Abstract
Plants possess large arsenals of immune receptors capable of recognizing all pathogen classes. To cause disease, pathogenic organisms must be able to overcome physical barriers, suppress or evade immune perception, and derive nutrients from host tissues. Consequently, to facilitate some of these processes, pathogens secrete effector proteins that promote colonization. This review covers recent advances in the field of effector biology, focusing on conserved cellular processes targeted by effectors from diverse pathogens. The ability of effectors to facilitate pathogen entry into the host interior, suppress plant immune perception, and alter host physiology for pathogen benefit is discussed. Pathogens also deploy effectors in a spatial and temporal manner, depending on infection stage. Recent advances have also enhanced our understanding of effectors acting in specific plant organs and tissues. Effectors are excellent cellular probes that facilitate insight into biological processes as well as key points of vulnerability in plant immune signaling networks.
Keywords: immunity, apoplastic effectors, intracellular effectors, pathogen, effector timing
INTRODUCTION
Although plants lack a global circulatory immune system, they possess a sophisticated innate immune system capable of recognizing all pathogen classes (17, 161). The coevolution between plants and pathogens over millions of years has culminated in large arsenals of immune receptors present in plant genomes. In order to gain entry into the plant interior, colonize diverse tissues, and cause disease, pathogens must be able to disable plant defense responses. A critical component required for pathogenesis is the secretion of pathogen proteins, called effectors, which modulate plant immunity and facilitate infection (76). In this review, we examine effectors that function in the apoplast as well as inside host cells.
Secreted effectors act in the apoplast or inside the cytoplasm of plant cells to manipulate their hosts. Not surprisingly, multiple effectors have been identified that suppress immune responses triggered by both extra- and intracellular receptors (41, 76). Immune receptors possessing extracellular domains, such as receptor-like proteins (RLPs) and receptor-like kinases (RLKs), are capable of perceiving conserved microbial features and eliciting pattern-triggered immunity (PTI) (161). These PTI receptors are termed pattern recognition receptors (PRRs). Intracellular immune receptors possess nucleotide-binding (NB) and leucine-rich repeat (LRR) domain architecture (17). These intracellular immune receptors specifically recognize pathogen effectors or effector activity, leading to the induction of effector-triggered immunity (ETI). A hallmark of ETI is the hypersensitive response (HR), a form of programmed cell death at the site of infection (17).
Plants can be infected by all pathogen classes, including viruses, fungi, oomycetes, bacteria, nematodes, and feeding insects. Plant viruses typically possess small genomes and utilize host transcriptional machinery to replicate inside host cells (21). Thus, most viral proteins can be conceptually considered as effectors. Bacteria possess multiple secretion systems that facilitate the secretion of effectors directly outside bacterial cells as well as inside host cells. The most well-characterized secretory pathway for Gram-negative bacterial effectors is the type III secretion system (T3SS), which delivers effectors inside host cells and is indispensable for pathogenesis (20). Nematode effectors can be directly secreted into host cells from the stylet or delivered to the plant apoplast by amphidial and hypodermal secretions (39). Insect pathogens, such as aphids and psyllids, can also deliver effectors during feeding through stylet secretions. Filamentous pathogens, such as fungi and oomycetes, secrete effectors via the general secretory pathway and through dedicated feeding and infection structures, such as haustoria and appressoria (102). Effectors secreted by filamentous pathogens possess an N-terminal signal peptide and can function extra- or intracellularly. Intracellular effectors may carry sequence motifs implicated in translocation inside the host cytoplasm, such as the RxLR dEER and the LxFLAK motifs present in the oomycete RxLR and Crinkler (CRN) effectors, respectively (102). In this review, we focus on key cellular processes that are targeted by diverse pathogen effectors with an emphasis on timing and specificity of infection.
KNOCKING DOWN THE GATE: EFFECTORS THAT ENHANCE PATHOGEN ENTRY
Initial Pathogen Invasion into the Plant Interior
Plant pathogens have evolved diverse mechanisms to invade the plant interior, from entry through natural openings or wounds to forced penetration through the epidermis. Pathogens must overcome physical barriers such as the plant cell wall and the waxy cuticular layer covering the epidermis of aerial plant organs (117). Fungi can sense cuticular components, resulting in stimulation of prepenetration processes and cutinase secretion (117). Some fungi can directly penetrate the leaf surface using specialized infection structures called appressoria, which adhere tightly to the leaf surface, facilitate localized secretion of plant cell wall–degrading enzymes, and generate the high turgor necessary for penetration through mechanical force (48). The rice blast fungus Magnaporthe oryzae, for example, develops a melanized appressorium whose turgor pressure enables the penetration peg to gain direct entry to plant epidermal cells (48). Other pathogens enter into the plant without damaging the cell wall through stomata and wounds.
Effectors from Filamentous and Insect Pathogens Facilitating Host Entry
Effectors from diverse pathogens have been identified that facilitate effective penetration and early invasion of host tissues. Ustilago maydis, the causal agent of smut disease in maize, secretes the Pep1 effector from fungal hyphae that is required for effective invasion of host tissue (27). Pep1 is a phylogenetically conserved effector across smut fungi infecting both dicots and monocots (45). U. maydis Δpep1 is not compromised in appressorial formation, but mutant hyphal growth is arrested immediately upon penetration of the plant epidermis (27). U. maydis Δpep1 elicits a strong plant defense response, and Pep1 inhibits plant peroxidases to suppress early maize defense responses (27, 44).
Piercing/sucking insects can cause direct damage during phloem feeding, and stylet-delivered effectors have been demonstrated to further facilitate effective penetration. The green peach aphid Myzus persicae feeds on host plants by inserting its stylet between cell layers until it reaches the phloem. A few salivary M. persicae effectors have been identified that inhibit plant defense responses and enhance feeding (89, 150). M. persicae secretes a salivary gland effector with homology to a macrophage migration inhibition factor (MIF) that enhances aphid fecundity on the host Vicia faba, enables feeding, and blocks plant defense responses (89). In vertebrates, MIFs act as cytokines, which function as critical modulators of innate immunity and inflammation (13). Future research investigating the role of pathogen-secreted cytokines for suppression of plant immune responses may reveal novel mechanisms facilitating pathogenesis in plant and animal systems.
Stomatal Manipulation by Bacterial Effectors
Stomata are the main port of entry for many pathogens. However, guard cells are active immune sensing cells and can rapidly induce stomatal closure upon perception of microbial features, thus blocking pathogen entry (84). Immunity-mediated stomatal closure requires the plant hormone salicylic acid (SA) (84). In contrast, the hormone jasmonic acid (JA) plays an antagonistic role with SA. Bacterial pathogens are able to manipulate stomatal opening through a variety of mechanisms. Some strains of Pseudomonas syringae pv. tomato use coronatine, a mimic of the bioactive plant hormone JA-isoleucine, to induce stomatal reopening on the leaf exterior (84). Recently, multiple T3SS bacterial effectors that manipulate stomatal apertures and thus enhance pathogen entry have been identified. The wildfire pathogen P. syringae pv. tabaci secretes the effector HopX1, which induces JA signaling (36). JA signaling is negatively regulated by JAZ transcriptional repressors (18). HopX1 is a cysteine protease (CP) that degrades multiple JAZ transcriptional repressors, leading to activation of JA-regulated genes (36). P. syringae strains possessing HopX1 do not produce coronatine but are nonetheless able to induce stomatal reopening, indicating thatHopX1 may be sufficient to induce JA signaling (36). The conserved P. syringae HopZ1a effector also targets JAZ proteins (52). HopZ1a is an acetyltransferase and acetylates both soybean and Arabidopsis JAZ proteins, leading to their degradation (52). Transgenic Arabidopsis plants expressing HopZ1a are also able to suppress stomatal defense (74). The P. syringae effector HopM1 suppresses stomatal defense by targeting the Arabidopsis 14-3-3 protein GRF8/AtMIN10 (68). Other bacterial effectors, such as AvrB and HopF2, also suppress stomatal immunity (49, 160). Collectively, these results highlight the importance of overcoming stomatal defenses for successful pathogen colonization. However, it is still unclear whether pathogen effectors can specifically target guard cells. Future investigation of specificity in effector delivery will significantly enhance our understanding of early pathogen entry events.
PLASMA MEMBRANE BATTLEGROUND: PATTERN RECOGNITION RECEPTORS AND ALLIES
The first line of active defense against invading microbes consists of plant PRRs at the plasma membrane, which can recognize conserved microbial features termed pathogen-associated molecular patterns (PAMPs) (161). Activation of PRR-mediated immunity results in stomatal closure, inhibiting pathogen proliferation at early stages of infection (84, 155). Therefore, adapted pathogens have evolved effectors that can inhibit PRR-mediated responses at multiple levels. Research in this area has significantly enhanced our understanding of plant immunity. The two most well-characterized PRRs are flagellin-sensing 2 (FLS2) and the elongation factor Tu receptor (EFR), which both recognize epitopes of conserved bacterial proteins. FLS2 and EFR, as well as their co-receptor BAK1, are targeted by several P. syringae effectors to suppress plant immune responses (76). For example, AvrPtoB is an E3 ubiquitin ligase that promotes degradation of several PRRs (37, 38). AvrPto inhibits the kinase activities of FLS2 and EFR to enhance bacterial virulence (152). HopF2, AvrPto, and AvrPtoB also target BAK1 to inhibit downstream immune responses (119, 159). The P. syringae effector HopAO1, a protein tyrosine phosphatase, dephosphorylates activated EFR to suppress immune responses (75).
Effectors can not only inhibit the activity of PRR complexes but also interfere with PRR translation. The P. syringae effector HopU1, a mono-ADP-ribosyltransferase, interferes with translation by targeting RNA-binding proteins, including the glycine-rich RNA-binding protein GRP7 (33). GRP7 binds translational components and RNA, including immunity-related transcripts (91). HopU1 inhibits the ability of GRP7 to bind PRR mRNAs, decreasing the amount of PRRs at the plasma membrane and interfering with immune responses (91).
Pathogen effectors are also known to interfere with components of PRR complexes as well as downstream responses. Members of the plant receptor-like cytoplasmic kinase (RLCK) subfamily VII have been implicated as positive regulators of immune responses (42, 71, 156). The BIK1 RLCK is a member of PRR complexes and can transphosphorylate primary immune receptors as well as the coreceptor BAK1 (71). Furthermore, BIK1 also transphosphorylates the NADPH oxidase RBOHD, leading to an oxidative burst that could directly act as an antimicrobial compound as well as reinforce the plant cell wall (56, 64). Multiple bacterial effectors have been identified that specifically target RLCKs. The P. syringae AvrPphB effector is a protease that directly cleaves PBS1, BIK1, and other RLCKs (120, 156). The Xanthomonas campestris effector AvrAC, also known as XopAC, is an uridylyl transferase that uridylylates several RLCKs, including BIK1 (31, 42). AvrAC-mediated uridylylation of RLCKs inhibits their kinase activity, interfering with downstream immune signaling and promoting bacterial virulence (31, 42). However, Arabidopsis has evolved the ability to detect AvrAC-mediated uridylylation of the PBL2 RLCK, inducing ETI (144). Downstream of PAMP perception, suites of mitogen-activated protein kinases (MAPKs) are activated, leading to transcriptional reprogramming in the nucleus (161). P. syringae effectors can also inhibit MAPKs to stop downstream signaling. For example, the phosphothreonine lyase effector HopAI1 irreversibly inactivates several MAPKs, including MPK3, MPK6, and MPK4; the AvrB effector targets the kinases MPK4 and RIPK to promote infection (22, 65, 157, 158). Pathogen effectors that interfere with later defense responses, such as vesicular trafficking and callose deposition, have also been identified (92).
THE BEST DEFENSE IS A GOOD OFFENSE: EFFECTOR-MEDIATED INHIBITION OF PAMP PERCEPTION
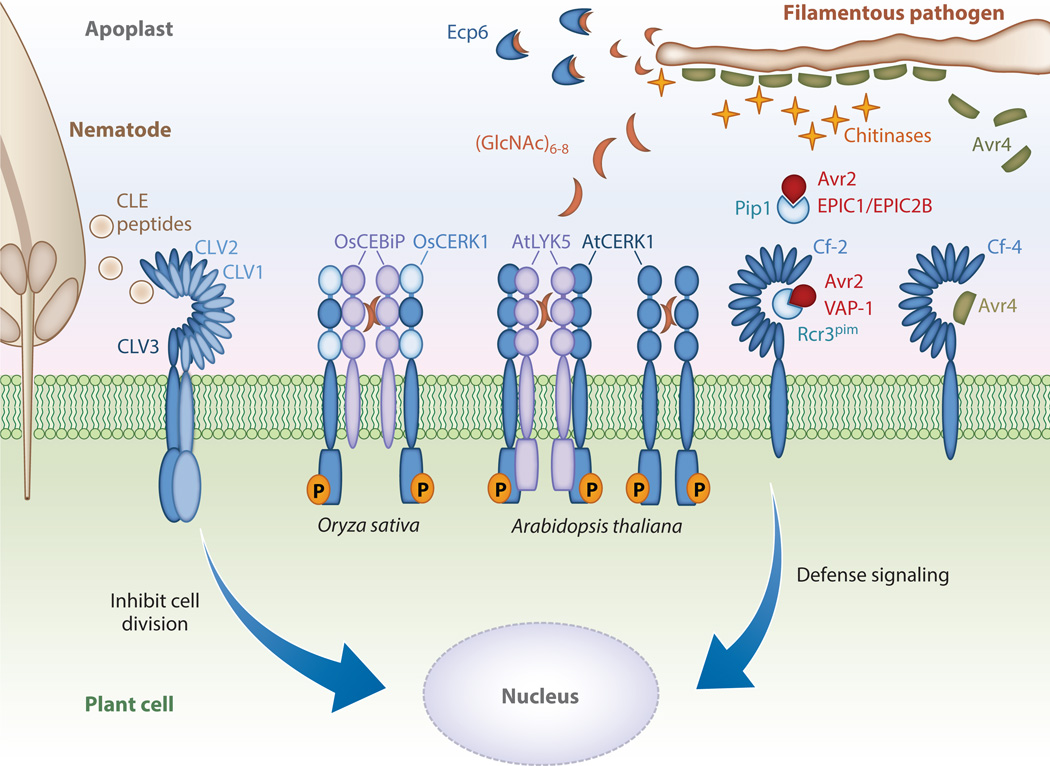
In addition to actively inhibiting PRR-mediated responses, some pathogens have evolved mechanisms to circumvent immune detection by sheltering or masking PAMPs perceived by PRRs (Figure 1). Oligosaccharides derived from the major polymer components of the bacterial (peptidoglycan), fungal (β-glucan, chitin, chitosan), and oomycete (β-glucan, cellulose) cell walls are potent elicitors of plant immune responses, resulting in the reinforcement of the plant cell wall with callose, the secretion of immune-related proteases, and the production of phytoalexins and pathogenesis-related proteins with antimicrobial activities (47, 116, 161). Some pathogenesis-related proteins are lytic enzymes that can destroy the integrity of the pathogen cell wall and inhibit growth (116). Consequently, several microbial effectors function to directly or indirectly protect against hydrolytic assaults or mask the detection of microbial features.
Figure 1.
The battle in the apoplast between plants and pathogens. Filamentous pathogens secrete multiple effectors to interfere with host immunity. The Cladosporium fulvum Avr4 effector shields and masks chitin present in the fungal cell walls from tomato chitinases during infection. The Avr2 effector binds to and inhibits tomato apoplastic proteases such as Rcr3 and PiP1. The plant-parasitic nematode Globodera rostochiensis secretes the VAP1 effector that blocks Rcr3’s active site. The Phytophthora infestans EPIC1 and EPIC2B effectors also inhibit apoplastic proteases. Perception of apoplastic effectors can be mediated by receptor-like proteins (RLPs). Avr2 results in a conformational change in Rcr3, which is perceived by the cognate tomato Cf-2 RLP. Perception of Avr4 by the tomato RLP Cf-4 is hypothesized to occur through direct effector binding. The LysM-domain containing effector Ecp6 interferes with host immunity by sequestering short chito-oligosaccharides that could be released by the fungus. Such short chito-oligosaccharides, usually six to eight oligomers in length [(GlcNAc)6–8], act as PAMPs (pathogen-associated molecular patterns) and are perceived by plant LsyM domain–containing immune receptors, including Arabidopsis thaliana CERK1 and LYK5, a receptor-like kinases (RLKs), and the Oryza sativa CEBiP. Cyst nematodes secrete effectors mimicking plant CLE (CLAVATA3/endosperm surrounding region-related) peptide hormones, which are perceived by CLAVATA RLKs. CLAVATA RLKs regulate stem cell maintenance, and it is hypothesized that nematode CLE effectors act to inhibit cell division to promote syncytium development. Not to scale.
Effector Inhibition of Plant Proteolytic Activity
Secreted CPs are widely distributed in plants and can function in defense against microbial pathogens as well as in pathogen recognition, signaling, and activation of immune responses (47). Multiple apoplastic effectors from filamentous pathogens are capable of inhibiting host proteases (51). Avr2 is a small, cysteine-rich, apoplastic effector protein from the tomato pathogen Cladosporium fulvum (syn. Passalora fulva), which binds and inhibits the papain-like CPs Rcr3 and PiP1 during infection of the host (118). Silencing Avr2 in C. fulvum leads to reduced pathogen virulence on tomato, whereas overexpression of Avr2 in Arabidopsis increases susceptibility to filamentous pathogens (141). Tomato plants possessing the Cf-2 RLP are able to perceive the conformational changes induced upon Rcr3 by binding to Avr2, leading to the activation of plant immune responses (111). It is hypothesized that Pip1, which accumulates to considerably higher levels than Rcr3 in the apoplast, is the primary target of Avr2, whereas Rcr3 acts as a decoy to facilitate immune perception of C. fulvum (118, 139) (Figure 1).
The EPIC1 and EPIC2B effectors from the oomycete pathogen Phytophthora infestans, the causal agent of late blight of potato and tomato, also target and inhibit the CPs Rcr3 and PiP1 in tomato as well as C14 in potato (57, 122, 133) (Figure 1). The plant-parasitic nematode Globodera rostochiensis secretes the VAP1 effector that perturbs Rcr3’s active site (70) (Figure 1). In the absence of the correspondingCf-2 immune receptor, Rcr3 from Solanum pimpinellifolium enhances G. rostochiensis virulence, implying that Rcr3 is a virulence target of VAP1 (70). VAP1, Avr2, EPIC1, and EPIC2B do not share sequence or structural homology, indicating that inhibition of host CPs is an important aspect of microbial pathogenesis that has independently evolved across phylogenetically distant microbial pathogens. In tomato, pathogen-secreted CP inhibitors are targeted for degradation by the serine protease P69B, which in turn is inhibited by the P. infestans EPI1 and EPI10 effectors that act as Kazal-like serine protease inhibitors (131, 132) (Figure 1). This is a compelling example of an apoplastic coevolutionary arms race between host and pathogen.
Stealthy Secretions: Effectors Masking Microbial Features
In addition to actively defending against proteolytic attacks, filamentous pathogens can also secrete effectors that offer passive protection (Figure 1). The C. fulvum apoplastic effector Avr4 possesses a carbohydrate-binding module of family 14 (CBM14) that specifically binds and protects fungal chitin from host-derived chitinases (138) (Figure 1). Several tomato chitinases accumulate in the vicinity of invading C. fulvum hyphae (151). Accordingly, Avr4 is specifically expressed during host colonization and accumulates on the surface of the intercellular hyphae; silencing of Avr4 in C. fulvum leads to attenuated virulence and growth on tomato (138, 140). Thus, Avr4 may promote pathogen virulence by protecting the pathogen from degradation and adverting the release of PAMPs that can be perceived by plant PRRs (Figure 1).
Functional orthologs of Avr4 have been identified in a number of fungal species within the Dothideomycete class, including the banana pathogen Pseudocercospora fijiensis, the tomato pathogen Pseudocercospora fuligena, and the pine tree pathogen Dothistroma, among others (24, 124).
Chitin-binding effectors can also play a critical role in preventing the activation of chitininduced host defenses. The C. fulvum Ecp6 effector is abundantly produced during colonization and possesses three lysine motif (LysM) domains with affinity for short chito-oligosaccharides (10). Unlike Avr4, binding of Ecp6 to chito-oligosaccharides does not fortify against chitinases. Instead, Ecp6 is able to directly compete for the same ligand chito-oligosaccharides with plant PRR chitin receptors, such as the Arabidopsis chitin elicitor receptor kinase 1 (AtCERK1) and the rice chitin elicitor binding protein (CEBiP) (112, 161) (Figure 1). The Ecp6 crystal structure revealed that it possesses a high affinity chitin-binding site composed of LysM1 and LysM3 in addition to LsyM2, which possesses lower binding affinity (23, 112). In contrast, the crystal structure of AtCERK1 indicates that only one of the three LysM domains binds chitin, which could explain the considerably lower affinity of this protein for chitin (67, 112). As with Avr4, functional orthologs of Ecp6 that are able to suppress chitin-induced plant immune responses in order to enable parasitic infection of the host are found in other fungal species, including M. oryzae and the wheat pathogen Zymoseptoria tritici (syn. Mycosphaerella graminicola) (81, 85). Taken together, these studies highlight elegant and diverse strategies employed by pathogen effectors to inhibit plant immune recognition.
ORCHESTRATING EFFECTOR EXPRESSION OVER TIME AND SPACE
Plant pathogens possess diverse lifestyles, ranging from stealthy obligate biotrophs that require robust immune suppression and cannot be maintained axenically outside their hosts to necrotrophs that feed off dead tissue and seek to activate cell death (48, 143). Depending on the stage of infection, pathogens can also adopt intermediate lifestyles. Hemibiotrophs initially exhibit biotrophy but switch to necrotrophy during later stages of infection (143). In the case of filamentous fungi and oomycetes, colonization of the plant is achieved through intracellular invasive hyphae that can grow from cell to cell as well as terminal feeding structures, such as haustoria (102). Below, we focus on advancements in effector biology with respect to timing of effector expression.
Waves of Effector Expression in Relation to Infection Stages
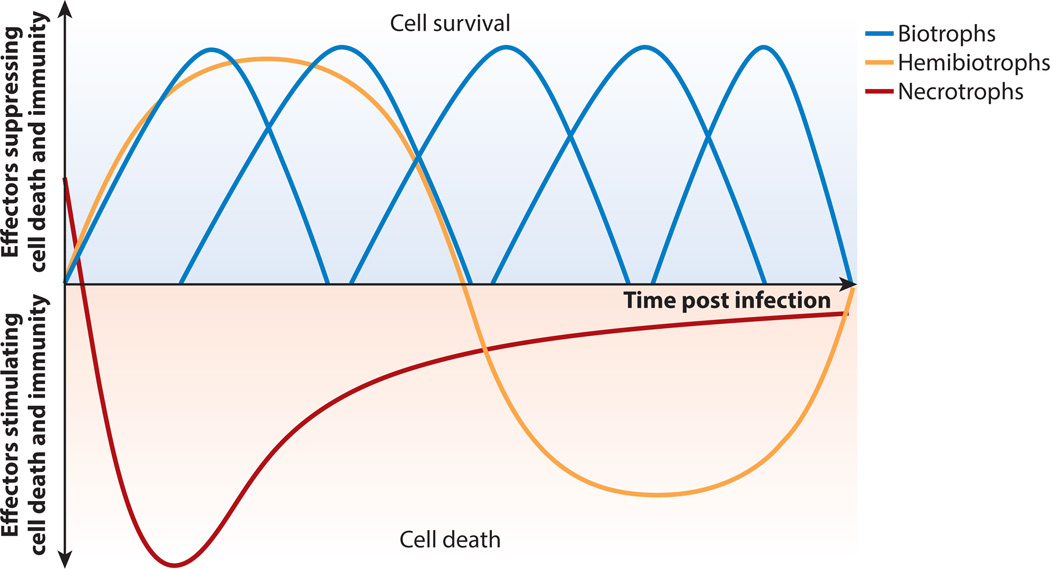
Filamentous pathogens can possess large arsenals of predicted secreted effectors. These effectors are differentially expressed during the course of infection or in a histologically specific manner (Figure 2). Hemibiotrophic Colletotrichum pathogens comprise more than 600 species that cause anthracnose disease on a wide variety of plants (88). Transcriptome analyses of Colletotrichum higginsianum during infection of Arabidopsis revealed three waves of effector expression (60, 94). Specific effectors were expressed during distinct pathogenic stages, suggesting that particular effectors function in each stage (Figure 2). Coordinated expression of specific effector sets occurred during appressorial penetration, biotrophic growth in host cells, and the conversion from biotrophy to necrotrophy (60). When transiently expressed in Nicotiana benthamiana, specific C. higginsianum effectors either inhibit or promote cell death (60). Consistent with their expression patterns, cell death–inhibiting effectors were expressed during the biotrophic phase, whereas cell death–inducing effectors were expressed during the switch to necrotrophy (60) (Figure 2).
Figure 2.
Waves of effector expression over time and pathogen type. Plant pathogens range from obligate biotrophs with narrow host ranges to hemibiotrophs and necrotrophs with broad host ranges. Effector expression patterns change over the course of infection and type of tissue infected. Obligate biotrophs secrete effectors that act to suppress immune recognition and promote cell survival. Hemibiotrophs initially secrete effectors promoting cell survival, but during later stages of infection secrete cell death–promoting effectors. Necrotrophic pathogens can also secrete effectors promoting cell survival during very early infection stages but predominantly secrete cell death–promoting effectors to facilitate colonization.
Phytophthora spp. also exhibit waves of effector expression during infection. Phytophthora sojae, the causal agent of root and stem rot of soybean, possesses ~400 effectors carrying the RxLRdEER motif (135). Using transient expression in N. benthamiana, the ability of 169 effectors to either elicit cell death or suppress INF1-mediated cell death was investigated (146). The most strongly expressed effectors during early to intermediate stages of infection were able to suppress cell death, whereas a second effector class was induced during the first 12 hours of infection and elicited cell death (146) (Figure 2).
Misexpression of stage-specific effectors significantly reduced P. sojae virulence (146). Other pathogens, such as Phytophthora capsici and Blumeria graminis f. sp. hordei, also exhibit successive waves of effector expression during infection (43, 55). These experiments highlight the importance of appropriate timing of effector expression to mediate different infection stages and differentially regulate plant responses (Figure 2).
Effector Specificity by Infection Stage and Plant Organ
U. maydis effector knockouts have revealed the importance of particular effectors for different stages of infection as well as infection of different plant organs (107, 114). After penetration of epidermal cells, fungal hyphae are surrounded by the host plasma membrane and can move throughout mesophyll tissue (27, 28). Fungal hyphae accumulate inside and surround vascular bundles, where they likely obtain nutrients (28). During later stages of infection, U. maydis induces tumor formation that is conducive to large-scale proliferation of the fungus (28). The U. maydis Pep1 effector is required for initial penetration of maize, whereas the Pit2 effector acts slightly later in infection and Pit2 knockouts are unable to maintain a biotrophic interaction (26, 27). The U. maydis effector Tin2 is induced during early infection, functions inside plants cells, and shifts plant metabolism in favor of the pathogen (129). Tin2 inhibits degradation of the maize kinase ZmTTK1, which controls activation of anthocyanin biosynthetic genes (129). In the absence of Tin2, vascular bundles exhibit strong lignification, resulting in the decreased ability of U. maydis to reach vascular tissue for nutrient acquisition (129).
In addition to U. maydis effectors acting in different stages of infection, effectors have been identified exhibiting plant organ specificity. Transcriptional profiling of infected maize seedlings, adult leaves, and tassels revealed specific plant and pathogen genes induced in an organ-specific manner (121). Subsequent functional analyses verified alterations in virulence in particular maize organs for effector knockouts, depending on their specificity of expression (107, 114, 121). For example, the See1 effector induces plant DNA synthesis required for tumor progression in leaf cells but not in immature tassels (107). Collectively, these studies highlight the importance of specific effector sets for enabling successful infections over time and in distinct plant organs. Despite clear evidence of waves of effector expression during infection, few studies have investigated how effectors are transcriptionally regulated in filamentous pathogens. Future research in this area will enable greater mechanistic insight into how effector expression is tightly controlled, including identification of specific transcription factors, in eukaryotic pathogens.
EFFECTORS THAT INDUCE CELL DEATH AND PROMOTE NECROTROPHY OF PLANT PATHOGENS
In contrast to strict biotrophs, effectors from necrotrophic fungi and other hemibiotrophs deliberately induce necrosis to promote virulence (Figure 2). These necrotrophic effectors can either be unspecific or act as host-selective toxins that contribute to pathogen host range (147). Ethylene (ET)-inducing peptide 1 (NEP1)-like proteins (NLPs) are a protein superfamily produced by bacteria, fungi, and oomycete plant pathogens (101). In many dicots, NLPs elicit cell death as well as strong immunity-associated responses (106). More than 70% of NLPs originate from pathogens exhibiting necrotrophic or hemibiotrophic lifestyles (106). It is plausible that these NLPs elicit immune responses to trigger cell death and nutrient acquisition. In Arabidopsis, NLPs are perceived as PAMPs, and a conserved 20 amino acid fragment is recognized by the RLP23 immune receptor and elicits classic PTI responses such as the extracellular ROS burst, ethylene production, MAPK activation, and defense gene expression (1, 97, 106). Crinkler (CRN) effectors also induce necrosis as well as a characteristic leaf crinkling phenotype (115). CRNs are abundantly distributed within oomycetes and possess a characteristic N-terminal LxFLAK translocation domain, and the C termini of several CRNs localize to the plant nuclei (115). The P. infestans CRN8 effector possesses kinase activity and localizes to plant nuclei, indicating that it interferes with host signaling pathways during infection (115, 137).
Although the necrotrophic effectors described above mostly lack host selectivity, others induce severe necrosis on particular plant genotypes possessing dominant sensitivity genes (127). Cloning of multiple sensitivity genes revealed that they resemble intracellular plant immune receptors, with NB sites and LRR domain architecture (127). Thus, necrotrophic effectors can exploit the activation of plant NB-LRRs to induce effector-triggered sensitivity as opposed to ETI. The pathogens Pyrenophora tritici-repentis and Stagonospora nodorum (syn. Parastagonospora nodorum) secrete multiple effectors that induce severe necrosis in wheat genotypes possessing corresponding sensitivity genes (96). The PtrToxA effector produced by P. tritici-repentis triggers necrosis or chlorosis in wheat genotypes containing Tsn1 (19). Tsn1 possesses NB-LRR domain architecture in addition to a serine/threonine protein kinase domain (30). Upon fungal secretion, the PtrToxA effector is internalized only in wheat genotypes possessing Tsn1, localizes to the chloroplast, and interacts with ToxAB1, which may be involved in thylakoid formation (79, 80). PtrToxA orthologs have now been identified in the sister species Stagonospora avenaria tritici as well as the maize pathogen Cochliobolus heterostrophus (73, 82). Notably, molecular phylogenetic analysis has suggested that the ToxA homolog present in P. triticirepentis is likely to have been horizontally acquired from P. nodorum, thus leading to the emergence of highly pathogenic populations that instigated new disease epidemics (32). At least eight more pairs of necrotrophic effectors and cognate plant sensitivity genes have been reported in the P. nodorum–wheat interaction, highlighting the importance of necrotrophic effector-triggered sensitivity (96).
EFFECTORS EXHIBIT DIVERSITY IN CELL-TO-CELL MOVEMENT AND LOCALIZATION
Not only can effectors target diverse cellular processes, but recent work has illustrated that effectors from the rice blast fungus M. oryzae canmove from cell to cell to enhance subsequent colonization (59). After entering the plant interior, M. oryzae initially colonizes rice in a biotrophic manner using intracellular invasive hyphae (48). Fungal transformants expressing fluorescently labeled effectors have enabled the investigation of effector expression, secretion, and movement during natural infection of rice. Recognized effectors and secreted proteins associated with the biotrophic phase accumulate in a structure called the biotrophic interfacial complex, and some effectors could be visualized inside host cells (59). In contrast, apoplastic effectors were uniformly expressed throughout invasive hyphae (59). Interestingly, some cytosolic effectors, such as PWL2and BAS1, were able to move up to four cells away from invading hyphae, possibly through plasmodesmata (59). Effectors that can move from cell to cell likely play a role in priming host cells for pathogen colonization.
In addition to the capability to move from cell to cell, effectors can also exhibit diverse subcellular localizations. Thus, effectors are excellent cellular probes, and specific effectors have been identified that localize to almost every plant subcellular compartment. Effectors from filamentous pathogens exhibiting focal accumulation surrounding the biotrophic interface have been identified. The cucumber pathogen Colletotrichum orbiculare secretes a variety of effectors that accumulate around the neck of biotrophic hyphae (50). C. orbiculare’s exocyst-related component SEC4 also localized to the same compartment, and disruption of SEC4 in the pathogen impaired effector delivery to the plant-pathogen interface (50). These results highlight the importance of focal effector secretion to enable targeted inhibition of plant immune responses.
EFFECTOR MANIPULATION OF VESICULAR TRAFFICKING
Endocytosis and exocytosis are key cellular processes that are involved in the accumulation of primary immune receptors at the plasma membrane as well as in focal protein delivery and secretion. Secretory vesicles deliver antimicrobial cargo to the sites of infection to inhibit pathogen proliferation (130). These antimicrobial molecules can be defense-related proteins, cell wall depositions, and phytotoxins (130). In addition, mutations affecting exocytosis and vesicular trafficking have compromised resistance to different pathogens. For instance, T-DNA mutants, Exo70B2 and Exo70H1, in two members of the Arabidopsis exocyst complex are more susceptible to P. syringae pv. maculicola, and exo70B2 exhibits increased abnormal papillae formation after inoculation with the barley powdery mildew pathogen B. graminis f. sp. hordei (100). PEN1/SYP121, a plasma membrane syntaxin involved in focal secretion and papillae formation, is also required for resistance to B. graminis f. sp. hordei, as the pen1 mutant allows enhanced penetration of this fungal pathogen (5). Taken together, these results highlight the importance of exocytosis for effective plant defense responses.
One well-characterized effector that interferes with secretion of immune cargo is the P. infestans RxLR effector AVRblb2 (11). During infection, AVRblb2 exhibits focal accumulation surrounding haustoria. Using transient expression in N. benthamiana, AVRblb2 was demonstrated to associate with and specifically inhibit secretion into the plant apoplast of the papain-like CP C14 (11). As described above, this class of immune proteases is involved in enhancing plant immune perception and is targeted by multiple apoplastic effectors (47). Silencing of C14 in N. benthamiana resulted in enhanced susceptibility, whereas overexpression of C14 resulted in enhanced resistance to P. infestans, thus highlighting the importance of this immune protease for inhibiting pathogen proliferation (11). AVRblb2 is under positive selection, and different allelic variants are maintained in global populations of P. infestans, indicating that specific alleles contribute differentially to virulence (95). Another P. infestans RxLR effector, AVR1, interacts with the exocyst component (29). In addition, the M. oryzae effector AVR-Pii interacts with two Exo70 components in rice (34). Overexpression of pathogen effectors in plants commonly inhibits secretion of callose and pathogenesis-related proteins (41). Future experiments investigating whether these are direct or indirect effects will help determine whether vesicular trafficking is a conserved process targeted by many effectors as well as shed light on the biological relevance of effector overexpression in plants.
Pathogen effectors that specifically disrupt intracellular vesicular trafficking have also been identified. The P. syringae effector HopM1 interferes with vesicular trafficking, is encoded in the conserved effector locus of several P. syringae strains, and significantly contributes to bacterial pathogenicity (2). HopM1 induces host proteasome-dependent degradation of AtMIN7, an adenine diphosphate ribosylation factor–guanine nucleotide exchange factor (ARF-GEF) involved in vesicular trafficking (92). Both HopM1 and AtMIN7 localize to the trans-Golgi network/early endosome compartment where AtMIN7 regulates endocytosis of plasma membrane proteins (93, 128). Furthermore, Arabidopsis atmin7 plants exhibit enhanced susceptibility to virulent and avirulent P. syringae (92, 93). AtMIN7 is not the only virulence target of HopM1, as this effector can suppress other plant immune responses independent of its association with AtMIN7 (35). The P. infestans AVR3a effector also disrupts internalization of activated FLS2 and may interfere with endocytosis of other PRRs through its association with the host dynamin-related protein 2, a GTPase involved in endocytosis and membrane trafficking (14). Thus, effectors can regulate specific aspects of exo- and endocytosis and interfere with immune receptor abundance at the plasma membrane.
EFFECTORS THAT MIMIC AND INTERFERE WITH PLANT HORMONES
Multiple aspects of plant growth, development, and defense are regulated by complex hormone signaling networks. SAis important for resistance to biotrophic and hemibiotrophic pathogens, and JA and ET are associated with resistance to necrotrophs and herbivorous insects (103). In addition, SA is also required for the induction of systemic acquired resistance. Signaling networks involving SA, JA, and ET are important for PTI and ETI responses (134). Pathogens have evolved innovative strategies to manipulate signaling of most plant hormones to enable pathogen proliferation. Below, we review plant hormones repeatedly targeted by diverse pathogen effectors.
SA is considered the canonical defense hormone that functions to inhibit biotrophic pathogens. Components of SA biosynthesis and perception are targeted by effectors from diverse pathogens. The U. maydis Cmu1 effector is a chorismate mutase (25). Chorismate mutases function in the shikimate pathway, which produces aromatic amino acids, precursors of secondary metabolites such as the hormones auxin and SA, cell wall products, and pigments (78). Maize plants infected with U. maydis Δ cmuI strains have reduced disease symptoms and increased SA levels compared with wild-type strains (25). Cmu effectors have also been identified in several nematodes, including the potato cyst nematodes G. rostochiensis and Globodera pallida, the soybean cyst nematode Heterodera glycines, and the sugar beet cyst nematode Heterodera schachtii (7, 53,72, 142). Thus, chorismate mutase effectors manipulate the shikimate pathway to interfere with immune responses and promote virulence. The filamentous pathogens P. sojae and Verticillium dahliae secrete the virulence promoting effectors Pslsc1 and Vdlscl, respectively. Pslsc1 and Vdlscl are isochorismatases, enzymes that hydrolyze the SA precursor isochorismate to disrupt SA metabolism (66).
Several pathogens induce JA signaling to promote pathogen virulence. JA is able to inhibit SA-mediated responses. As discussed above, P. syringae effectors activate the JA pathway by directly or indirectly mediating degradation of JAZ proteins, thereby suppressing SA signaling and facilitating pathogen entry (36, 52, 160). The Arabidopsis downy mildew pathogen Hyaloperonospora arabidopsidis is an obligate biotroph that secretes the RxLR effector HaRxL44 during infection. HaRxL44 induces the degradation of the Mediator subunit 19a (12). Mediator is a conserved multiprotein complex in eukaryotes that bridges RNA polymerase II with diverse transcription factors (154). HaRxL44-mediated degradation of Med19a results in elevated JA levels and decreased SA responses (12). Arabidopsis plants lacking MED19a or overexpressing HaRxL44 are more susceptible to H. arabidopsidis, whereas plants overexpressing MED19a are more resistant, indicating a role of MED19a in immunity (12). The C2 protein from Tomato yellow leaf curl virus is conserved across geminiviruses and targets a component of the COP9 signalosome CSN5, resulting in suppression of JA-mediated responses (69). Thus, effectors have evolved to disrupt JA signaling at multiple levels, highlighting the importance of manipulating JA signaling to promote pathogenicity.
In addition to interfering with plant hormone perception and signaling, pathogens can also produce mimics of plant hormones. For example, some strains of P. syringae produce coronatine, a JA-isoleucine mimic (8). Cyst nematodes secrete effectors that mimic plant CLAVATA3/endosperm surrounding region-related (CLE) peptide hormones (Figure 1). Plant CLE peptides are secreted, are mobile, and are perceived by RLKs (61). In plants, CLE perception is critical for cell-to-cell communication and controls multiple developmental processes (61). CLE-like effectors are posttranslationally processed, revealing a 12–13 amino acid mature peptide ligand with similarity to plant CLE peptides (16). Overexpression of CLE-like effectors in Arabidopsis results in premature meristem termination and phenocopies plants overexpressing CLE peptides (145). RNAi-mediated gene silencing of cyst nematode CLE-like genes results in decreased parasitism, demonstrating the importance of CLE-like effectors for virulence (6, 99). CLE-like effectors are recognized by several plant RLKs, which are required for nematode-induced syncytium (giant multinucleated cell) development and successful nematode infection (109, 110). It is hypothesized that cyst nematodes secrete CLE-like effectors to reprogram plant tissues as feeding sites for their benefit. Pathogen effectors are also able to alter other plant hormones like auxin and gibberellins to favor pathogenicity (103). However, the amplitude and timing of pathogen hormone manipulation are still not well understood. Future research in this area will enhance our understanding of plant hormone dynamics during pathogen infection.
REPROGRAMMING THE HOST: EFFECTORS MANIPULATING HOST GENE EXPRESSION
Plant perception of microbial pathogens involves transcriptional reprogramming toward defense. Transcriptome analyses have revealed that 44% of Arabidopsis genes are differentially regulated after inoculation with P. syringae (63). Similarly, U. maydis induces transcriptional reprogramming of up to 21% of maize genes (28). Pathogens have evolved diverse mechanisms to manipulate host gene expression by targeting host transcription factors, inducing the expression of susceptibility genes and interfering with host gene silencing (Figure 3).
Figure 3.
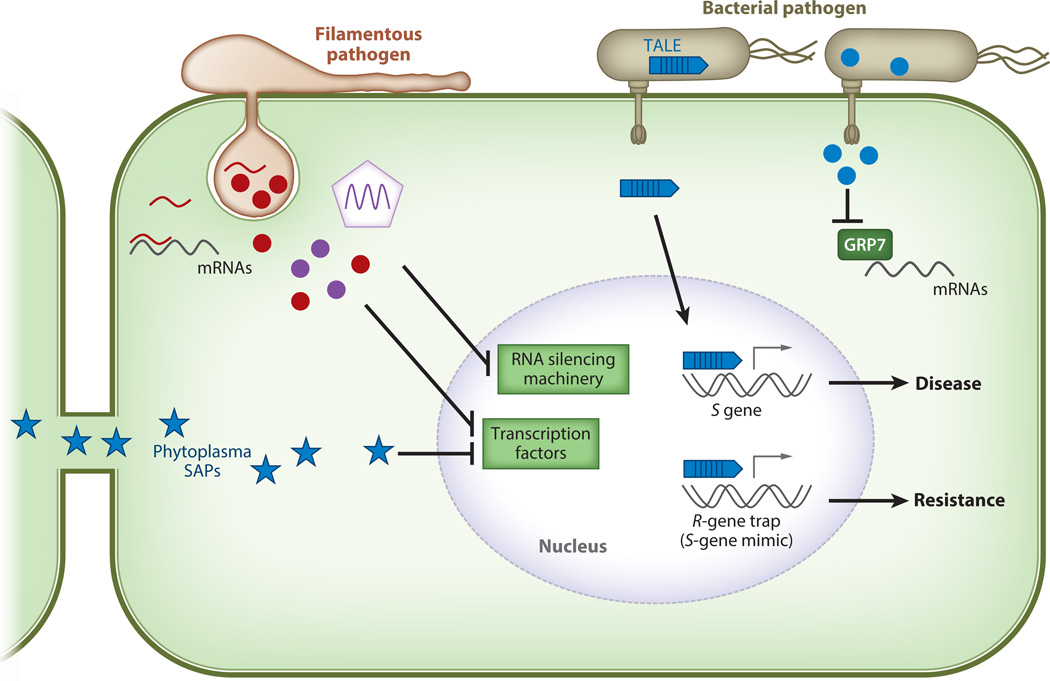
Pathogen effectors manipulate host gene transcription. Bacterial effectors are depicted in blue, effectors from filamentous pathogens are depicted in red, and viral effectors are depicted in purple. Transcriptional activator-like effectors (TALEs) from Xanthomonas and Ralstonia bacterial pathogens are secreted through the type III secretion system (T3SS) into host cells. TALEs bind to promoter regions of target genes to activate transcription of susceptibility (S) genes and promote virulence. However, some plant genotypes possess resistance (R) genes with TALE-binding promoter regions and act as decoys to activate immune responses. The bacterial pathogen Pseudomonas syringae delivers the T3SS effector HopU1 (blue circle) that targets the RNA-binding protein GRP7. HopU1 interferes with GRP7’s ability to bind messenger RNAs, including immunity-related RNAs. Phytoplasmas are insect-transmitted, phloem-limited bacterial pathogens that secrete SAP effectors (blue star). SAP effectors are able to move throughout the plant via plasmodesmata, and some, such as SAP11 and SAP54, manipulate host transcription factors to induce plant developmental changes favoring insect colonization. Filamentous pathogens (oomycetes and fungi) as well as viruses deliver effectors to suppress plant RNA silencing and thus interfere with plant transcription factors and promote virulence. Botrytis cinerea is able to deliver fungal small RNAs into the host cells that suppress gene expression (red line).
Effector Modulation of Plant Transcription Factors
Phytoplasmas are a class of phloem-limited obligate plant pathogens and are insect transmitted (126). Phytoplasmas cause a variety of diseases and significantly impact host development. They belong to the class Mollicutes, lack an outer membrane, and do not possess a T3SS (40). However, phytoplasmas are able to directly secrete effectors outside the bacterial cell that can move throughout infected plants and interface with host transcription factors in the nucleus (126). The Aster Yellow phytoplasma strain Witches’ Broom (AY-WB) secretes the effector SAP11 that binds and destabilizes the transcription factors Cincinnata (CIN)-related teosinte branched1, cycloidea, proliferating cell factors 1 (TCP1), and TCP2, which are involved in plant development (125). SAP11 reduces the abundance of CIN-TCP transcription factors, including TCP4, which regulates the expression of lipoxygenase 2 (LOX2) involved in JA biosynthesis. Arabidopsis plants expressing SAP11 have reduced expression of LOX2 and produce less JA, indicating that SAP11 downregulates LOX2 expression and JA signaling by destabilizing TCP4 (125). Overexpression of SAP11 in Arabidopsis phenocopies AY-WB–infected plants, resulting in increased stem number and enhanced fecundity of the insect vector Macrosteles quadrilineatus. Silencing members of the Arabidopsis CIN-TCP family resulted in similar phenotypes (125). Plants infected with AY-WB also exhibit altered flower development, resulting in leaf-like flowers (phyllody), which is thought to enhance M. quadrilineatus attraction and fecundity. The effector SAP54 induces degradation of type II MADS-domain transcription factors involved in floral development, resulting in phyllody (77). Thus, phytoplasmas regulate plant development through degradation of transcription factors to favor insect vector colonization (Figure 3).
Host transcription factors appear to be hubs targeted by multiple pathogen effectors in diverse ways (87, 149). Effectors can directly target transcription factors or transcriptional repressors by altering their stability and subcellular localization, and by blocking their activity. For example, the RxLR effector Pi03192 from P. infestans and the coat protein of Turnip crinkle virus interact with host NAC transcription factors, preventing their localization to the nucleus, thereby affecting their function (83, 108) (Figure 3). Furthermore, TCP transcription factors appear to be key points of vulnerability in plant immune signaling and are targeted by diverse pathogens, including phytoplasmas (described above) as well as Phytophthora (123). JAZ proteins, which transcriptionally repress JA signaling, are targeted by multiple pathogen effectors, representing another vulnerable node in plant immune signaling (36, 52). WRKY transcription factors, known for their DNA-binding domain with a WRKYGQK sequence motif, are also key proteins regulating defense responses to several pathogens (98). Arabidopsis also possesses RRS1-R, a canonical intracellular immune receptor possessing an additional WRKY domain that mimics WRKY transcription factors targeted by pathogen effectors that function as a decoy to activate immunity (62, 113). Nematode effectors also interfere with host transcription. The 10A07 effector from the sugar beet nematode H. schachtii is phosphorylated by the plant kinase IPK, facilitating its nuclear localization and interaction with the auxin-responsive transcription factor INDOLE-3-ACETIC ACIDINDUCIBLE16(IAA16) (46). Thus, 10A07 interferes with auxin signaling by its interaction with IAA16 transcription factor.
Glorious TALEs: Effectors Acting as Plant Transcription Factors
Pathogen effectors can directly act as transcription factors and induce the expression of host susceptibility genes (Figure 3). These T3SS effectors are found in multiple Xanthomonas and Ralstonia bacterial pathogens and are called transcriptional activator-like effectors (TALEs). The elucidation of how TALEs specifically bind the promoters of their target genes has facilitated the identification of specific effector targets (9, 86). Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of rice bacterial blight, delivers the TALE PthXo1, which binds to the promoter region of OsSWEET11, a sucrose transporter gene, to induce its expression and promote bacterial pathogenicity (153). Another SWEET gene, OsSWEET14, is induced by Xoo TALEs AvrXa7 and PthXo3 (4). Thus, the virulence targets of multiple TALEs are sugar transporters in the SWEET gene family, likely facilitating sugar export for bacterial consumption (15) (Figure 3). Deciphering TALE DNA-binding specificity has enabled the prediction of target genes that confer bacterial susceptibility as well as targets present in resistant plants that drive the expression of defense genes (Figure 3). This knowledge has also enabled the generation of synthetic TALEs fused to nucleases, which have been heavily used for genome editing in plant, animal, and human cells (54). The widespread use of TALEs for genome editing illustrates the importance of investigating the mechanistic basis of effector function.
Effectors Targeting RNA Silencing Machinery
RNA silencing (or posttranscriptional gene silencing) is a conserved regulatory mechanism in eukaryotes. Plants also use the RNA silencing machinery as a defense mechanism against viral pathogens (21). Many plant viruses have an RNA genome and replicate via a double-stranded RNA intermediate (21). Double-stranded RNA is a potent trigger for RNA silencing, and enables the host to specifically target viral RNA for degradation. To overcome this defense response, multiple plant viruses have evolved suppressors of RNA silencing that are required for viral proliferation (21) (Figure 3). One of the first characterized viral suppressors of RNA silencing is the helper component proteinase P1/HC-Pro protein from Tobacco etch potyvirus (3, 58). P1/HC-Pro has been implicated in several functions, including viral genome amplification, polyprotein processing, cell-to-cell movement, and aphid transmission (136). However, all these functions are associated with its RNA silencing suppression activity. Several viral suppressors of RNA silencing target different components of the RNA silencing pathway, including 2b from Cucumber mosaic virus and P19 from Tomato bushy stunt virus (21). These suppressors are key effectors required for viral disease progression in plants.
Aside from viruses, other pathogen classes possess effectors that interfere with posttranscriptional processes. P. sojae delivers the RxLR effectors PSR1 and PSR2 (Phytophthora suppressor of RNA silencing 1 and 2), which act as suppressors of RNA silencing (Figure 3). PSR2-silenced P. sojae is less virulent on soybean plants (104). Expression of PSR1 in N. benthamiana increases pathogenicity of the Potato virus X and P. infestans, demonstrating the importance of this effector for pathogen proliferation (104). Arabidopsis plants expressing PSR1 have reduced accumulation of small RNAs, including precursor microRNAs (miRNAs) and small interfering RNAs (siRNAs) (104). PSR1 targets PSR1-interacting protein 1, a nuclear protein with an RNA helicase domain that regulates accumulation of miRNAs and siRNAs (105). Thus, PSR1 affects biogenesis of small RNAs, most likely in a Dicer-like dependent manner. PSR2 suppresses different components of RNA silencing and reduces the accumulation of specific trans-acting siRNAs (104). P. syringae can deliver effectors that interfere with the miRNA pathway, but it is not known whether this is a direct effect (90). Fungal pathogens can also manipulate RNA silencing components. The Botrytis cinerea fungus expresses small RNAs targeting the host silencing machinery to impair the expression of immune-related genes (148) (Figure 3). These data demonstrate that altering host transcriptional processes is a common mechanism utilized by pathogen effectors to suppress plant immune responses and facilitate pathogen proliferation.
CONCLUDING REMARKS
The ability to robustly deliver effectors enables pathogens to successfully colonize their hosts. Over the past two decades, research in effector biology has revealed diverse host targets and has significantly impacted our understanding of both pathogen and host biological processes. Diverse pathogens are able to target common plant immune components. For example, stealthy pathogens secrete cell death and immune suppressing effectors, whereas necrotrophic pathogens target the same components for promotion of cell death and immunity. Initial investigations have also revealed that effectors are deployed in complex spatial and temporal manners. Future research in effector biology will undoubtedly reveal novel strategies employed by pathogen effectors that can be exploited to control plant disease.
SUMMARY POINTS.
Effectors are excellent cellular probes to dissect diverse biological processes and points of vulnerability in plant immune signaling.
Effectors can modify plant gene expression by modulating transcription factors, directly regulating transcription, and interfering with plant RNA silencing machinery.
Filamentous pathogens express effectors in waves during infection.
Effectors from diverse pathogens differentially target similar cellular processes in order to achieve opposite outcomes.
Manipulation of PRR complexes, hormone signaling, vesicular trafficking, and gene expression are common strategies used by diverse plant pathogens.
FUTURE ISSUES.
Do effectors play roles beyond plant-microbe interactions?
Do effectors from different pathogens act in synergistic or antagonistic manners in mixed plant infections?
How are effectors from diverse pathogens delivered inside host cells?
What mechanisms (e.g., transcription factors) are regulating the expression of eukaryotic pathogen effectors over time, space, and plant organ?
Are there correlations between effector conservation, expression, and contribution to virulence?
Are there differences in expression and hierarchy of delivery for bacterial type III effectors?
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants. 2015;1:15140. doi: 10.1038/nplants.2015.140. [DOI] [PubMed] [Google Scholar]
- 2.Alfano JR, Charkowski AO, Deng W, Badel JL, Petnicki-Ocwieja T, et al. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. PNAS. 2000;97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, et al. A viral suppressor of gene silencing in plants. PNAS. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antony G, Zhou J, Huang S, Li T, Liu B, et al. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 2010;22:3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell. 2004;15:5118–5129. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakhetia M, Urwin PE, Atkinson HJ. QPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Mol. Plant-Microbe Interact. 2007;20:306–312. doi: 10.1094/MPMI-20-3-0306. [DOI] [PubMed] [Google Scholar]
- 7.Bekal S, Niblack TL, Lambert KN. A chorismate mutase from the soybean cyst nematode Heterodera glycines shows polymorphisms that correlate with virulence. Mol. Plant-Microbe Interact. 2003;16:439–446. doi: 10.1094/MPMI.2003.16.5.439. [DOI] [PubMed] [Google Scholar]
- 8.Bender CL, Alarcón-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 10.Bolton MD, van Esse HP, Vossen JH, de Jonge R, Stergiopoulos I, et al. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 2008;69:119–136. doi: 10.1111/j.1365-2958.2008.06270.x. [DOI] [PubMed] [Google Scholar]
- 11.Bozkurt TO, Schornack S, Win J, Shindo T, Ilyas M, et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. PNAS. 2011;108:20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD. A downy mildew effector attenuates salicylic acid–triggered immunity in Arabidopsis by interacting with the host mediator complex. PLOS Biol. 2013;11:e1001732. doi: 10.1371/journal.pbio.1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat. Rev. Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaparro-Garcia A, Schwizer S, Sklenar J, Yoshida K, Petre B, et al. Phytophthora infestans RXLR-WY effector AVR3a associates with dynamin-related protein 2 required for endocytosis of the plant pattern recognition receptor FLS2. PLOS ONE. 2015;10:e0137071. doi: 10.1371/journal.pone.0137071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Hou B, Lalonde S, Takanaga H, Hartung ML, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–534. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Lang P, Chronis D, Zhang S, De Jong WS, et al. In planta processing and glycosylation of a nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-like effector and its interaction with a host CLAVATA2-like receptor to promote parasitism. Plant Physiol. 2015;167:262–272. doi: 10.1104/pp.114.251637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang Y-H, Coaker G. Effector triggered immunity: NLR immune perception and downstream defense responses. Arabidopsis Book. 2015;11:e0183. [Google Scholar]
- 18.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 19.Ciuffetti LM, Manning VA, Pandelova I, Betts MF, Martinez JP. Host-selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis–wheat interaction. New Phytol. 2010;187:911–919. doi: 10.1111/j.1469-8137.2010.03362.x. [DOI] [PubMed] [Google Scholar]
- 20.Cornelis GR. The type III secretion injectisome, a complex nanomachine for intracellular “toxin” delivery. Biol. Chem. 2010;391:745–751. doi: 10.1515/BC.2010.079. [DOI] [PubMed] [Google Scholar]
- 21.Csorba T, Kontra L, Burgyan J. Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology. 2015;479–480:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 22.Cui H, Wang Y, Xue L, Chu J, Yan C, et al. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–175. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, et al. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science. 2010;329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- 24.de Wit PJGM, van der Burgt A, Okmen B, Stergiopoulos I, Abd-Elsalam KA, et al. The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLOS Genet. 2012;8:e1003088. doi: 10.1371/journal.pgen.1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, et al. Metabolic priming by a secreted fungal effector. Nature. 2011;478:395–398. doi: 10.1038/nature10454. [DOI] [PubMed] [Google Scholar]
- 26.Doehlemann G, Reissmann S, Assmann D, Fleckenstein M, Kahmann R. Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis–induced tumour formation. Mol. Microbiol. 2011;81:751–766. doi: 10.1111/j.1365-2958.2011.07728.x. [DOI] [PubMed] [Google Scholar]
- 27.Doehlemann G, van der Linde K, Assmann D, Schwammbach D, Hof A, et al. Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLOS Pathog. 2009;5:e1000290. doi: 10.1371/journal.ppat.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, et al. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2008;56:181–195. doi: 10.1111/j.1365-313X.2008.03590.x. [DOI] [PubMed] [Google Scholar]
- 29.Du Y, Mpina MH, Birch PRJ, Bouwmeester K, Govers F. Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 2015;169:1975–1990. doi: 10.1104/pp.15.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faris JD, Zhang ZC, Lu HJ, Lu SW, Reddy L, et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. PNAS. 2010;107:13544–13549. doi: 10.1073/pnas.1004090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng F, Yang F, Rong W, Wu X, Zhang J, et al. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485:114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- 32.Friesen TL, Stukenbrock EH, Liu ZH, Meinhardt S, Ling H, et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 2006;38:953–956. doi: 10.1038/ng1839. [DOI] [PubMed] [Google Scholar]
- 33.Fu ZQ, Guo M, Jeong B, Tian F, Elthon TE, et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–289. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 34.Fujisaki K, Abe Y, Ito A, Saitoh H, Yoshida K, et al. Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J. 2015;83:875–887. doi: 10.1111/tpj.12934. [DOI] [PubMed] [Google Scholar]
- 35.Gangadharan A, Sreerekha M, Whitehill J, Ham JH, Mackey D. The Pseudomonas syringae pv. tomato type III effector HopM1 suppresses Arabidopsis defenses independent of suppressing salicylic acid signaling and of targeting AtMIN7. PLOS ONE. 2013;8:e82032. doi: 10.1371/journal.pone.0082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gimenez-Ibanez S, Boter M, Fernández-Barbero G, Chini A, Rathjen JP, Solano R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLOS Biol. 2014;12:e1001792. doi: 10.1371/journal.pbio.1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 2009;19:1–7. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 38.Gohre V, Spallek T, Haweker H, Mersmann S, Mentzel T, et al. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 2008;18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- 39.Goverse A, Smant G. The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 2014;52:243–265. doi: 10.1146/annurev-phyto-102313-050118. [DOI] [PubMed] [Google Scholar]
- 40.Gundersen DE, Lee IM, Rehner SA, Davis RE, Kingsbury DT. Phylogeny of mycoplasma like organisms (phytoplasmas): a basis for their classification. J. Bacteriol. 1994;176:5244–5254. doi: 10.1128/jb.176.17.5244-5254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo M, Tian F, Wamboldt Y, Alfano JR. The majority of the type III effector inventory of Pseudomonas syringae pv tomato DC3000 can suppress plant immunity. Mol. Plant-Microbe Interact. 2009;22:1069–1080. doi: 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guy E, Lautier M, Chabannes M, Roux B, Lauber E, et al. xopAC-triggered immunity against Xanthomonas depends on Arabidopsis receptor-like cytoplasmic kinase genes PBL2 and RIPK. PLOS ONE. 2013;8:e73469. doi: 10.1371/journal.pone.0073469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hacquard S, Kracher B, Maekawa T, Vernaldi S, Schulze-Lefert P, Ver Loren van Themaat E. Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. PNAS. 2013;110:E2219–E2228. doi: 10.1073/pnas.1306807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, Doehlemann G. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLOS Pathog. 2012;8:e1002684. doi: 10.1371/journal.ppat.1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hemetsberger C, Mueller AN, Matei A, Herrberger C, Hensel G, et al. The fungal core effector Pep1 is conserved across smuts of dicots and monocots. New Phytol. 2015;206:1116–1126. doi: 10.1111/nph.13304. [DOI] [PubMed] [Google Scholar]
- 46.Hewezi T, Juvale PS, Piya S, Maier TR, Rambani A, et al. The cyst nematode effector protein 10A07 targets and recruits host posttranslational machinery to mediate its nuclear trafficking and to promote parasitism in Arabidopsis. Plant Cell. 2015;27:891–907. doi: 10.1105/tpc.114.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horger AC, van der Hoorn RA. The structural basis of specific protease-inhibitor interactions at the plant-pathogen interface. Curr. Opin. Struct. Biol. 2013;23:842–850. doi: 10.1016/j.sbi.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Howard RJ, Valent B. Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 1996;50:491–512. doi: 10.1146/annurev.micro.50.1.491. [DOI] [PubMed] [Google Scholar]
- 49.Hurley B, Lee D, Mott A, Wilton M, Liu J, et al. The Pseudomonas syringae type III effectorHopF2 suppresses Arabidopsis stomatal immunity. PLOS ONE. 2014;9:e114921. doi: 10.1371/journal.pone.0114921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irieda H, Maeda H, Akiyama K, Hagiwara A, Saitoh H, et al. Colletotrichum orbiculare secretes virulence effectors to a biotrophic interface at the primary hyphal neck via exocytosis coupled with SEC22-mediated traffic. Plant Cell. 2014;26:2265–2281. doi: 10.1105/tpc.113.120600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jashni MK, Mehrabi R, Collemare J, Mesarich CH, de Wit PJGM. The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci. 2015;6:584. doi: 10.3389/fpls.2015.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang S, Yao J, Ma KW, Zhou H, Song J, et al. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLOS Pathog. 2013;9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones JT, Furlanetto C, Bakker E, Banks B, Blok V, et al. Characterization of a chorismate mutase from the potato cyst nematode Globodera pallida. Mol. Plant Pathol. 2003;4:43–50. doi: 10.1046/j.1364-3703.2003.00140.x. [DOI] [PubMed] [Google Scholar]
- 54.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jupe J, Stam R, Howden AJ, Morris JA, Zhang R, et al. Phytophthora capsici–tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 2013;14:R63. doi: 10.1186/gb-2013-14-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 57.Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, et al. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010;154:1794–1804. doi: 10.1104/pp.110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasschau KD, Carrington JC. A counter defensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 59.Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park S-Y, et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell. 2010;22:1388–1403. doi: 10.1105/tpc.109.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleemann J, Rincon-Rivera LJ, Takahara H, Neumann U, Ver Loren van Themaat E, et al. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLOS Pathog. 2012;8:e1002643. doi: 10.1371/journal.ppat.1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leasure CD, He ZH. CLE and RGF family peptide hormone signaling in plant development. Mol. Plant. 2012;5:1173–1175. doi: 10.1093/mp/sss082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Roux C, Huet G, Jauneau A, Camborde L, Tremousaygue D, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 63.Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, et al. Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell. 2015;27:3038–3064. doi: 10.1105/tpc.15.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L, Li M, Yu L, Zhou Z, Liang X, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15:329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, Elmore JM, Lin ZD, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu T, Song T, Zhang X, Yuan H, Su L, et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014;5:4686. doi: 10.1038/ncomms5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu TT, Liu ZX, Song CJ, Hu YF, Han ZF, et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336:1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 68.Lozano-Durán R, Bourdais G, He SY, Robatzek S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014;202:259–269. doi: 10.1111/nph.12651. [DOI] [PubMed] [Google Scholar]
- 69.Lozano-Durán R, Rosas-Diaz T, Gusmaroli G, Luna AP, Taconnat L, et al. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCFE3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell. 2011;23:1014–1032. doi: 10.1105/tpc.110.080267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lozano-Torres JL, Wilbers RHP, Gawronski P, Boshoven JC, Finkers-Tomczak A, et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. PNAS. 2012;109:10119–10124. doi: 10.1073/pnas.1202867109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. PNAS. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu SW, Tian D, Borchardt-Wier HB, Wang X. Alternative splicing: a novel mechanism of regulation identified in the chorismate mutase gene of the potato cyst nematode Globodera rostochiensis. Mol. Biochem. Parasitol. 2008;162:1–15. doi: 10.1016/j.molbiopara.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 73.Lu SW, Turgeon BG, Edwards MC. A ToxA-like protein from Cochliobolus heterostrophus induces light-dependent leaf necrosis and acts as a virulence factor with host selectivity on maize. Fungal Genet. Biol. 2015;81:12–24. doi: 10.1016/j.fgb.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 74.Ma K-W, Jiang S, Hawara E, Lee D, Pan S, et al. Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 2015;208:1157–1168. doi: 10.1111/nph.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science. 2014;343:1509–1512. doi: 10.1126/science.1248849. [DOI] [PubMed] [Google Scholar]
- 76.Macho AP, Zipfel C. Targeting of plant pattern recognition receptor triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 2015;23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 77.MacLean AM, Orlovskis Z, Kowitwanich K, Zdziarska AM, Angenent GC, et al. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLOS Biol. 2014;12:e1001835. doi: 10.1371/journal.pbio.1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012;63:73–105. doi: 10.1146/annurev-arplant-042811-105439. [DOI] [PubMed] [Google Scholar]
- 79.Manning VA, Ciuffetti LM. Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell. 2005;17:3203–3212. doi: 10.1105/tpc.105.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manning VA, Hardison LK, Ciuffetti LM. Ptr ToxA interacts with a chloroplast-localized protein. Mol. Plant-Microbe Interact. 2007;20:168–177. doi: 10.1094/MPMI-20-2-0168. [DOI] [PubMed] [Google Scholar]
- 81.Marshall R, Kombrink A, Motteram J, Loza-Reyes E, Lucas J, et al. Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 2011;156:756–769. doi: 10.1104/pp.111.176347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McDonald MC, Oliver RP, Friesen TL, Brunner PC, McDonald BA. Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol. 2013;199:241–251. doi: 10.1111/nph.12257. [DOI] [PubMed] [Google Scholar]
- 83.McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, et al. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLOS Pathog. 2013;9:e1003670. doi: 10.1371/journal.ppat.1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 85.Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell. 2012;24:322–335. doi: 10.1105/tpc.111.092957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moscou M, Bogdanove A. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 87.Mukhtar MS, Carvunis A, Dreze M, Epple P, Steinbrenner J, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Munch S, Lingner U, Floss DS, Ludwig N, Sauer N, Deising HB. The hemibiotrophic lifestyle of Colletotrichum species. J. Plant Physiol. 2008;165:41–51. doi: 10.1016/j.jplph.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 89.Naessens E, Dubreuil G, Giordanengo P, Baron OL, Minet-Kebdani N, et al. A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr. Biol. 2015;25:1898–1903. doi: 10.1016/j.cub.2015.05.047. [DOI] [PubMed] [Google Scholar]
- 90.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nicaise V, Joe A, Jeong B, Korneli C, Boutrot F, et al. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013;32:701–712. doi: 10.1038/emboj.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nomura K, DebRoy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 93.Nomura K, Mecey C, Lee Y, Imboden LA, Chang JH, He SY. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. PNAS. 2011;108:10774–10779. doi: 10.1073/pnas.1103338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012;44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oliva RF, Cano LM, Raffaele S, Win J, Bozkurt TO, et al. A recent expansion of the RXLR effector gene Avrblb2 is maintained in global populations of Phytophthora infestans indicating different contributions to virulence. Mol. Plant-Microbe Interact. 2015;28:901–912. doi: 10.1094/MPMI-12-14-0393-R. [DOI] [PubMed] [Google Scholar]
- 96.Oliver RP, Friesen TL, Faris JD, Solomon PS. Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 2012;50:23–43. doi: 10.1146/annurev-phyto-081211-173019. [DOI] [PubMed] [Google Scholar]
- 97.Oome S, Raaymakers TM, Cabral A, Samwel S, Bohm H, et al. Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. PNAS. 2014;111:16955–16960. doi: 10.1073/pnas.1410031111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Patel N, Hamamouch N, Li C, Hussey R, Mitchum M, et al. Similarity and functional analyses of expressed parasitism genes in Heterodera schachtii and Heterodera glycines. J. Nematol. 2008;40:299–310. [Google Scholar]
- 100.Pecenkova T, Hala M, Kulich I, Kocourkova D, Drdova E, et al. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 2011;62:2107–2116. doi: 10.1093/jxb/erq402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pemberton CL, Salmond GPC. The Nep1-like proteins: a growing family of microbial elicitors of plant necrosis. Mol. Plant Pathol. 2004;5:353–359. doi: 10.1111/j.1364-3703.2004.00235.x. [DOI] [PubMed] [Google Scholar]
- 102.Petre B, Kamoun S. How do filamentous pathogens deliver effector proteins into plant cells? PLOS Biol. 2014;12:e1001801. doi: 10.1371/journal.pbio.1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pieterse CM, Van derDoes D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 104.Qiao Y, Liu L, Xiong Q, Flores C, Wong J, et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013;45:330–333. doi: 10.1038/ng.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qiao Y, Shi J, Zhai Y, Hou Y, Ma W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. PNAS. 2015;112:5850–5855. doi: 10.1073/pnas.1421475112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qutob D, Kemmerling B, Brunner F, Kufner I, Engelhardt S, et al. Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell. 2006;18:3721–3744. doi: 10.1105/tpc.106.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Redkar A, Hoser R, Schilling L, Zechmann B, Krzymowska M, et al. A secreted effector protein of Ustilago maydis guides maize leaf cells to form tumors. Plant Cell. 2015;27:1332–1351. doi: 10.1105/tpc.114.131086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ren T, Qu F, Morris TJ. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to Turnip crinkle virus. Plant Cell. 2000;12:1917–1925. doi: 10.1105/tpc.12.10.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Replogle A, Wang J, Bleckmann A, Hussey RS, Baum TJ, et al. Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 2011;65:430–440. doi: 10.1111/j.1365-313X.2010.04433.x. [DOI] [PubMed] [Google Scholar]
- 110.Replogle A, Wang J, Paolillo V, Smeda J, Kinoshita A, et al. Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis. Mol. Plant-Microbe Interact. 2013;26:87–96. doi: 10.1094/MPMI-05-12-0118-FI. [DOI] [PubMed] [Google Scholar]
- 111.Rooney HCE, van’t Klooster JW, van der Hoorn RAL, Joosten MHAJ, Jones JDG, de Wit PJGM. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- 112.Sanchez-Vallet A, Saleem-Batcha R, Kombrink A, Hansen G, Valkenburg DJ, et al. Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife. 2013;2:e00790. doi: 10.7554/eLife.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 114.Schilling L, Matei A, Redkar A, Walbot V, Doehlemann G. Virulence of the maize smut Ustilago maydis is shaped by organ-specific effectors. Mol. Plant Pathol. 2014;15:780–789. doi: 10.1111/mpp.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schornack S, vanDamme M, Bozkurt TO, Cano LM, Smoker M, et al. Ancient class of translocated oomycete effectors targets the host nucleus. PNAS. 2010;107:17421–17426. doi: 10.1073/pnas.1008491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sels J, Mathys J, De Coninck BMA, Cammue BPA, De Bolle MFC. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 2008;46:941–950. doi: 10.1016/j.plaphy.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 117.Serrano M, Coluccia F, Torres M, L’Haridon F, Metraux JP. The cuticle and plant defense to pathogens. Front. Plant Sci. 2014;5:274. doi: 10.3389/fpls.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shabab M, Shindo T, Gu C, Kaschani F, Pansuriya T, et al. Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell. 2008;20:1169–1183. doi: 10.1105/tpc.107.056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shan L, He P, Li J, Heese A, Peck SC, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 121.Skibbe DS, Doehlemann G, Fernandes J, Walbot V. Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science. 2010;328:89–92. doi: 10.1126/science.1185775. [DOI] [PubMed] [Google Scholar]
- 122.Song J, Win J, Tian MY, Schornack S, Kaschani F, et al. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. PNAS. 2009;106:1654–1659. doi: 10.1073/pnas.0809201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stam R, Motion G, Boevink PC, Huitema E. A conserved oomycete CRN effector targets and modulates tomato TCP14-2 to enhance virulence. BioRxiv. 2013 doi: 10.1094/MPMI-06-20-0172-R. doi: http://dx.doi.org/10.1101/001248. [DOI] [PubMed] [Google Scholar]
- 124.Stergiopoulos I, van den Burg HA, Okmen B, Beenen HG, van Liere S, et al. Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. PNAS. 2010;107:7610–7615. doi: 10.1073/pnas.1002910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. PNAS. 2011;108:E1254–E1263. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sugio A, MacLean AM, Kingdom HN, Grieve VM, Manimekalai R, Hogenhout SA. Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu. Rev. Phytopathol. 2011;49:175–195. doi: 10.1146/annurev-phyto-072910-095323. [DOI] [PubMed] [Google Scholar]
- 127.Tan KC, Oliver RP, Solomon PS, Moffat CS. Proteinaceous necrotrophic effectors in fungal virulence. Funct. Plant Biol. 2010;37:907–912. [Google Scholar]
- 128.Tanaka H, Kitakura S, De Rycke R, De Groodt R, Friml J. Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 2009;19:391–397. doi: 10.1016/j.cub.2009.01.057. [DOI] [PubMed] [Google Scholar]
- 129.Tanaka S, Brefort T, Neidig N, Djamei A, Kahnt J, et al. A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife. 2014;3:e01355. doi: 10.7554/eLife.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Teh OK, Hofius D. Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J. Exp. Bot. 2014;65:1297–1312. doi: 10.1093/jxb/ert441. [DOI] [PubMed] [Google Scholar]
- 131.Tian MY, Benedetti B, Kamoun S. A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 2005;138:1785–1793. doi: 10.1104/pp.105.061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tian MY, Huitema E, da Cunha L, Torto-Alalibo T, Kamoun S. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 2004;279:26370–26377. doi: 10.1074/jbc.M400941200. [DOI] [PubMed] [Google Scholar]
- 133.Tian MY, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 2007;143:364–377. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLOS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RHY, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 136.Urcuqui-Inchima S, Haenni AL, Bernardi F. Potyvirus proteins: a wealth of functions. Virus Res. 2001;74:157–175. doi: 10.1016/s0168-1702(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 137.van Damme M, Bozkurt TO, Cakir C, Schornack S, Sklenar J, et al. The Irish potato famine pathogen Phytophthora infestans translocates the CRN8 kinase into host plant cells. PLOS Pathog. 2012;8:e1002875. doi: 10.1371/journal.ppat.1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van den Burg HA, Harrison SJ, Joosten MHAJ, Vervoort J, de Wit PJGM. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant-Microbe Interact. 2006;19:1420–1430. doi: 10.1094/MPMI-19-1420. [DOI] [PubMed] [Google Scholar]
- 139.van der Hoorn RAL, Kamoun S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van Esse HP, Bolton MD, Stergiopoulos I, de Wit PJGM, Thomma BPHJ. The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant-Microbe Interact. 2007;20:1092–1101. doi: 10.1094/MPMI-20-9-1092. [DOI] [PubMed] [Google Scholar]
- 141.van Esse HP, van’t Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, et al. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell. 2008;20:1948–1963. doi: 10.1105/tpc.108.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vanholme B, Kast P, Haegeman A, Jacob J, Grunewald W, Gheysen G. Structural and functional investigation of a secreted chorismate mutase from the plant-parasitic nematode Heterodera schachtii in the context of related enzymes from diverse origins. Mol. Plant Pathol. 2009;10:189–200. doi: 10.1111/j.1364-3703.2008.00521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vleeshouwers VGAA, Oliver RP. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant-Microbe Interact. 2014;27:196–206. doi: 10.1094/MPMI-10-13-0313-IA. [DOI] [PubMed] [Google Scholar]
- 144.Wang G, Roux B, Feng F, Guy E, Li L, et al. The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe. 2015;18:285–295. doi: 10.1016/j.chom.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 145.Wang J, Replogle A, Hussey R, Baum T, Wang X, et al. Identification of potential host plant mimics of CLAVATA3/ESR (CLE)-like peptides from the plant-parasitic nematode Heterodera schachtii. Mol. Plant Pathol. 2011;12:177–186. doi: 10.1111/j.1364-3703.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Q, Han C, Ferreira AO, Yu X, Ye W, et al. Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell. 2011;23:2064–2086. doi: 10.1105/tpc.111.086082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang XL, Jiang N, Liu JL, Liu WD, Wang GL. The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence. 2014;5:722–732. doi: 10.4161/viru.29798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wessling R, Epple P, Altmann S, He Y, Yang L, et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16:364–375. doi: 10.1016/j.chom.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Will T, Tjallingii WF, Thonnessen A, van Bel AJ. Molecular sabotage of plant defense by aphid saliva. PNAS. 2007;104:10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wubben JP, Lawrence CB, de Wit PJGM. Differential induction of chitinase and 1,3-β-glucanase gene expression in tomato by Cladosporium fulvum and its race-specific elicitors. Physiol. Mol. Plant Pathol. 1996;48:105–116. [Google Scholar]
- 152.Xiang T, Zong N, Zou Y, Wu Y, Zhang J, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 2008;18:1–7. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 153.Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. PNAS. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang Y, Li L, Qu LJ. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 2015;58:106–118. doi: 10.1111/jipb.12377. [DOI] [PubMed] [Google Scholar]
- 155.Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010;153:1188–1198. doi: 10.1104/pp.110.157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang J, Li W, Xiang T, Liu Z, Laluk K, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 157.Zhang J, Shao F, Li Y, Cui H, Chen L, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 158.Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, et al. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe. 2012;11:253–263. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 159.Zhou J, Wu S, Chen X, Liu C, Sheen J, et al. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 2014;77:235–245. doi: 10.1111/tpj.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhou Z, Wu Y, Yang Y, Du M, Zhang X, et al. An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell. 2015;27:2032–2041. doi: 10.1105/tpc.15.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014;35:345–351. doi: 10.1016/j.it.2014.05.004. [DOI] [PubMed] [Google Scholar]