun 7: 11383 10.1038/ncomms11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR (2014) APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 7: 1833–1841. 10.1016/j.celrep.2014.05.012 [DOI] [PubMed] [Google Scholar]
- 21.Periyasamy M, Patel H, Lai CF, Nguyen VT, Nevedomskaya E, et al. (2015) APOBEC3B-Mediated Cytidine Deamination Is Required for Estrogen Receptor Action in Breast Cancer. Cell Rep 13: 108–121. 10.1016/j.celrep.2015.08.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebhandl S, Huemer M, Greil R, Geisberger R (2015) AID/APOBEC deaminases and cancer. Oncoscience 2: 320–333. 10.18632/oncoscience.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieuwerts AM, Willis S, Burns MB, Look MP, Meijer-Van Gelder ME, et al. (2014) Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm Cancer 5: 405–413. 10.1007/s12672-014-0196-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Land AM, Wang J, Law EK, Aberle R, Kirmaier A, et al. (2015) Degradation of the cancer genomic DNA deaminase APOBEC3B by SIV Vif. Oncotarget 6: 39969–39979. 10.18632/oncotarget.5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Diest PJ (2002) No consent should be needed for using leftover body material for scientific purposes. For. Bmj 325: 648–651. [PubMed] [Google Scholar]
- 26.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, et al. (2014) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 138: 241–256. 10.5858/arpa.2013-0953-SA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breast Cancer Guideline, NABON 2012.
- 28.Tsuboi M, Yamane A, Horiguchi J, Yokobori T, Kawabata-Iwakawa R, et al. (2016) APOBEC3B high expression status is associated with aggressive phenotype in Japanese breast cancers. Breast Cancer 23: 780–788. 10.1007/s12282-015-0641-8 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Delahanty R, Guo X, Zheng W, Long J (2015) Integrative genomic analysis reveals functional diversification of APOBEC gene family in breast cancer. Hum Genomics 9: 34 10.1186/s40246-015-0056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoefnagel LD, van de Vijver MJ, van Slooten HJ, Wesseling P, Wesseling J, et al. (2010) Receptor conversion in distant breast cancer metastases. Breast Cancer Res 12: R75 10.1186/bcr2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoefnagel LD, Moelans CB, Meijer SL, van Slooten HJ, Wesseling P, et al. (2012) Prognostic value of estrogen receptor alpha and progesterone receptor conversion in distant breast cancer metastases. Cancer 118: 4929–4935. 10.1002/cncr.27518 [DOI] [PubMed] [Google Scholar]
- 32.Hoefnagel LD, van der Groep P, van de Vijver MJ, Boers JE, Wesseling P, et al. (2013) Discordance in ERalpha, PR and HER2 receptor status across different distant breast cancer metastases within the same patient. Ann Oncol 24: 3017–3023. 10.1093/annonc/mdt390 [DOI] [PubMed] [Google Scholar]
- 33.Schrijver WA, Jiwa LS, van Diest PJ, Moelans CB (2015) Promoter hypermethylation profiling of distant breast cancer metastases. Breast Cancer Res Treat 151: 41–55. 10.1007/s10549-015-3362-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiwa LS, van Diest PJ, Hoefnagel LD, Wesseling J, Wesseling P, et al. (2014) Upregulation of Claudin-4, CAIX and GLUT-1 in distant breast cancer metastases. BMC Cancer 14: 864 10.1186/1471-2407-14-864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cescon DW, Haibe-Kains B, Mak TW (2015) APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci U S A 112: 2841–2846. 10.1073/pnas.1424869112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper. There are no ethical or legal restrictions to any of the data used for this study.