Abstract
Phosphorylation of the aquaporin-2 (AQP2) water channel at four COOH-terminal serines plays a central role in the regulation of water permeability of the renal collecting duct. The level of phosphorylation at these sites is determined by a balance between phosphorylation by protein kinases and dephosphorylation by phosphatases. The phosphatases that dephosphorylate AQP2 have not been identified. Here, we use large-scale data integration techniques to identify serine-threonine phosphatases likely to interact with AQP2 in renal collecting duct principal cells. As a first step, we have created a comprehensive list of 38 S/T phosphatase catalytic subunits present in the mammalian genome. Then we used Bayes’ theorem to integrate available information from large-scale data sets from proteomic and transcriptomic studies to rank the known S/T phosphatases with regard to the likelihood that they interact with AQP2 in renal collecting duct cells. To broaden the analysis, we have generated new proteomic data (LC-MS/MS) identifying 4538 distinct proteins including 22 S/T phosphatases in cytoplasmic fractions from native inner medullary collecting duct cells from rats. The official gene symbols corresponding to the top-ranked phosphatases (common names in parentheses) were: Ppp1cb (PP1-β), Ppm1g (PP2C), Ppp1ca (PP1-α), Ppp3ca (PP2-B or calcineurin), Ppp2ca (PP2A-α), Ppp1cc (PP1-γ), Ppp2cb (PP2A-β), Ppp6c (PP6C), and Ppp5c (PP5). This ranking correlates well with results of prior reductionist studies of ion and water channels in renal collecting duct cells.
Keywords: systems biology, collecting duct, kidney, vasopressin, LC-MS/MS
aquaporin-2 (AQP2) is an integral membrane protein that allows water molecules to cross the apical plasma membrane of collecting duct cells in the renal tubule of the mammalian kidney (22). In the kidney, its regulation is mainly controlled by the peptide hormone vasopressin, which is synthesized in the hypothalamus, then stored and secreted from the posterior pituitary. Vasopressin controls AQP2 in at least two ways (22): regulation of AQP2 levels in the plasma membrane via trafficking of AQP2 containing membrane vesicles, and control of the overall abundance of AQP2 present in the principal cells by regulating transcription of the Aqp2 gene.
A common process by which transporters are regulated involves the control of the level of phosphorylation. The putting on and taking off of phosphates at regulatory sites is performed, respectively, by protein kinases, which add phosphate groups, and protein phosphatases, which remove them. The overall level of phosphorylation, then, depends on the net activity of these two types of enzymes. There are at least four vasopressin-regulated phosphorylation sites in the AQP2 protein at Serine residues 256, 261, 264, and 269 (7, 11). Phosphorylation at these sites governs the trafficking of AQP2 into and out of the collecting duct apical plasma membrane (2, 12, 14, 21, 28). The phosphatases that reverse phosphorylation at these sites belong to the Serine/Threonine (S/T) phosphatase subgroup. Our goal here is to identify which of the serine/threonine (S/T) protein phosphatases coded by the mammalian genome are most likely to interact with AQP2 in the renal collecting duct. This analysis uses prior transcriptomic and proteomic data from inner medullary collecting ducts (IMCDs) isolated from rat kidneys (31), from cultured mouse mpkCCD cells (a vasopressin-responsive cell culture model of cortical collecting duct) (19, 35), from microdissected rat collecting duct segments (16), as well as new data presented in this paper profiling the cytoplasmic proteome of rat IMCDs.
The present paper has three aims: 1) to curate a list of S/T phosphatase catalytic subunits present in the mammalian genome and make the information available via a publicly accessible webpage; 2) to use Bayes’ rule to integrate available information from large-scale data sets to predict what S/T phosphatases may interact with AQP2 in renal collecting duct cells; and 3) to relate the phosphatase predictions made via Bayesian analysis to the reductionist literature on regulation of transport in collecting duct principal cells.
METHODS
Assembly of a Database of Mammalian Protein Phosphatase Catalytic Subunits
We assembled a list of the known serine/threonine (S/T) phosphatase catalytic subunits in human, mouse, and rat genomes. The original list was taken from DEPOD (the human DEPhOsphorylation Database at http://www.koehn.embl.de/depod/), and the amino acid sequences of the catalytic regions of representative members of each major family were used in multiple BLAST searches of proteomic (https://hpcwebapps.cit.nih.gov/ESBL/Database/IMCD_Proteome/index.html) and transcriptomic data (https://hpcwebapps.cit.nih.gov/ESBL/Database/Transcriptomic/IMCDdatabase.html) to identify other members relevant to collecting duct transport. Human gene symbols were converted to mouse symbols using the Automated Bioinformatics Extractor (ABE) tool (https://hpcwebapps.cit.nih.gov/ESBL/ABE/). The data were curated on an Excel spreadsheet, which was converted to an .html file, edited with Notepad++, and posted on a permanent web server to make it available for public access at https://hpcwebapps.cit.nih.gov/ESBL/Database/Phosphatases/. This database was used as a beginning point in the identification of the S/T phosphatases most likely to interact with AQP2 in the renal collecting duct. The database excludes regulatory subunits.
Use of Bayes’ Theorem to Predict AQP2-Phosphatase Interactions
To determine the S/T phosphatases most likely to dephosphorylate AQP2 at any of its four vasopressin-regulated phosphorylation sites, we ranked the S/T phosphatases using Bayes’ theorem (1, 20) as illustrated in Fig. 1A. Here the prior probability vector, P(A), and the vector of likelihoods from any new data set, P(B|A), are used to calculate a new posterior probability vector P(A|B). The term P(B) is calculated as the scalar sum of probabilities of B over all A. For the calculations in this study, the dimension of the vectors is 38, reflecting all 38 S/T phosphatase catalytic subunits found in mammalian genomes. As shown in Fig. 1B, the application of each step using Bayes’ rule can be represented as a so-called "Bayesian operator" that receives two vectors of equal length as inputs and produces a new posterior probability vector of the same length. This operator can be applied multiple times in series to integrate multiple data sets. The posterior probability vector from each step is used as the prior probability vector for the next. To initiate the process on the first step, we set all values for the prior probability vector to 1/38, i.e., a uniform probability distribution over all S/T phosphatases.
Fig. 1.
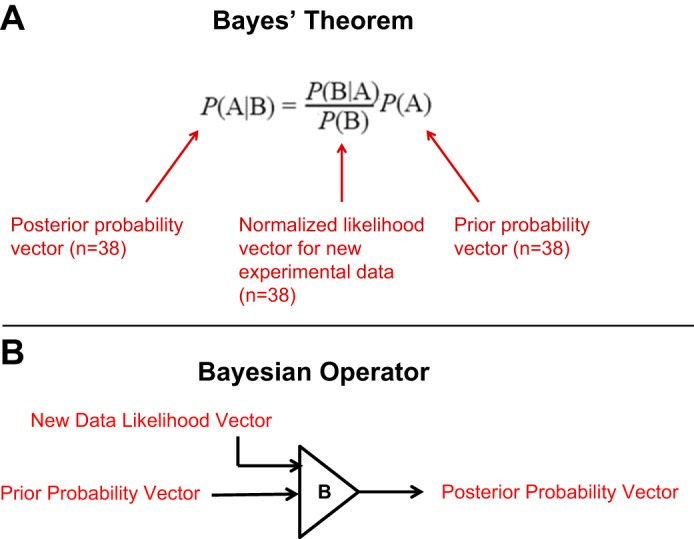
Bayes’ rule is used to integrate multiple data sets. A: Bayes’ rule integrates 2 probability vectors (dimension = 38 for family of S/T phosphatases): a prior probability vector and a “likelihood vector” derived from values in a particular large-scale data set. The result is a single posterior probability vector that can be used as a new prior probability vector to integrate the next data set. B: the application of Bayes’ rule in this setting can be viewed as application of a “Bayesian operator” that converts 2 equal-length vectors to obtain a new vector of the same length. Use of this operator can be repeated multiple times to integrate different data sets into the overall calculation, using the posterior probability vector of 1 step as the prior probability vector of the next step.
Table 1 lists the data sets used and the methods for estimating the likelihood vector for each. Where possible we use the complement of the minimum Bayes’ factor as described by Goodman (10) to estimate likelihood values from quantitative data. Table 1 also includes the rationale for use of each data set. The rationale for use of data sets 1, 3, and 4 (Table 1) is that for a phosphatase to interact with AQP2 in collecting duct cells, its gene must be expressed. The assignment of likelihood values recognizes that "not found" does not necessarily mean "not expressed," so no absolute zero likelihood values are assigned for any S/T phosphatase. The rationale for application of data set 2 (Table 1) is “If a phosphatase plays a critical role in regulation of AQP2, we expect it to be expressed in all collecting duct segments.” This is simply application of the principle of Ockham’s razor or the Law of Parsimony (https://en.wikipedia.org/wiki/Occam's_razor). Specifically, when one considers the ways that AQP2 phosphorylation can be regulated, the simplest view is that the same phosphatases are involved in all AQP2-expressing renal tubule segments. The general rationale for data sets 5–8 (Table 1) is that for a phosphatase to interact with AQP2, it must be present in the same subcellular compartments as AQP2. This question is addressed in different ways using data from proteomic analysis of differential centrifugation fractions in mpkCCD cells (database 5), data from surface biotinylation in mpkCCD cells (database 6), proteomic analysis of cytoplasmic fractions from mpkCCD cells (database 7), and proteomic analysis of an IMCD cytoplasmic fraction (database 8). In this paper, we use the term “cytoplasmic fraction” to refer to all nonnuclear elements of the cells including membrane components, cytosol, and organelles. Since AQP2 is localized to the cytoplasmic fraction, we argue that interacting proteins must also reside in the cytoplasmic fraction.
Table 1.
Data sets used in Bayes' calculations in this study
| Data Set | Rationale | Assignment of Likelihood Values: P(B|A) | Noise Threshold | URL |
|---|---|---|---|---|
| Rat IMCD transcriptome (data set 1) | The ability of a phosphatase to dephosphorylate aquaporin-2 depends on whether or not it is expressed in collecting duct cells. | Complements of minimal Bayes' factors (z* = value/noise) | 0.4 | https://esbl.nhlbi.nih.gov/IMCD-transcriptome/ |
| Rat RNA-Seq (data set 2) | If a phosphatase plays a critical role in regulation of aquaporin-2, we expected it to be expressed in all collecting duct segments. | Complements of minimal Bayes' factors derived from χ2 statistic | https://hpcwebapps.cit.nih.gov/ESBL/Database/NephronRNAseq/All_transcripts.html | |
| Mouse mpkCCD transcriptome (data set 3) | The ability of a phosphatase to dephosphorylate aquaporin-2 depends on whether or not it is expressed in collecting duct cells. | Complements of Minimal Bayes' Factors (z* = value/noise) | 0.4 | https://esbl.nhlbi.nih.gov/mpkCCD-transcriptome/ |
| Mouse mpkCCD proteome (data set 4) | The ability of a phosphatase to dephosphorylate aquaporin-2 depends on whether or not it is expressed in collecting duct cells. | Complements of minimal Bayes' factors (z* = value/noise) | 10th percentile value | https://hpcwebapps.cit.nih.gov/ESBL/Database/mpkCCD_Protein_Abundances/ |
| Dot products from mpkCCD subcellular fraction proteomics (data set 5) | The ability of a phosphatase to dephosphorylate aquaporin-2 depends on whether or not it is found in the same subcellular compartment. | Complements of minimal Bayes' factors (z* = value/noise) | median dot product | https://hpcwebapps.cit.nih.gov/ESBL/Database/mpkFractions/proteomic_fractions_linear.html |
| Apical mpkCCD proteome (data set 6) | The ability of a phosphatase to dephosphorylate aquaporin-2 is greater if it is found in the apical region of the cell. | Complements of minimal Bayes' factors derived from χ2 statistic | http://sbel.mc.ntu.edu.tw/mpkCCDqAMP/qAMP.htm#SamePlace | |
| mpkCCD cytoplasmic fraction proteome (Data set 7) | The ability of a phosphatase to dephosphorylate aquaporin-2 depends on whether or not it is found in the cytoplasm. | If found in cytoplasm fraction, P = 0.95; if found only in nuclear fraction, P = 0.2; if not found, P = 0.4 | https://hpcwebapps.cit.nih.gov/ESBL/Database/mNPD/index.html | |
| Rat IMCD cytoplasmic fraction proteome (data set 8) | The ability of a phosphatase to dephosphorylate aquaporin-2 depends on whether or not it is found in the cytoplasm. | If normalized signal >7, P = 0.95; if signal between 0 and 7, P = 0.75; if signal = 0, P = 0.5 | This paper |
Protein Mass Spectrometry
We used liquid chromatography–tandem mass spectrometry (LC-MS/MS) to analyze the cytoplasmic fraction of IMCD cells isolated from rat kidneys as described by Pickering et al. (23) and summarized as follows. Thirty male Sprague-Dawley rats were euthanized according to an approved National Heart, Lung, and Blood Institute Animal Care and Use Committee protocol (H-0110R3). IMCD suspensions were prepared as previously described (4). The kidney inner medullas were dissected, minced, and digested into suspensions by incubation at 37°C for 70–90 min in digestion solution (250 mM sucrose, 10 mM triethanolamine, pH 7.6) containing collagenase B (3 mg/ml; Roche, Indianapolis, IN) and hyaluronidase (3 mg/ml; Worthington, Lakewood, NJ). The resulting inner medullary suspension (whole IM) was subjected to three low-speed centrifugations (at 70 g, 20 s) to separate the IMCD-enriched fraction in the pellet from the non-IMCD fraction in the supernatant. The supernatants containing non-IMCD elements were discarded. The final pellet was resuspended in tubule suspension solution containing (in mM) 118 NaCl, 5 KCl, 4 Na2HPO4, 25 NaHCO3, 2 CaCl2, 1.2 MgSO4, 5.5 glucose, and 5 sodium acetate (300 mOsm) split in half and exposed to either the vasopressin analog dDAVP (1 nM) or its vehicle for 30 min. This method has been previously shown to successfully isolate IMCD cells that are viable (6), vasopressin responsive (3), and largely free of non-IMCD cells (31). From this point, subcellular fractionation procedures followed Tchapyjnikov et al. (30) using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Life Technologies, #78835) to obtain fractions three fractions, the NE (nuclear extract), the NP (nuclear pellet), and the cytoplasm. Total protein concentrations were measured (BCA assay, Life Technologies, #23227). Mass spectrometric analyses of the NE and NP fractions were previously reported by Pickering et al. (23). For this study, we performed protein mass spectrometry of the cytoplasmic fraction only. Based on this procedure, we use the term "cytoplasm" in this paper to refer to all nonnuclear elements of the cells including membrane components, cytosol, and organelles.
The cytoplasmic fraction was concentrated using Amicon centrifugal filters, Ultracel 3K (Millipore) to ~400 μg in 110 μl of solution. A 5× concentrate of Laemmli solution (7.5% SDS, 30% glycerol, 50 mM Tris, pH 6.8) with bromophenol blue (1 part 5× concentrate: 4 parts protein sample) was added to the samples. One-dimensional SDS-PAGE was performed using 12% polyacrylamide SDS-Tris-Glycine gels (Bio-Rad). Molecular weight markers (Precision Plus, Bio-Rad) were run in separate lanes. The gels were run for 45 min at 3.00 A and 200 V and were then stained with Imperial protein stain (Thermo Scientific). The gels were destained for 30 min in MS-grade water (JT Baker #JTB-9831–02) and then sliced into 39 equal-sized pieces. Each gel piece was diced into 1.5 mm3 blocks using a razor blade. These samples underwent reduction with DTT, alkylation, and in gel trypsinization as described (24) except for the use of 12.5 ng/μl Trypsin Gold (Promega).
After digestion, the peptides were extracted from the gel pieces using four successive washes in 50% ACN/0.5% formic acid. Samples were dried and peptides were then suspended in 0.1% formic acid in MS-grade water. Samples were desalted using C-18 spin columns (Thermo Scientific, # 89870) according to the protocol provided with the kit. The resulting samples were dried (Speed-Vac) for storage. Immediately before mass spectrometry analysis, the samples were re-dissolved in 0.1% formic acid in MS-grade water.
LC-MS/MS.
The samples were analyzed on a nanoflow LC system (Eksigent, Dublin, CA) coupled to a tandem mass spectrometer (Orbitrap Velos Pro Hybrid; Thermo Scientific, San Jose, CA). The sample loading onto a peptide trap cartridge (Agilent Technologies, Palo Alto, CA) occurred at a flow rate of 6 μl/min. The trapped peptides were then fractionated with a reversed-phase PicoFrit column (New Objective, Woburn, MA) using a linear gradient of 5–35% ACN in 0.1% FA. The gradient time was 45 min at a flow rate of 0.25 μl/min. Precursor mass spectra (MS1) were acquired in the Orbitrap at 60,000 resolution, and product mass spectra (MS2) were acquired with the ion trap.
To maximize the number of peptide identifications, three algorithms were used to match spectra to peptides, viz. those coded by Mascot (27), SEQUEST (34) and InsPecT (29). The posttranslational modifications allowed were a fixed carbamidomethyl modification on cysteine, variable deamination modifications on asparagine and glutamine, and a variable oxidation modification on methionine. False discovery rate (FDR) at a peptide level was set to 0.01 based on target-decoy analysis (8). To identify ambiguous identifications, we used an in-house program (coded in Java) called ProMatch (30) (https://esbl.nhlbi.nih.gov/Bioinformatic%20Tools.htm). Peptides that only matched to one gene symbol were extracted as a “unique” identification. The peptides that matched to more than one gene symbol were separated as “multiple” identifications. To reconcile the multiple identifications, transcriptomic data from Affymetrix array profiling of rat renal IMCD transcripts was used (31). (Database available at: https://dir.nhlbi.nih.gov/papers/lkem/imcdtr/.). If a given gene was not expressed, based on the transcriptomic data (median normalized value <0.4), its protein product was dropped from consideration. The SEQUEST and Mascot searches were executed within Proteome Discoverer, and peptides with FDR < 0.01 and peptide rank = 1 were retained for further analyses. The InsPecT search was carried on the Biowulf Linux Cluster at the National Institutes of Health (https://hpc.nih.gov/systems). Raw files (35.5 GB), search results, and all spectra have been uploaded to the ProteomeXchange Consortium (33) via the PRIDE partner repository with the data set identifier PXD005488. These data are accessible at http://www.ebi.ac.uk/pride/archive/.
Spectral counting.
The number of peptide matches for a given gene symbol were counted and designated as the “spectral count” for that gene symbol. This was the sum of all spectra matching to the gene symbol in all gel slices. The spectral counts in the control samples and the dDAVP samples were compared using the Fisher exact test with the contingency table consisting of the control count, dDAVP count, total count of all peptides in control, and total count of all peptides in dDAVP. For this analysis, peptides that matched to more than one gene symbol were assigned to the gene symbol with the most abundant transcript in the IMCD based on prior data (31). Spectral counting data were used in this study for the Bayes’ theorem-based analysis by extracting data for all S/T phosphatases and mapping the spectral counting values to likelihood values as indicated in Table 1.
RESULTS
Database of Mammalian S/T Protein Phosphatase Catalytic Subunits
The goal was to identify S/T phosphatases that are candidates for roles in the regulation of the AQP2 water channel as well as other proteins involved in renal collecting duct transport. For this, we used Bayes’ rule to integrate data from multiple sources to rank all S/T phosphatases with regard to probability of interacting with aquaporin-2. To do such an analysis, a list of all S/T phosphatase catalytic subunits present in mammalian genomes was needed. However, because of the lack of a readily accessible, fully curated list of known S/T phosphatases, we compiled our own list (methods) containing 38 mammalian S/T phosphatase catalytic subunits. This data set has been made available as a freely accessible database located at https://hpcwebapps.cit.nih.gov/ESBL/Database/Phosphatases/. A link is provided to allow users to download the data into an electronic spreadsheet. The webpage can be searched using the browser’s intrinsic search command.
LC/MS-MS Identification of Cytoplasmic Proteome in Rat IMCD
LC-MS/MS analysis of the cytoplasmic fraction isolated from freshly isolated rat IMCD suspensions identified 47,916 distinct peptides from proteins coded by 4,538 distinct genes. We provide the data as a publically accessible web page from which the information can be downloaded (https://hpcwebapps.cit.nih.gov/ESBL/Database/IMCDCytoplasm/). From this, we extracted spectral counting data for all 38 S/T phosphatases to be used in the Bayes’ theorem-based analysis (Table 2). In brief, we found 22 out of the 38 S/T phosphatases in the rat IMCD cytoplasmic fraction. None of these showed a significant change in abundance in response to the vasopressin analog dDAVP. Interestingly, of the 85 proteins that did show significant changes in abundance (Table 3), eight were ubiquitin E3 ligases (P < 0.0001, χ2 = 51.0 vs. 46 E3 ligases among 4,492 unregulated proteins). Those that were decreased in abundance were Herc1, Herc2, Mycbp2, Rnf213, and Ubr4. Those that were increased were Trim25, Rnf31, and Ube3c.
Table 2.
S/T phosphatases found in cytoplasmic fraction of rat IMCD by protein mass spectrometry
| Gene Symbol | Amino Acids, n | Spectral Counts in Control | Relative Abundance* | Annotation | dDAVP: Control Ratio | P (Fisher exact) |
|---|---|---|---|---|---|---|
| Ppp1cb | 327 | 18 | 55.05 | serine/threonine-protein phosphatase PP1-beta catalytic subunit | 0.94 | 0.5384 |
| Ppp2cb | 309 | 12 | 38.83 | serine/threonine-protein phosphatase 2A catalytic subunit beta isoform | 1.08 | 0.4675 |
| Ppm1g | 305 | 8 | 26.23 | protein phosphatase 1G | 1.67 | 0.2080 |
| Pdp1 | 563 | 10 | 17.76 | [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, mitochondrial isoform d | 1.10 | 0.4701 |
| Ppp6c | 119 | 2 | 16.81 | serine/threonine-protein phosphatase 6 catalytic subunit | 0.88 | 0.5254 |
| Ppp5c | 307 | 5 | 16.29 | serine/threonine-protein phosphatase 5 | 1.50 | 0.3572 |
| Pptc7 | 261 | 4 | 15.33 | protein phosphatase PTC7 homolog | 0.80 | 0.5199 |
| Ppp3ca | 330 | 4 | 12.12 | serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | 1.00 | 0.6543 |
| Ppp1ca | 337 | 4 | 11.87 | serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 1.25 | 0.4801 |
| Ctdsp1 | 542 | 6 | 11.07 | carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 | 0.75 | 0.5177 |
| Ppm1k | 372 | 4 | 10.75 | protein phosphatase 1K, mitochondrial | 0.75 | 0.5177 |
| Ppp3cb | 499 | 4 | 8.02 | serine/threonine-protein phosphatase 2B catalytic subunit beta isoform | 1.50 | 0.4848 |
| Ppp1cc | 521 | 4 | 7.68 | serine/threonine-protein phosphatase PP1-gamma catalytic subunit | 1.00 | 0.6543 |
| Ilkap | 307 | 2 | 6.51 | integrin-linked kinase-associated serine/threonine phosphatase 2C | 1.00 | 0.6995 |
| Ppm1a | 465 | 3 | 6.45 | protein phosphatase 1A | 1.00 | 0.6995 |
| Ppm1b | 382 | 2 | 5.24 | protein phosphatase 1B | 0.67 | 0.5152 |
| Ppm1f | 392 | 2 | 5.10 | protein phosphatase 1F | 2.00 | 0.4879 |
| Ctdp1 | 244 | 1 | 4.10 | RNA polymerase II subunit A C-terminal domain phosphatase | 2.00 | 0.4879 |
| Ppp4c | 525 | 2 | 3.81 | serine/threonine-protein phosphatase 4 catalytic subunit | 1.50 | 0.4848 |
| Ctdnep1 | 309 | 1 | 3.24 | CTD nuclear envelope phosphatase 1 | 1.00 | 0.7580 |
| Ctdspl | 450 | 1 | 2.22 | CTD small phosphatase-like protein | 1.00 | 0.6995 |
| Ppp2ca | 969 | 1 | 1.03 | serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform | 2.00 | 0.4879 |
Number of spectra in control (non-dDAVP) normalized by number of amino acids and multiplied by 1,000.
Table 3.
Proteins that change in of abundance in cytoplasmic fraction of rat IMCDs in response to dDAVP for 30 min (P < 0.1 by Fisher exact test)
| Gene Symbol | Control Spectral Counts | dDAVP Spectral Counts | dDAVP: Control Ratio | P (Fisher exact) | Annotation |
|---|---|---|---|---|---|
| Aftph | 0 | 5 | ∞ | 0.0288 | aftiphilin |
| Tcaf2 | 0 | 5 | ∞ | 0.0288 | TRPM8 channel-associated factor 2 |
| Rnf31 | 0 | 5 | ∞ | 0.0288 | E3 ubiquitin-protein ligase RNF31 |
| Krt84 | 0 | 4 | ∞ | 0.0586 | keratin, type II cuticular Hb4 |
| Arhgef1 | 1 | 6 | 6.00 | 0.0574 | rho guanine nucleotide exchange factor 1 |
| Trim25 | 4 | 15 | 3.75 | 0.0079 | E3 ubiquitin/ISG15 ligase TRIM25 |
| Ube3c | 2 | 7 | 3.50 | 0.0821 | ubiquitin-protein ligase E3C |
| Spag9 | 5 | 16 | 3.20 | 0.0110 | C-Jun-amino-terminal kinase-interacting protein 4 |
| Ctnnbl1 | 3 | 9 | 3.00 | 0.0655 | beta-catenin-like protein 1 |
| Nt5c2 | 4 | 10 | 2.50 | 0.0803 | 5′-nucleotidase, cytosolic II |
| Rufy1 | 4 | 10 | 2.50 | 0.0803 | RUN and FYVE domain-containing protein 1 |
| Htatsf1 | 5 | 12 | 2.40 | 0.0630 | HIV Tat-specific factor 1 homolog |
| Ptpn23 | 5 | 11 | 2.20 | 0.0937 | tyrosine-protein phosphatase non-receptor type 23 |
| Atp6v1h | 9 | 17 | 1.89 | 0.0722 | V-type proton ATPase subunit H |
| Capn5 | 8 | 15 | 1.88 | 0.0915 | calpain-5 |
| Cul5 | 8 | 15 | 1.88 | 0.0915 | cullin-5 |
| Ap2m1 | 14 | 23 | 1.64 | 0.0783 | AP-2 complex subunit mu |
| Eps8l1 | 13 | 21 | 1.62 | 0.0974 | epidermal growth factor receptor kinase substrate 8-like 1 |
| Pfas | 13 | 21 | 1.62 | 0.0974 | phosphoribosylformylglycinamidine synthase |
| Actn1 | 20 | 30 | 1.50 | 0.0824 | alpha-actinin-1 |
| Fbn1 | 65 | 80 | 1.23 | 0.0871 | fibrillin-1 |
| Plec | 267 | 208 | 0.78 | 0.0102 | plectin |
| Ahnak | 230 | 179 | 0.78 | 0.0156 | neuroblast differentiation-associated protein AHNAK |
| Lrba | 124 | 91 | 0.73 | 0.0255 | lipopolysaccharide-responsive and beige-like anchor protein |
| Utrn | 156 | 112 | 0.72 | 0.0089 | utrophin |
| Tln2 | 49 | 34 | 0.69 | 0.0819 | talin-2 |
| Dync1h1 | 280 | 191 | 0.68 | 0.0001 | cytoplasmic dynein 1 heavy chain 1 |
| Prpf8 | 49 | 33 | 0.67 | 0.0650 | pre-mRNA-processing-splicing factor 8 |
| Tpr | 54 | 35 | 0.65 | 0.0391 | nucleoprotein TPR |
| Vps13a | 58 | 35 | 0.60 | 0.0164 | vacuolar protein sorting-associated protein 13A |
| Akap12 | 40 | 24 | 0.60 | 0.0398 | A-kinase anchor protein 12 |
| Mtor | 37 | 22 | 0.59 | 0.0441 | serine/threonine-protein kinase mTOR |
| Acsl1 | 24 | 14 | 0.58 | 0.0863 | long-chain-fatty-acid–CoA ligase 1 |
| Anxa3 | 24 | 14 | 0.58 | 0.0863 | annexin A3 |
| Piezo1 | 35 | 20 | 0.57 | 0.0379 | piezo-type mechanosensitive ion channel component 1 |
| Syne2 | 137 | 75 | 0.55 | 0.0000 | nesprin-2 |
| Macf1 | 200 | 105 | 0.53 | 0.0000 | microtubule-actin cross-linking factor 1 |
| Ubr4 | 139 | 71 | 0.51 | 0.0000 | E3 ubiquitin-protein ligase UBR4 |
| Map1b | 32 | 16 | 0.50 | 0.0193 | microtubule-associated protein 1B |
| Mycbp2 | 15 | 7 | 0.47 | 0.0773 | E3 ubiquitin-protein ligase MYCBP2 |
| Erc1 | 13 | 6 | 0.46 | 0.0949 | ELKS/Rab6-interacting/CAST family member 1 |
| Dst | 104 | 42 | 0.40 | 0.0000 | dystonin |
| Igf2r | 35 | 14 | 0.40 | 0.0027 | cation-independent mannose-6-phosphate receptor |
| RGD1307100 | 26 | 10 | 0.38 | 0.0074 | fragile site-associated protein homolog |
| Sorl1 | 36 | 13 | 0.36 | 0.0010 | sortilin-related receptor |
| Htt | 37 | 13 | 0.35 | 0.0007 | huntingtin |
| Dync2h1 | 15 | 5 | 0.33 | 0.0245 | cytoplasmic dynein 2 heavy chain 1 |
| Birc6 | 53 | 17 | 0.32 | 0.0000 | baculoviral IAP repeat-containing protein 6 |
| Pnn | 10 | 3 | 0.30 | 0.0521 | pinin |
| Fryl | 27 | 8 | 0.30 | 0.0013 | protein furry homolog-like |
| Rnf213 | 51 | 13 | 0.25 | 0.0000 | E3 ubiquitin-protein ligase RNF213 |
| Nup133 | 16 | 4 | 0.25 | 0.0072 | nuclear pore complex protein Nup133 |
| Herc2 | 12 | 3 | 0.25 | 0.0204 | E3 ubiquitin-protein ligase HERC2 |
| Mrpl40 | 8 | 2 | 0.25 | 0.0606 | 39S ribosomal protein L40, mitochondrial |
| Zzef1 | 10 | 2 | 0.20 | 0.0220 | zinc finger ZZ-type and EF-hand domain-containing protein 1 |
| Tacc2 | 24 | 4 | 0.17 | 0.0001 | transforming acidic coiled-coil-containing protein 2 |
| Dmxl1 | 18 | 3 | 0.17 | 0.0010 | dmX-like protein 1 |
| RGD1562629 | 12 | 2 | 0.17 | 0.0076 | neurobeachin |
| Mdn1 | 6 | 1 | 0.17 | 0.0680 | midasin |
| Neb | 6 | 1 | 0.17 | 0.0680 | nebulin |
| Prkdc | 6 | 1 | 0.17 | 0.0680 | DNA-dependent protein kinase catalytic subunit |
| Usp10 | 6 | 1 | 0.17 | 0.0680 | ubiquitin carboxyl-terminal hydrolase 10 |
| Vps13b | 6 | 1 | 0.17 | 0.0680 | vacuolar protein sorting-associated protein 13B |
| Herc1 | 19 | 3 | 0.16 | 0.0006 | E3 ubiquitin-protein ligase HERC1 |
| Scfd2 | 7 | 1 | 0.14 | 0.0388 | sec1 family domain-containing protein 2 |
| Zw10 | 7 | 1 | 0.14 | 0.0388 | centromere/kinetochore protein zw10 homolog |
| Fry | 15 | 2 | 0.13 | 0.0015 | protein furry homolog |
| Wdfy3 | 16 | 2 | 0.13 | 0.0008 | WD repeat and FYVE domain-containing protein 3 |
| Lrp2 | 21 | 1 | 0.05 | 0.0000 | low-density lipoprotein receptor-related protein 2 |
| Dhx30 | 10 | 0 | 0.00 | 0.0011 | putative ATP-dependent RNA helicase DHX30 |
| Lrp1 | 10 | 0 | 0.00 | 0.0011 | prolow-density lipoprotein receptor-related protein 1 |
| Upf2 | 6 | 0 | 0.00 | 0.0172 | regulator of nonsense transcripts 2 |
| Acss2 | 4 | 0 | 0.00 | 0.0666 | acetyl-coenzyme A synthetase, cytoplasmic |
| Ankrd17 | 4 | 0 | 0.00 | 0.0666 | ankyrin repeat domain-containing protein 17 |
| Atrx | 4 | 0 | 0.00 | 0.0666 | transcriptional regulator ATRX |
| Cntrl | 4 | 0 | 0.00 | 0.0666 | centriolin |
| Dmxl2 | 4 | 0 | 0.00 | 0.0666 | dmX-like protein 2 |
| Hal | 4 | 0 | 0.00 | 0.0666 | histidine ammonia-lyase |
| Kidins220 | 4 | 0 | 0.00 | 0.0666 | kinase D-interacting substrate of 220 kDa |
| Krt17 | 4 | 0 | 0.00 | 0.0666 | keratin, type I cytoskeletal 17 |
| LOC100294508 | 4 | 0 | 0.00 | 0.0666 | dyslexia-associated protein KIAA0319-like protein homolog |
| Snx13 | 4 | 0 | 0.00 | 0.0666 | sorting nexin-13 |
| Tmem126b | 4 | 0 | 0.00 | 0.0666 | complex I assembly factor TMEM126B, mitochondrial |
| Wdr18 | 4 | 0 | 0.00 | 0.0666 | WD repeat-containing protein 18 |
| Xpo5 | 4 | 0 | 0.00 | 0.0666 | exportin-5 |
Bayes’ Theorem-Based Analysis of S/T Phosphatase Catalytic Subunits
Which of the 38 S/T phosphatases in the mammalian genome could be responsible for regulation of AQP2 and other transporters in the renal collecting duct? Using Bayes’ rule, we started with equal a priori probabilities for each phosphatase in the curated database (Table 4, column 1) equal to 1/38 and sequentially updated all probabilities (columns 2–9) using the large-scale data sets listed in Table 1. After integration of all data sets, we obtained a final ranked list of S/T phosphatases (Table 4, column 10) most likely to interact with AQP2 in the renal collecting duct, headed by (in order) Ppp1cb (tied for 1st), Ppm1g (tied for 1st), Ppp1ca, Ppp3ca, Ppp2ca, Ppp1cc, Ppp2cb, Ppp6c, and Ppp5c. A listing of the key characteristics of these S/T phosphatases is given in Table 5. These are discussed further in the discussion.
Table 4.
Ranking of S/T phosphatase catalytic subunits with regard to likelihood of interacting with aquaporin-2 in renal collecting duct
| Column 1 | Column 2 | Column 3 | Column 4 | Column 5 | Column 6 | Column 7 | Column 8 | Column 9 | Column 10 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Name | Mouse Gene Symbol | Initial Probability | Rat Transcriptome | Rat RNA-Seq | Mouse Transcriptome | Mouse Proteome | Proteomics (dot product) | Apical mpkCCD Proteome | mpkCCD Cytoplasm | Rat IMCD Cytoplasm | Final |
| serine/threonine-protein phosphatase PP1-beta catalytic subunit | Ppp1c b | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0629 | 0.0906 | 0.1446 | 0.2420 | 0.2482 | 0.2482 |
| protein phosphatase 1G | Ppm1 g | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0629 | 0.0906 | 0.1446 | 0.2420 | 0.2482 | 0.2482 |
| serine/threonine-protein phosphatase PP1-alpha catalytic subunit | Ppp1ca | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0629 | 0.0906 | 0.1179 | 0.1974 | 0.2024 | 0.2024 |
| serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform isoform 2 | Ppp3ca | 0.0263 | 0.0440 | 0.0462 | 0.0522 | 0.0560 | 0.0807 | 0.1051 | 0.0741 | 0.0759 | 0.0759 |
| serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform | Ppp2ca | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0629 | 0.0906 | 0.0428 | 0.0717 | 0.0580 | 0.0580 |
| serine/threonine-protein phosphatase PP1-gamma catalytic subunit | Ppp1 ml | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0629 | 0.0906 | 0.1446 | 0.0510 | 0.0522 | 0.0522 |
| serine/threonine-protein phosphatase 2A catalytic subunit beta isoform | Ppp2cb | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0629 | 0.0906 | 0.1179 | 0.0416 | 0.0426 | 0.0426 |
| serine/threonine-protein phosphatase 6 catalytic subunit | Ppp6c | 0.0263 | 0.0440 | 0.0518 | 0.0577 | 0.0619 | 0.0892 | 0.0422 | 0.0149 | 0.0152 | 0.0152 |
| serine/threonine-protein phosphatase 5 | Ppp5c | 0.0263 | 0.0403 | 0.0475 | 0.0456 | 0.0490 | 0.0445 | 0.0210 | 0.0148 | 0.0152 | 0.0152 |
| protein phosphatase 1B isoform 4 | Ppm1b | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0629 | 0.0356 | 0.0169 | 0.0119 | 0.0096 | 0.0096 |
| protein phosphatase 1A | Ppm1a | 0.0263 | 0.0368 | 0.0386 | 0.0437 | 0.0468 | 0.0670 | 0.0317 | 0.0112 | 0.0090 | 0.0090 |
| serine/threonine-protein phosphatase 4 catalytic subunit | Ppp4c | 0.0263 | 0.0384 | 0.0329 | 0.0372 | 0.0399 | 0.0574 | 0.0272 | 0.0096 | 0.0077 | 0.0077 |
| serine/threonine-protein phosphatase 2B catalytic subunit beta isoform isoform 3 | Ppp3cb | 0.0263 | 0.0440 | 0.0377 | 0.0426 | 0.0458 | 0.0530 | 0.0251 | 0.0088 | 0.0071 | 0.0071 |
| carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 | Ctdsp1 | 0.0263 | 0.0440 | 0.0518 | 0.0586 | 0.0628 | 0.0094 | 0.0045 | 0.0031 | 0.0032 | 0.0032 |
| integrin-linked kinase-associated serine/threonine phosphatase 2C | Ilkap | 0.0263 | 0.0437 | 0.0515 | 0.0551 | 0.0592 | 0.0120 | 0.0100 | 0.0035 | 0.0029 | 0.0029 |
| CTD small phosphatase-like protein | Ctdspl | 0.0263 | 0.0440 | 0.0462 | 0.0522 | 0.0560 | 0.0062 | 0.0030 | 0.0021 | 0.0021 | 0.0021 |
| protein phosphatase PTC7 homolog | Pptc7 | 0.0263 | 0.0044 | 0.0052 | 0.0059 | 0.0056 | 0.0003 | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
| protein phosphatase 1D | Ppm1d | 0.0263 | 0.0440 | 0.0462 | 0.0473 | 0.0051 | 0.0007 | 0.0003 | 0.0001 | 0.0001 | 0.0001 |
| protein phosphatase 1F | Ppm1f | 0.0263 | 0.0044 | 0.0038 | 0.0010 | 0.0011 | 0.0002 | 0.0001 | 0.0001 | 0.0000 | 0.0000 |
| [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, mitochondrial isoform d | Pdp1 | 0.0263 | 0.0044 | 0.0024 | 0.0003 | 0.0003 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| protein phosphatase 1H isoform 1 | Ppm1 h | 0.0263 | 0.0415 | 0.0436 | 0.0049 | 0.0053 | 0.0001 | 0.0001 | 0.0000 | 0.0000 | 0.0000 |
| ubiquitin-like domain-containing CTD phosphatase 1 | Ublcp1 | 0.0263 | 0.0440 | 0.0462 | 0.0500 | 0.0536 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| pyruvate dehydrogenase [acetyl-transferring]-phosphatase 2, mitochondrial | Pdp2 | 0.0263 | 0.0440 | 0.0242 | 0.0027 | 0.0003 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| protein phosphatase 1M isoform 1 | Ppm1m | 0.0263 | 0.0044 | 0.0013 | 0.0009 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| CTD small phosphatase-like protein 2 isoform c | Ctdspl2 | 0.0263 | 0.0044 | 0.0013 | 0.0015 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| protein phosphatase 1K, mitochondrial precursor | Ppm1k | 0.0263 | 0.0044 | 0.0024 | 0.0003 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| CTD nuclear envelope phosphatase 1 | Ctdnep1 | 0.0263 | 0.0044 | 0.0052 | 0.0006 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform isoform 3 | Ppp3 ml | 0.0263 | 0.0044 | 0.0013 | 0.0002 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| PH domain leucine-rich repeat-containing protein phosphatase 1 | Phlpp1 | 0.0263 | 0.0044 | 0.0024 | 0.0003 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| serine/threonine-protein phosphatase with EF-hands 1 | Ppef1 | 0.0263 | 0.0044 | 0.0013 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| PH domain leucine-rich repeat-containing protein phosphatase 2 | Phlpp2 | 0.0263 | 0.0044 | 0.0013 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| protein phosphatase 1E | Ppm1e | 0.0263 | 0.0044 | 0.0013 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| serine/threonine-protein phosphatase with EF-hands 2 | Ppef2 | 0.0263 | 0.0044 | 0.0013 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| protein phosphatase 1J | Ppm1j | 0.0263 | 0.0044 | 0.0013 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| probable protein phosphatase 1N | Ppm1n | 0.0263 | 0.0044 | 0.0013 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| RNA polymerase II subunit A C-terminal domain phosphatase | Ctdp1 | 0.0263 | 0.0292 | 0.0160 | 0.0096 | 0.0103 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 isoform a | Ctdsp2 | 0.0263 | 0.0044 | 0.0013 | 0.0015 | 0.0003 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| protein phosphatase 1L | Ppm1l | 0.0263 | 0.0394 | 0.0217 | 0.0171 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
Chart gives posterior probabilities after indicated elements of data from Table 1 were assimilated into the Bayes’ theorem-based analysis. Bayes’ rule operator is commutative.
Table 5.
Characteristics of top-ranked S/T phosphatases in collecting duct
| Gene Symbol | Common Name | Functional Annotation (UniProt) | Evidence in Collecting Ducts from -Omic Studies |
|---|---|---|---|
| Ppp1cb | PP1-β | Protein phosphatase that associates with over 200 regulatory proteins to form highly specific holoenzymes which dephosphorylate hundreds of biological targets. | Ppp1cb protein has a half-life of 16.1 h in mouse mpkCCD cells in the absence of vasopressin and 9.8 h in presence of vasopressin (26). It was identified as attached to the apical plasma membrane by surface biotinylation in cultured mouse mpkCCD cells (19). |
| Ppm1g | PP2C | Member of the PP2C family of Ser/Thr protein phosphatases that are known to be negative regulators of cell stress response pathways. | Based on single-tubule RNA-Seq, the mRNA level for Ppm1g is highest in the CCD (cortical collecting duct) (16). Ppm1g protein has a half-life of 22.8 h in mouse mpkCCD cells in the presence and absence of vasopressin (26). It was identified in or attached to the apical plasma membrane by surface biotinylation in cultured mouse mpkCCD cells (19). |
| Ppp1ca | PP1-α | Protein phosphatase that associates with over 200 regulatory proteins to form highly specific holoenzymes which dephosphorylate hundreds of biological targets. | Ppp1ca protein has a half-life of 17.8 h in mouse mpkCCD cells in the absence of vasopressin and 12.9 h in presence of vasopressin (26). It was identified as attached to the apical plasma membrane by surface biotinylation in cultured mouse mpkCCD cells (19). |
| Ppp3ca | PP2B | Calcium-dependent, calmodulin-stimulated protein phosphatase. Many of the substrates contain a PxIxIT motif. (Also called “Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform”). | Ppp3ca protein has a half-life of 35.5 h in mouse mpkCCD cells in the absence of vasopressin (26). It was identified as attached to the apical plasma membrane by surface biotinylation in cultured mouse mpkCCD cells (19). |
| Calcineurin | |||
| Ppp2ca | PP2A-α | PP2A is the major phosphatase for microtubule-associated proteins (MAPs). PP2A can modulate the activity of phosphorylase B kinase, casein kinase 2, mitogen-stimulated S6 kinase, and MAP-2 kinase. Activates RAF1 by dephosphorylating it at "Ser-259." | Based on single-tubule RNA-Seq, the mRNA level for Ppp2ca protein is highest in the OMCD (outer medullary collecting duct) (16). Ppp2ca protein has a half-life of 20.4 h in mouse mpkCCD cells in the absence of vasopressin and is essentially unchanged by vasopressin (26). |
| Ppp1cc | PP1-γ | Protein phosphatase that associates with over 200 regulatory proteins to form highly specific holoenzymes which dephosphorylate hundreds of biological targets. | Based on single-tubule RNA-Seq, the mRNA level for Ppp1cc protein is highest in the CCD (cortical collecting duct) (16). Ppp1cc protein has a half-life of 11.2 h in mouse mpkCCD cells in the absence of vasopressin and is essentially unchanged by vasopressin (26). It was identified as attached to the apical plasma membrane by surface biotinylation in cultured mouse mpkCCD cells (19). |
| Ppp2cb | PP2A-β | PP2A can modulate the activity of phosphorylase B kinase, casein kinase 2, mitogen-stimulated S6 kinase, and MAP-2 kinase. | Ppp2cb protein has a half-life of 67.1 h in mouse mpkCCD cells in the absence of vasopressin and only 4.1 h in the presence of vasopressin (26). |
| Ppp6c | PP6C | A component of a signaling pathway regulating cell cycle progression. NH2-terminal domain restricts G1 to S phase progression in cancer cells, in part through control of cyclin D1. Downregulates MAP3K7 kinase activation by dephosphorylation of MAP3K7. | Ppp6c protein has a half-life of 36 h in mouse mpkCCD cells in the absence of vasopressin (26). |
| Ppp5c | PP5 | Serine/threonine-protein phosphatase that dephosphorylates a myriad of proteins. May modulate TGF-beta signaling pathway by the regulation of SMAD3 phosphorylation and protein expression levels. | Ppp5c protein has a half-life of 25.6 h in mouse mpkCCD cells in the absence of vasopressin and is essentially unchanged by vasopressin (26). |
Sensitivity analysis.
To test the relative information content of each data set used for the ranking of S/T phosphatases with regard to likelihood of interacting with AQP2 in collecting duct cells, we performed a sensitivity analysis in which we dropped each data set from the analysis in turn and recalculated the rankings (Table 6). For all analyses with dropped data sets (except for small effects of set 7 removal), the top five phosphatases were unchanged relative to the ranking using all data sets. This indicates that no one data set had undue influence on the rankings. Set 7 is the result of proteomic profiling of the cytoplasmic fraction isolated from mouse mpkCCD cells. When this set was dropped, Ppp3ca and Ppp2ca fell slightly in ranking, while Ppp1cc and Ppp2cb rose slightly.
Table 6.
Sensitivity analysis
| Gene Symbol | All Sets | Remove Set 1 | Remove Set 2 | Remove Set 3 | Remove Set 4 | Remove Set 5 | Remove Set 6 | Remove Set 7 | Remove Set 8 |
|---|---|---|---|---|---|---|---|---|---|
| Ppp1cb | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ppm1g | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 |
| Ppp1ca | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 4 | 3 |
| Ppp3ca | 4 | 4 | 4 | 4 | 4 | 4 | 5 | 6 | 4 |
| Ppp2ca | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 8 | 5 |
| Ppp1cc | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 2 | 6 |
| Ppp2cb | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 5 | 7 |
| Ppp6c | 8 | 9 | 8 | 9 | 8 | 13 | 8 | 7 | 8 |
| Ppp5c | 9 | 8 | 9 | 8 | 9 | 11 | 9 | 11 | 9 |
| Ppm1b | 10 | 11 | 13 | 10 | 10 | 10 | 10 | 13 | 10 |
Rankings of phosphatases obtained by dropping the indicated data sets and repeating the analysis. The set designator corresponds to values given parenthetically in first column of Table 1.
Comparison of S/T phosphatase rankings from rat data alone vs. mouse data alone.
The Bayes’ theorem-based ranking of S/T phosphatases with regard to likelihood of interacting with AQP2 in collecting duct cells combined data from rat and mouse studies. We asked the question: “How well do the rat data alone or the mouse data alone match the ranking obtained with the combined rat and mouse data?” As shown in Table 7, although there is some divergence among lower ranked S/T phosphatases, the top three S/T phosphatases from rat data alone or the mouse data alone matched the top three phosphatases from the Bayes’ analysis using the combined data. These three phosphatases were Ppp1cb, Ppm1g, and Ppp1ca. Since, the rat data and the mouse data were acquired independently, the agreement of species-separated analyses attests to the robustness of the overall method. It also supports the view that the mpkCCD cell line (source of mouse data) is an appropriate cell model for native collecting ducts (source of rat data) with regard to the S/T phosphatases that interact with AQP2.
Table 7.
Comparison of Bayes’ theorem-based rankings for S/T phosphatases using rat data alone vs. mouse data alone
| Protein Name | Gene Symbol | Posterior Prob. (all data) | Rank (all data) | Posterior Prob. (rat alone) | Rank (rat alone) | Posterior Prob. (mouse alone) | Rank (mouse alone) |
|---|---|---|---|---|---|---|---|
| Serine/threonine-protein phosphatase PP1-beta catalytic subunit | Ppp1cb | 0.2482 | 1 | 0.1283 | 1 | 0.2359 | 1 |
| Protein phosphatase 1G | Ppm1g | 0.2482 | 2 | 0.1283 | 3 | 0.2359 | 2 |
| Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | Ppp1ca | 0.2024 | 3 | 0.1283 | 1 | 0.1923 | 3 |
| Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Ppp3ca | 0.0759 | 4 | 0.0482 | 8 | 0.0809 | 4 |
| Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform | Ppp2ca | 0.0580 | 5 | 0.1013 | 4 | 0.0699 | 5 |
| Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | Ppp1cc | 0.0522 | 6 | 0.0270 | 10 | 0.0497 | 6 |
| Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform | Ppp2cb | 0.0426 | 7 | 0.0270 | 10 | 0.0405 | 7 |
| Serine/threonine-protein phosphatase 6 catalytic subunit | Ppp6c | 0.0152 | 8 | 0.0270 | 10 | 0.0145 | 11 |
| Serine/threonine-protein phosphatase 5 | Ppp5c | 0.0152 | 9 | 0.0495 | 6 | 0.0158 | 8 |
| Protein phosphatase 1B | Ppm1b | 0.0096 | 10 | 0.0426 | 9 | 0.0116 | 13 |
| Protein phosphatase 1A | Ppm1a | 0.0090 | 11 | 0.0159 | 16 | 0.0146 | 10 |
| Serine/threonine-protein phosphatase 4 catalytic subunit | Ppp4c | 0.0077 | 12 | 0.0135 | 18 | 0.0147 | 9 |
| Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform | Ppp3cb | 0.0071 | 13 | 0.0155 | 17 | 0.0118 | 12 |
Table includes only the top 13 phosphatases from the full analysis using data from both species.
DISCUSSION
In this study, we used Bayes’ rule to integrate several large-scale data sets from proteomics and transcriptomics studies to identify serine/threonine phosphatase catalytic subunits most likely to interact with AQP2 and possibly other transport proteins in the renal collecting duct. The analysis did not consider tyrosine phosphatases or dual-specificity phosphatases. The properties of the nine top-ranked S/T phosphatases are shown in Table 5. In the following, we discuss these phosphatases with respect to the reductionist literature. One problem with organizing such a discussion is the unfortunate discordance between the commonly used terminology for phosphatases and that used for official gene symbols. To facilitate the discussion, we have included the common terms used in most of the prior papers in Table 5 along with their associated gene symbols.
Several prior studies have examined the roles of S/T phosphatases in the regulation of the AQP2 water channel. Valenti et al. (32) found that the PP2A-selective phosphatase inhibitor okadaic acid promoted a 60% increase in AQP2 phosphorylation at Ser256 and increased AQP2 translocation to the apical plasma membrane in cultured collecting duct cells. PP2A corresponds to the gene symbols Ppp2ca (rank 5) and Ppp2cb (rank 7) and is calcium insensitive. Similarly, calyculin (a nonselective PP1 and PP2A inhibitor, increased AQP2 phosphorylation at Ser256 and Ser264 (without a significant effect on Ser261 and Ser269) and also increased the abundance AQP2 in the plasma membrane (25). Jo et al. (13) discovered a protein complex in endosomes from the renal IMCD consisting of an AKAP, the regulatory subunit of protein kinase A RII, protein kinase C zeta, and the calcium-calmodulin regulated phosphatase PP2B (also called calcineurin; gene symbol: Ppp3ca; rank 4 in Table 5). AQP2 present in these endosomes was found to be dephosphorylated in vitro by addition of exogenous PP2B. Gooch et al. (9) studied AQP2 phosphorylation and trafficking in calcineurin knockout mice, showing that vasopressin-mediated phosphorylation of AQP2 at Ser256 was paradoxically decreased compared with wild-type littermates. However, in the calcineurin knockout mice, there was a striking lack of apical accumulation of AQP2 in response to vasopressin, indicating that AQP2 exocytosis or endocytosis may have been affected. Consistent with this finding, a calcineurin inhibitor, FK-506, did not alter apical plasma membrane accumulation of AQP2 in native rat IMCD cells, despite increasing AQP2 phosphorylation at Ser261 and Ser264 (25). The water permeability response in isolated perfused collecting ducts is markedly reduced by inhibitors of calmodulin (5, 6), and a portion of this effect could have been mediated by loss of calcineurin action. Li et al. (17) showed that hypertonicity promotes the nuclear translocation of calcineurin-regulated NFATc proteins (in addition to the calcineurin insensitive Nfat5) with the subsequent induction of AQP2 expression. Possible roles in AQP2 regulation for three top ranked phosphatases in our Bayesian analysis (Ppp1cb, Ppm1g, Ppp1ca) have not, to our knowledge, been investigated. However, work by Kubokawa et al. (15) suggests a role for PP-2C, corresponding to the second ranked phosphatase Ppm1g, in regulation of the Kcnj1 potassium channel (ROMK) in the renal cortical collecting duct. Furthermore, Lin et al. (18) have implicated PP1 [corresponding to Ppp1cb (rank 1) and Ppp1ca (rank 3)] in the regulation of ROMK in the cortical collecting duct. Specifically, they found that c-Src regulates the interaction between WNK4 and SGK1 by increasing PP1 binding to WNK4, thereby decreasing WNK4 phosphorylation and regulating ROMK activity in cortical collecting duct. Finally, several of the top-ranked phosphatases were found in a comprehensive surface-biotinylation/mass-spectrometry study of mpkCCD cells to be apically located peripheral-membrane proteins (Table 5) (19), i.e., vicinal to AQP2, ROMK, and other apical transporters. These are Ppp1ca, Ppp1cb, Ppp1cd, Ppm1g, and calcineurin.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
S.L., C.M.P., C.-L.C., E.N.U., and M.A.K. conception and design of research; S.L., C.M.P., and E.N.U. performed experiments; S.L., V.R., C.R.G., C.M.P., C.-L.C., and M.A.K. analyzed data; S.L., V.R., C.M.P., C.-L.C., and M.A.K. interpreted results of experiments; S.L. and M.A.K. prepared figures; S.L. and M.A.K. drafted manuscript; S.L., V.R., C.R.G., C.M.P., C.-L.C., E.N.U., and M.A.K. edited and revised manuscript; S.L., V.R., C.R.G., C.M.P., C.-L.C., E.N.U., and M.A.K. approved final version of manuscript.
ACKNOWLEDGMENTS
The work was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute (NHLBI) (project ZO1-HL-001285, M. A. Knepper). LC-MS/MS studies were performed in the NHLBI Proteomics Core Facility (Director, Marjan Gucek). Sophia LeMaire was supported by the NHLBI Summer Internship Program (Herbert Geller, Director). C. M. Pickering is an undergraduate student from the Department of Chemical Engineering at Auburn University and was a member of the Biomedical Engineering Student Internship Program (BESIP) supported by the National Institute for Biomedical Imaging and Bioengineering (June–August, 2014). E. Umejiego was supported by Biomedical Research Training Program for Individuals from Underrepresented Groups of the NHLBI under the leadership of Dr. Helena Mishoe.
REFERENCES
- 1.Bradford D, Raghuram V, Wilson JL, Chou CL, Hoffert JD, Knepper MA, Pisitkun T. Use of LC-MS/MS and Bayes’ theorem to identify protein kinases that phosphorylate aquaporin-2 at Ser256. Am J Physiol Cell Physiol 307: C123–C139, 2014. doi: 10.1152/ajpcell.00377.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown D. The ins and outs of aquaporin-2 trafficking. Am J Physiol Renal Physiol 284: F893–F901, 2003. doi: 10.1152/ajprenal.00387.2002. [DOI] [PubMed] [Google Scholar]
- 3.Chou CL, DiGiovanni SR, Luther A, Lolait SJ, Knepper MA. Oxytocin as an antidiuretic hormone. II. Role of V2 vasopressin receptor. Am J Physiol Renal Physiol 269: F78–F85, 1995. [DOI] [PubMed] [Google Scholar]
- 4.Chou CL, Rapko SI, Knepper MA. Phosphoinositide signaling in rat inner medullary collecting duct. Am J Physiol Renal Physiol 274: F564–F572, 1998. [DOI] [PubMed] [Google Scholar]
- 5.Chou CL, Yip KP, Knepper MA. Role of Ca/calmodulin in vasopressin-stimulated aquaporin-2 trafficking in rat collecting duct (abstract). J Am Soc Nephrol 10: 13A, 1999.9890304 [Google Scholar]
- 6.Chou CL, Yip KP, Michea L, Kador K, Ferraris JD, Wade JB, Knepper MA. Regulation of aquaporin-2 trafficking by vasopressin in the renal collecting duct. Roles of ryanodine-sensitive Ca2+ stores and calmodulin. J Biol Chem 275: 36839–36846, 2000. doi: 10.1074/jbc.M005552200. [DOI] [PubMed] [Google Scholar]
- 7.Christensen BM, Zelenina M, Aperia A, Nielsen S. Localization and regulation of PKA-phosphorylated AQP2 in response to V(2)-receptor agonist/antagonist treatment. Am J Physiol Renal Physiol 278: F29–F42, 2000. [DOI] [PubMed] [Google Scholar]
- 8.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214, 2007. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 9.Gooch JL, Guler RL, Barnes JL, Toro JJ. Loss of calcineurin Aα results in altered trafficking of AQP2 and in nephrogenic diabetes insipidus. J Cell Sci 119: 2468–2476, 2006. doi: 10.1242/jcs.02971. [DOI] [PubMed] [Google Scholar]
- 10.Goodman SN. Toward evidence-based medical statistics. 2: The Bayes factor. Ann Intern Med 130: 1005–1013, 1999. doi: 10.7326/0003-4819-130-12-199906150-00019. [DOI] [PubMed] [Google Scholar]
- 11.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 103: 7159–7164, 2006. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW, Yu MJ, Pisitkun T, Chen F, Knepper MA. Vasopressin-stimulated increase in phosphorylation at Ser269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem 283: 24617–24627, 2008. doi: 10.1074/jbc.M803074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo I, Ward DT, Baum MA, Scott JD, Coghlan VM, Hammond TG, Harris HW. AQP2 is a substrate for endogenous PP2B activity within an inner medullary AKAP-signaling complex. Am J Physiol Renal Physiol 281: F958–F965, 2001. [DOI] [PubMed] [Google Scholar]
- 14.Kamsteeg EJ, Heijnen I, van Os CH, Deen PM. The subcellular localization of an aquaporin-2 tetramer depends on the stoichiometry of phosphorylated and nonphosphorylated monomers. J Cell Biol 151: 919–930, 2000. doi: 10.1083/jcb.151.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubokawa M, Kojo T, Komagiri Y, Nakamura K. Role of calcineurin-mediated dephosphorylation in modulation of an inwardly rectifying K+ channel in human proximal tubule cells. J Membr Biol 231: 79–92, 2009. doi: 10.1007/s00232-009-9207-z. [DOI] [PubMed] [Google Scholar]
- 16.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol 26: 2669–2677, 2015. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li SZ, McDill BW, Kovach PA, Ding L, Go WY, Ho SN, Chen F. Calcineurin-NFATc signaling pathway regulates AQP2 expression in response to calcium signals and osmotic stress. Am J Physiol Cell Physiol 292: C1606–C1616, 2007. doi: 10.1152/ajpcell.00588.2005. [DOI] [PubMed] [Google Scholar]
- 18.Lin DH, Yue P, Rinehart J, Sun P, Wang Z, Lifton R, Wang WH. Protein phosphatase 1 modulates the inhibitory effect of With-no-Lysine kinase 4 on ROMK channels. Am J Physiol Renal Physiol 303: F110–F119, 2012. doi: 10.1152/ajprenal.00676.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loo CS, Chen CW, Wang PJ, Chen PY, Lin SY, Khoo KH, Fenton RA, Knepper MA, Yu MJ. Quantitative apical membrane proteomics reveals vasopressin-induced actin dynamics in collecting duct cells. Proc Natl Acad Sci USA 110: 17119–17124, 2013. doi: 10.1073/pnas.1309219110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medvar B, Raghuram V, Pisitkun T, Sarkar A, Knepper MA. Comprehensive database of human E3 ubiquitin ligases: application to aquaporin-2 regulation. Physiol Genomics 48: 502–512, 2016. doi: 10.1152/physiolgenomics.00031.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moeller HB, Knepper MA, Fenton RA. Serine 269 phosphorylated aquaporin-2 is targeted to the apical membrane of collecting duct principal cells. Kidney Int 75: 295–303, 2009. doi: 10.1038/ki.2008.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen S, Agre P. The aquaporin family of water channels in kidney. Kidney Int 48: 1057–1068, 1995. doi: 10.1038/ki.1995.389. [DOI] [PubMed] [Google Scholar]
- 23.Pickering CM, Grady C, Medvar B, Emamian M, Sandoval PC, Zhao Y, Yang CR, Jung HJ, Chou CL, Knepper MA. Proteomic profiling of nuclear fractions from native renal inner medullary collecting duct cells. Physiol Genomics 48: 154–166, 2016. doi: 10.1152/physiolgenomics.00090.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA 101: 13368–13373, 2004. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren H, Yang B, Ruiz JA, Efe O, Ilori TO, Sands JM, Klein JD. Phosphatase inhibition increases AQP2 accumulation in the rat IMCD apical plasma membrane. Am J Physiol Renal Physiol 311: 2016, 2016. doi: 10.1152/ajprenal.00150.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoval PC, Slentz DH, Pisitkun T, Saeed F, Hoffert JD, Knepper MA. Proteome-wide measurement of protein half-lives and translation rates in vasopressin-sensitive collecting duct cells. J Am Soc Nephrol 24: 1793–1805, 2013. doi: 10.1681/ASN.2013030279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savitski MM, Lemeer S, Boesche M, Lang M, Mathieson T, Bantscheff M, Kuster B. Confident phosphorylation site localization using the Mascot Delta Score. Mol Cell Proteomics 10: M110.003830, 2011. doi: 10.1074/mcp.M110.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamma G, Robben JH, Trimpert C, Boone M, Deen PM. Regulation of AQP2 localization by S256 and S261 phosphorylation and ubiquitination. Am J Physiol Cell Physiol 300: C636–C646, 2011. doi: 10.1152/ajpcell.00433.2009. [DOI] [PubMed] [Google Scholar]
- 29.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77: 4626–4639, 2005. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 30.Tchapyjnikov D, Li Y, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. Proteomic profiling of nuclei from native renal inner medullary collecting duct cells using LC-MS/MS. Physiol Genomics 40: 167–183, 2010. doi: 10.1152/physiolgenomics.00148.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA. Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genomics 32: 229–253, 2008. doi: 10.1152/physiolgenomics.00201.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valenti G, Procino G, Carmosino M, Frigeri A, Mannucci R, Nicoletti I, Svelto M. The phosphatase inhibitor okadaic acid induces AQP2 translocation independently from AQP2 phosphorylation in renal collecting duct cells. J Cell Sci 113: 1985–1992, 2000. [DOI] [PubMed] [Google Scholar]
- 33.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32: 223–226, 2014. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yates JR III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67: 1426–1436, 1995. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 35.Yu MJ, Miller RL, Uawithya P, Rinschen MM, Khositseth S, Braucht DW, Chou CL, Pisitkun T, Nelson RD, Knepper MA. Systems-level analysis of cell-specific AQP2 gene expression in renal collecting duct. Proc Natl Acad Sci USA 106: 2441–2446, 2009. doi: 10.1073/pnas.0813002106. [DOI] [PMC free article] [PubMed] [Google Scholar]


