Abstract
Irritable bowel syndrome (IBS) is one of the most common gastrointestinal (GI) disorders. Despite its prevalence, the pathophysiology of IBS is not well understood although multiple peripheral and central factors are implicated. Recent studies suggest a role for alterations in gut microbiota in IBS. Significant advances in next-generation sequencing technology and bioinformatics and the declining cost have now allowed us to better investigate the role of gut microbiota in IBS. In the following review, we propose gut microbiota as a unifying factor in the pathophysiology of IBS. We first describe how gut microbiota can be influenced by factors predisposing individuals to IBS such as host genetics, stress, diet, antibiotics, and early life experiences. We then highlight the known effects of gut microbiota on mechanisms implicated in the pathophysiology of IBS including disrupted gut brain axis (GBA), visceral hypersensitivity (VH), altered GI motility, epithelial barrier dysfunction, and immune activation. While there are several gaps in the field that preclude us from connecting the dots to establish causation, we hope this overview will allow us to identify and fill in the voids.
Keywords: irritable bowel syndrome, IBS, gut microbiota, visceral hypersensitivity, VH, gut brain axis, GBA
irritable bowel syndrome (IBS) is a chronic heterogeneous gastrointestinal (GI) disorder of multifactorial (genetic, physiological, psychosocial and environment) origin, which afflicts nearly 11% of the world population (20). Recently revised Rome criteria (Rome IV) define IBS as “recurrent abdominal pain on average at least one day a week in the last 3 mo associated with two or more of the following: 1) related to defecation, 2) associated with a change in frequency of stool, and 3) associated with a change in form (consistency) of stool; symptoms should have persisted for at least six months” (https://ibsimpact.wordpress.com/2016/06/09/new-rome-iv-diagnostic-criteria-for-irritable-bowel-syndrome-ibs-unveiled-may-2016/). IBS was thought to be more prevalent in Western countries than in developing non-Western countries (20, 25, 48, 114, 149), although a recent study refutes this claim (24). In North America, up to 20% of the middle-aged population are affected while the reported prevalence is ~7% in South Asia (20, 114). Interestingly, the prevalence of IBS is higher in affluent Asian cities than in the poorer regions, which raises suspicion on the role of Western influence in these regions (42).
The human GI tract is home to trillions of microbes referred to as the gut microbiota. The role of gut microbiota in modulating GI physiology has been described in early gnotobiotic studies, which highlight the mutualistic relationship between gut microbiota and the host. Gut microbiota are involved in vital processes such as development of the host immune system (83, 111), maintenance of normal GI physiology (124), and fermentation of undigested carbohydrates (81). Alterations in gut microbiota structure and composition have been implicated in various GI tract disorders including IBS (50, 107). In fact, studies estimate that nearly 10% of IBS cases begin after an episode of gastroenteritis and leads to postinfectious IBS (PI-IBS), whose microbial signature closely resembles to that of diarrhea-predominant IBS (IBS-D) patients (49). Futhermore, a recent study has shown that symptom severity in IBS is negatively associated with microbial richness and a distinct microbial signature (132). Although several studies that have compared gut microbiota composition in patients with IBS to healthy patients have failed to provide consistent results (summarized in Table 1), some key findings include an increase in Firmicutes to Bacteroidetes ratio (50, 65, 107), decrease in Lactobacilli and Bifidobacteria population (8, 60, 79), and increase in Streptococci and Ruminococcus species (45, 57, 107, 117). A more consistent finding has been decreased alpha diversity (within individual differences); however, this may be nonspecific as decreased alpha diversity has also been reported in other metabolic and GI diseases such as obesity, type 2 diabetes, inflammatory bowel disease (Crohn’s disease, ulcerative colitis), and necrotizing enterocolitis (31, 80, 85, 87, 137). The variability in findings among studies is possibly due to the heterogeneous nature of the disease, differences in study design, differing methods for sample collection and data analysis, small sample size, and lack of longitudinal data. In addition to predominantly correlational studies, recent studies have evaluated the association of individual members of gut microbiota with symptoms paving the way for future mechanistic studies (112, 117). In spite of the need for better characterization of the gut microbiota in larger cohorts with longitudinal sampling, the above findings highlight an important role for this forgotten organ in IBS.
Table 1.
Summary of studies that report alteration in gut microbiota composition in patients with IBS
| Study No. | Study (Ref.) | Participants (n) | Diagnostic Criteria | Samples | Method of Detection | Major Findings |
|---|---|---|---|---|---|---|
| 1 | Balsari et al. 1982 (8) | IBS (20) Control (20) |
— | Fecal sample | Culture | Decreased Lactobacillus spp. |
| Decreased Bifidobacterium spp. | ||||||
| Decreased coliforms | ||||||
| 2 | Si et al. 2004 (122) | IBS (25) Control (25) |
Rome II | Fecal sample | Culture | Decreased Bifidobacterium spp. |
| Increased Enterobacteriaceae spp. | ||||||
| 3 | Malinen et al. 2005 (79) | IBS (27) Control (22) |
Rome II | Fecal sample | RT-PCR | Decreased Bifidobacterium spp. |
| Decreased Clostridium spp.in IBS-D | ||||||
| Decreased Lactobacillus spp. | ||||||
| Increased Veillonella spp. in IBS-C | ||||||
| 4 | Kassinen et al. 2007 (57) | IBS (24) Control (23) |
Rome II | Fecal sample | %G + C profiling, 16S rRNA sequencing | Decreased Collinsella |
| Decreased Coprococcus | ||||||
| 5 | Kerckhoffs et al., 2009 (60) | IBS (41) Control (26) |
Rome II | Fecal sample, duodenal mucosa | FISH, PCR | Decreased Bifidobacterium spp. |
| 6 | Krogius-kurikka et al. 2009 (65) | IBS (10) Control (23) |
Rome II | Fecal sample | %G + C profiling, 16S rRNA sequencing | Increased Proteobacteria |
| Increased Firmicutes | ||||||
| Decreased Actinobacteria | ||||||
| Decreased Bacteroidetes | ||||||
| 7 | Rajilić-Stojanović et al. 2011 (107) | IBS (62) Control (42) |
Rome II | Fecal sample | 16A rRNA sequencing, RT-PCR | Increased Firmicutes/Bacteroidetes ratio |
| Decreased Bifidobacterium and Faecalibacterium spp. | ||||||
| Increased Dorea, Ruminococcus, and Clostridium spp. | ||||||
| 8 | Saulnier et al. 2011 (117) | IBS (22) Control (22) |
Rome III | Fecal sample | 16S rRNA sequencing | Increased γ-proteobacteria spp. |
| 9 | Jeffery et al. 2012 (50) | IBS (37) Control (20) |
Rome II | Fecal sample | 16S rRNA sequencing | Increased Firmicutes/Bacteroidetes ratio |
| 10 | Tap et. 2016 (132) |
IBS (110) Control (39) |
Rome III | Fecal sample, colonic biopsy | 16S rRNA sequencing | No difference in α- or β-diversity between healthy and IBS |
| Decreased microbial richness | ||||||
| Increased Bacteroides | ||||||
| Decreased Prevotella and Methanobacteriales with IBS severity |
IBS, irritable bowel syndrome; IBS-D, diarrhea-predominant IBS; IBS-C, constipation-predominant IBS; FISH, fluorescence in situ hybridization; %G+C, percent guanine + cytosine.
Below we first present an overview of evidence, supporting the effect of risk factors such as host genetics, stress, diet, antibiotics, and early childhood experience known to be associated with IBS on gut microbiota composition and function.
Host Genetics Shape Gut Microbiota
The role of host genetics in IBS is highlighted by studies that show familial clustering of IBS. Although the familial pattern could result from a multitude of factors including host genetics, and environmental factors such as social learning and childhood experiences (68, 69, 71, 98), studies show that the relative contribution of host genetics is at least as significant as the environment (61, 68, 91, 126). Interestingly, both host genetics and the environment can shape and influence the gut microbiota (14). Using 16S rRNA based sequencing of stool samples from monozygotic twins and marital partner and unrelated individuals that were living in the same environment and had comparable feeding habits, Zoetendal et. al. (148a) found greater similarity among gut microbiota profile in monozygotic twins than unrelated individuals. Furthermore, findings from a recent study suggest that the heritable fraction of gut microbiota is temporally stable, being shaped mostly by host physiology and less by environmental factors (41). In another five separate studies that evaluated concordance (probability that 2 people with shared gene will develop similar disease) of IBS between twin pairs from various geographical regions found that the heritability (proportion of phenotypic variance attributable to genetic variance) contribution ranged anywhere between 0 and 57% (67, 68, 89, 91, 113), supporting a genetic contribution to IBS. In summary, these studies show that host genetics plays a prominent role in predisposition to IBS. Further host genetics also shape the gut microbiota composition. While gut microbiota changes are heritable and dependent on host physiology, future studies are needed to determine if gut microbiota changes as a result of host gene variations play a role in IBS pathogenesis.
Stress Modulates Gut Microbiota Composition
Chronic psychological stress influences the onset, duration, and severity of symptoms in IBS (26, 96, 132, 145). In a recent study authors found that anxiety and depression was correlated with the severity of IBS and this was reflected on the microbial signature (132). The authors had also previously reported a correlation between Actinomycetales and depression in patients with IBS. In rodents, chronic stress models such as maternal separation, prolonged restraint stress, and social disruption either early in life or during adulthood has been shown to significantly alter gut microbiota composition (6, 9, 96) via various mechanisms, which include 1) increasing circulating proinflammatory cytokines level (6), 2) disrupting the intestinal barrier (116), and 3) increasing the activity of the hypothalamus-pituitary-adrenal (HPA) axis (3, 28, 63). In humans, depression due to chronic stressful life events is associated with overrepresentation of bacteria from Enterobacteriaceae family (52), while psychological stress is associated with reduction in Lactobacilli spp. and increase in Escherichia coli and Pseudomonas spp. (73). These gut bacteria have been implicated in the development of IBS (59, 133, 134). A recent study shows that stress-induced changes in gut microbiota can also be transferred to offspring. In monkeys, for example prenatal stress is associated with decrease in beneficial Bifidobacterium and Lactobacillus in the GI tract of the offspring (7). Together, these findings suggest that both prenatal stress and postnatal stress modulate gut microbiota and support the potential role of gut microbiota as a mediator of stress in development of IBS.
Diet Alters Gut Microbiota Composition
Dietary intolerance is common in IBS patients with symptoms being triggered by specific foods, such as dairy products, grains, and fat (93). In fact, population-based studies have estimated that up to 70% of IBS patients suffer from symptoms of perceived food intolerance (72, 90, 93). While the mechanisms by which dietary ingredients trigger symptoms remain unclear, both long-term and short-term dietary changes have a significant impact on the gut microbiota, raising the possibility that gut microbes mediate the effect of diet on IBS symptoms. Long-term dietary patterns have been shown to shape the gut microbial composition. For example, long-term protein and animal fat intake is associated with increase in Bacteroides, while a long-term carbohydrate intake is associated with increase in Prevotella spp. (147). Similar enrichment in Prevotella at the expense of Bacteroides is also noted in children from Burkina Faso in rural Africa consuming primarily an agrarian diet compared with European children that consume modern Western diet rich in animal fat and protein (30). Short-term dietary changes are also associated with alterations in gut microbiota, which can be seen as early as a day after diet change and are mostly reversible. Interestingly, patients with IBS experience intermittent short-lasting exacerbations and it is tempting to hypothesize that short-term diet-related microbial changes might in part be responsible for the waxing and waning of symptoms (29). Microbial metabolism of dietary ingredients can lead to generation of metabolites such as organic acids, ammonia, methane, and hydrogen sulfide (108, 131), which may contribute to IBS symptoms. These dietary metabolites also support a dysbiotic microbiota with expansion of members within Gammaproteobacteria and at the same time suppress the growth of healthy gut microbiome (108). Dietary modifications such as low FODMAP (fermentable oligosaccharides, disaccharides, monosaccharides, and polyols) diet and other forms of dietary intervention including conventional dietary advice (16) are associated with improvement in symptoms and change in gut microbiota composition (43, 44), suggesting that while dietary ingredients and their metabolism by gut microbiota represent a mechanism via which diet influences IBS symptoms, the amount and timing of consumption of different dietary ingredients possibly play a vital role in symptom generation.
Antibiotics Administration Causes Short-Term and Long-Term Changes in Gut Microbiota Diversity
Antibiotic consumption is associated with development of IBS (82, 86, 139, 143). Adults and infants who consume antibiotics for non-GI diseases are at a greater risk of reporting bowel symptoms such as recurrent abdominal pain (139, 142). In fact, a study shows that patients who consume antibiotics for non-GI disorder are three times as likely to report functional bowel symptoms 4 mo later than those who did not take antibiotics (82). Gut microbiota are an obvious target for antibiotics taken for non-GI illnesses. Antibiotics in addition to affecting target pathogens also cause short-term and long-term changes in gut microbial diversity (51, 103, 128). Microbiota alterations due to antibiotic administration depend on several factors, including the spectrum of the agent, dosage, and the duration of treatment. A recent study showed that intravenous injection of β-lactam antibiotics, ampicillin/sulbactam, and cefazolin in a single patient for 14 days not only led to large short-term oscillatory fluctuations in microbial diversity but also long-term change in microbial diversity and composition (103). Bacterial genera such as Enterococcus, Blautia, Faecalibacterium, and Akkermansia bloomed at different stages of antibiotics treatment while taxa such as Actinobacteria, Betaproteobacteria, Streptococcaceae, Lachnospiraceae, Porphyromonadaceae, and Clostridiales were completely lost and did not reappear even 40 days after cessation of the antibiotic treatment (40, 103). Together these studies highlight the role of antibiotics in shaping gut microbiota composition.
Early Childhood Experiences Alter Gut Microbiota Composition Later in Life
Early life events such as privileged childhood living conditions, emotional and sexual abuse are associated with IBS (86, 115). Bradford et al. (17) in a recent study found that IBS patients reported early adverse life events more often than control patients. Interestingly, early childhood experiences also have a significant impact on gut microbiota. Since bacterial colonization during and shortly after birth has a significant impact on the composition of gut microbiota later in life, events such as mode of delivery (C-section or natural delivery), feeding (formula feeding or breast feeding), hygiene, and pet exposure have a significant influence on microbiota assembly. Infants that are delivered via C-section have microbiota composition predominated by Staphylococci, Coryneabacterium, and Propionibacterium spp, while vaginally born infants are colonized mostly by Lactobacillus, Bacteroides, Bifidobacterium, Prevotella, and Sneathia (5, 36, 97). Furthermore, breast-fed newborns possess gut microbiota dominated by Bifidobacterium (15), which is considered a beneficial microbe compared with formula fed, who have higher level of proinflammatory Gammaproteobacteria (15, 97). These studies raise the possibility that early childhood experiences impact gut microbiota diversity and composition. Further studies are however needed to determine if changes in gut microbiota resulting from early life adverse events contribute to development of IBS later in life.
In summary, these findings suggest that the predominant risk factors underlying IBS can affect gut microbiota composition and function. In the next part of the review, we will highlight the effect of gut microbiota on pathophysiological mechanisms underlying IBS, including the gut brain axis (GBA), GI motility, visceral sensation, epithelial barrier function, and immune activation.
Effect of Gut Microbiota on the GBA
The GBA is a bidirectional communication network that works through neural, endocrine, and immune pathways between the emotional and cognitive centers of the brain and the GI tract. GBA dysregulation is a common feature in the pathogenesis of IBS (64). Emerging evidence suggests that gut microbiota and their products can modulate the GBA (28). In a seminal study, Sudo et al. (127) found that postnatal gut microbial colonization programs the development of the HPA axis to regulate stress response. They found that germ-free (GF) mice exhibited significantly higher corticosterone and lower brain-derived neurotrophic factor (BDNF) levels in the cortex and the hippocampus compared with specific pathogen-free (SPF) mice, when subjected to restraint stress. Interestingly, monocolonization of GF mice with Bifidobacterium infantis reversed these effects highlighting the influence of individual gut microbial members on HPA axis regulation (127). Additionally, a recent study in healthy volunteers found that consumption of a fermented milk product containing Bifidobacterium, Lactobacilli, and Streptococcus thermophiles alters brain connectivity and function (135), supporting the effect of gut microbes on the GBA. One way by which microbiota alter brain function and behavior is via free fatty acid production (FAA) (140). As an example, propionic acid produced by gut bacteria readily crosses the blood-brain barrier and influences brain function and behavior in animals (120, 140). Besides FAA, gut microbes such as Lactobacilli and Bifidobacteria can also generate γ-amino butyric acid (GABA), an inhibitory neurotransmitter in the human brain (33). These studies highlight the important role of gut microbiota and their metabolites in modulating the GBA.
Effect of Gut Microbiota on Visceral Sensation
Visceral hypersensitivity (VH), a prominent feature in IBS patients, is characterized by reduced pain threshold to a painful stimuli. In a recent study, Crouzet et al. (27) found that GF rats colonized with gut microbiota from IBS patients displayed reduced pain threshold to colonic distension, when compared with GF rats that were colonized with gut microbiota from healthy individuals. Although the mechanisms by which microbiota transfer leads to VH in rats remain unclear, there was a significant decrease in Bifidobacteria and an increase in Enterobacteriaceae and other sulfate-reducing bacteria (27). Concomitantly, there was higher hydrogen excretion and sulfide production, the two principal components of pronociceptive gasotransmitter hydrogen sulfide (H2S) (27, 96). Excitatory acid-sensitive nociceptors called transient receptor potential vanilloid type 1 (TRPV1) channel proteins that are found in the vagal and spinal afferents are important mediators of VH in IBS (23, 53, 88, 146). Interestingly probiotic Lactobacillus reuteri exerts an antinociceptive effect via TRPV1 highlighting a potential role for gut microbiota/microbial products in VH (102).
Effect of Gut Microbiome on GI Motility
GI motility requires complex coordination among neurons, interstitial cells of Cajal, smooth muscle, and the immune cells to facilitate digestion of nutrients and movement of unwanted waste along the length of the GI tract. Alteration in GI motility is a hallmark of IBS. Gut microbiota and its metabolites can influence GI motility by affecting one of several pathways involving enteric neurons, glia, or enteric muscularis macrophages. For example, gut microbiota-derived lipopolysaccharide (LPS) and microbiota products such as the short-chain fatty acids (SCFAs) promote enteric neuronal survival (4, 125). In addition SCFAs also affect neurotransmitter release and influence the cross talk between enteric neurons, smooth muscles and muscularis macrophages to regulate GI motility (56, 92, 125). Microbiota and their products also affect the development, maturation, and generation of mucosal enteric glial cells, which might play a role in regulating GI motility (12, 54). Recently, gut microbiota bile acid metabolism has been implicated in GI motility (37) and their interaction with the enteric nervous system (32).
The role of gut microbiota in regulating GI motility in IBS is further supported by interventional studies using probiotics. For example, a fermented dairy product containing Bifidobacterium lactis has been found to accelerate GI transit and improve symptoms in patients with constipation-predominant IBS (2), while a probiotic mixture containing various strains of Lactobacillus and Bifidobacterium has been shown to improve stool consistency and overall symptoms in IBS-D patients (62). Overall, these studies not only highlight the importance of gut microbiota in regulating GI motility but also underscore how probiotics can be therapeutically used to treat GI dysmotility in IBS.
Effect of Gut Microbiota on Intestinal Barrier Dysfunction
Integrity of the intestinal epithelial barrier is essential for maintaining adequate nutrient transport and providing a barrier against pathogens residing in the gut lumen. While the gut microbiota and their metabolites play an important role in maintaining the integrity of the epithelial barrier, alterations in gut microbiota in turn can disrupt the epithelial barrier (58). Increased intestinal permeability is a feature of IBS-D and has been linked to symptom generation (19, 38, 148). Bacterial derived SCFAs help in maintenance of intestinal barrier structure and function (100, 101, 129). Butyrate, a bacterial derived SCFA, for example, prevents bacterial translocation by increasing expression of tight junction proteins such as claudin, occludin, and zonula occludens proteins (105). This is particularly interesting given the fact that reduction in butyrate producing gut bacteria has been observed in patients with IBS (106). Furthermore, Lactobacillus rhamnosus GG, a probiotic strain has been shown to induce claudin expression in neonatal mice suggesting that exposure to bacteria early in life promotes the maturation and development of the epithelial barrier (55, 99).
In addition to affecting tight junction proteins along the epithelial barrier, gut microbiota and their metabolites also regulate mucus layer (35, 123). Mucus layer forms the barrier between the lumen and the epithelial cells and prevents pathogen access to the epithelial surface (136). Changes in the amount and/or the composition of mucus may lead to inflammatory responses (70). Mucin degraders such as Ruminococcus torques and Ruminococcus gnavus are associated with severity of bowel symptoms in IBS (75, 78, 130, 133). Furthermore, a multispecies probiotic formulation containing a combination of Lactobacillus rhamnosus GG, L. rhamnosus Lc705, Propionibacterium freudenreichii ssp. Shermanii JS, and Bifidobacterium breve Bb99 decreases mucolytic Ruminococcus torques in IBS possibly by upregulating cell-surface mucin secretion and preventing its adherence to the epithelial layer (74, 76, 77, 94).
Effect of Gut Microbiota on Immune Activation
Immune activation underlying IBS has been an important area of investigation with several studies showing low-grade inflammation and infiltration of inflammatory cells notably the mast cells (MCs) and macrophages in the intestinal mucosa of IBS patients (10, 11, 22, 144). MCs regulate innate immunity through expression of pattern recognition receptors such as Toll-like receptors (TLRs). Notable examples of TLR expression in MCs associated with IBS include TLR2 and TLR4 (13, 18). Bacterial components in the gut such as flagellin and LPS can act as TLR ligands. Interestingly, an increased serum level of antibodies specific to flagellin and LPS concomitant with increased TLR (TLR4 and TLR5) expression has been observed in IBS patients (34). Activation of mast cells through TLR ligands causes release of inflammatory mediators such as histamine, tryptase, and prostaglandin E2 (PGE2). Release of these inflammatory mediators is also increased in IBS patients (11). These mediators acting through a G-coupled receptor called protease-activated receptors (PAR) can influence excitability of enteric neurons and potentially lead to VH in IBS (21). Together these studies suggest increased innate immune system activity can be driven by gut bacteria or their components. The resulting mast cell activation can potentially play a role in symptom generation in IBS.
Besides increased innate immune system activity, immunoregulatory molecules including pro- and anti-inflammatory cytokines are also altered in patients with IBS. For example, an increase in proinflammatory cytokines [tumor necrosis factor-α (TNF-α) and IL 1β, IL-6, and IL-8] level (46, 118, 121) and a decrease in anti-inflammatory cytokine (IL-10) have been reported in in both plasma and peripheral blood mononuclear cell (PBMC) of IBS patients (95, 118, 141; reviewed in Ref. 47). Probiotics have been investigated for their ability to restore cytokine balance. A placebo-controlled randomized clinical trial in patients with IBS showed that a 12-wk course of B. infantis resulted in an increase in the IL-10/IL-12 ratio similar to levels found in healthy patients (95). Furthermore, probiotic Bifidobacterium longum administration reduced proinflammatory TNF-α and probiotic combination of Streptococcus thermophilus, Lactobacillus acidophilus, Bifidobacterium bifidum, and Lactobacillus bulgaricus decreased TNF-α, interferon-γ (IFN-γ), and PGE2 levels in mice (109, 138). Together, these studies suggest that probiotics modulate both innate and adaptive immunity to potentially impart a therapeutic benefit and treat immune dysregulation in IBS (18, 34, 84, 119). In addition to probiotics, microbial metabolites of dietary nutrients have been shown to be anti-inflammatory. Two recent studies describe the role of gut microbial metabolites of dietary tryptophan, which act as aryl hydrocarbon receptor (AHR) ligands and can improve inflammation in peripheral GI tissues as well as the central nervous system highlighting the importance of microbial metabolites as mediators of their effect on the immune system (66, 110).
Summary and Perspective
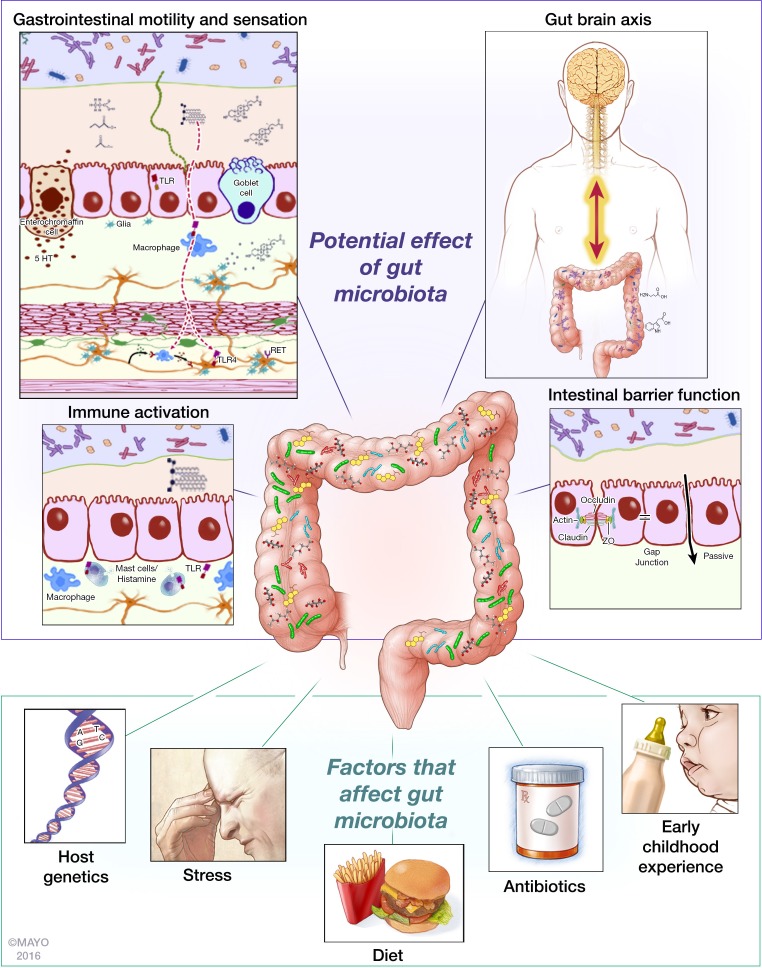
In this review, we have highlighted the effect of factors implicated in development of IBS on gut microbiota composition and function and also the role of gut microbiota in the pathophysiology of IBS (Fig. 1). The recent finding of the efficacy of rifaximin in patients with IBS without constipation, providing significant relief of IBS symptoms, bloating, abdominal pain, and loose or watery stools, further supports the role of gut microbiota in pathogenesis of IBS (1, 104), although the mechanism by which rifaximin exerts this effect still remains to be elucidated. It is tempting to propose gut microbiota as a unifying factor in development of IBS based on the evidence presented in this review. While several pieces of data as highlighted in the review support this notion, we realize there are still significant gaps that need to be filled before we can establish causality. The advances in technology and the emerging studies, however, provide a robust framework to more effectively identify and fill these gaps. Longitudinal study designs, with well-annotated clinical metadata allowing better characterization of gut microbiota and its relationship to diet and symptoms; systems approaches in understanding host-microbe interactions; and the use of gnotobiotic models to advance mechanistic paradigms will prove crucial as we embark on the next phase of this scientific journey. Our current approach of single or combination biotherapeutic products will need to be replaced with more targeted approaches. The use of engineered biotherapeutics with capability of releasing metabolites of interest constitutively and in sufficient amounts at sites of interest and the use of defined microbial communities with ability to better establish itself in the context of an existing community will likely replace the current approach. Similarly dietary approaches targeting the microbiome will need to be personalized based on an individual’s microbiome and its metabolic capacity. Overall the outlook is optimistic and we now have the necessary tools and the knowledge as we embark on developing effective microbiota targeted therapies for IBS.
Fig. 1.
Gut microbiota is a common denominator in pathophysiology of irritable bowel syndrome (IBS). Gut microbiota modulates pathophysiological mechanisms underlying IBS such as gastrointestinal motility and sensation, gut brain axis, immune activation, and intestinal barrier function. Gut microbiota composition is affected by risk factors underlying IBS such as host genetics, stress, diet, antibiotics usage and early childhood experience. 5-HT, 5-hydroxytryptamine (serotonin); TLR, Toll-like receptor; ZO, zonula occludens.
GRANTS
This work was made possible by funding from National Institute of Diabetes and Digestive and Kidney Diseases Grant K08-DK-100638 and Global Probiotic Council (to P. C. Kashyap) and Center for Individualized Medicine (CIM; Mayo Clinic; to P. C. Kashyap).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Y.B. and P.C.K. prepared figures; Y.B., D.A.M.P., and P.C.K. drafted manuscript; Y.B. and P.C.K. edited and revised manuscript; P.C.K. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Michael King for help with the illustration and Kristy Zodrow for administrative assistance.
REFERENCES
- 1.Acosta A, Camilleri M, Shin A, Linker Nord S, O’Neill J, Gray AV, Lueke AJ, Donato LJ, Burton DD, Szarka LA, Zinsmeister AR, Golden PL, Fodor A. Effects of rifaximin on transit, permeability, fecal microbiome, and organic acid excretion in irritable bowel syndrome. Clin Transl Gastroenterol 7: e173, 2016. doi: 10.1038/ctg.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrawal A, Houghton LA, Morris J, Reilly B, Guyonnet D, Goupil Feuillerat N, Schlumberger A, Jakob S, Whorwell PJ. Clinical trial: the effects of a fermented milk product containing Bifidobacterium lactis DN-173 010 on abdominal distension and gastrointestinal transit in irritable bowel syndrome with constipation. Aliment Pharmacol Ther 29: 104–114, 2009. doi: 10.1111/j.1365-2036.2008.03853.x. [DOI] [PubMed] [Google Scholar]
- 3.Ait-Belgnaoui A, Durand H, Cartier C, Chaumaz G, Eutamene H, Ferrier L, Houdeau E, Fioramonti J, Bueno L, Theodorou V. Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology 37: 1885–1895, 2012. doi: 10.1016/j.psyneuen.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 143: 1006–16.e4, 2012. doi: 10.1053/j.gastro.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J, Jun W. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17: 690–703, 2015. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun 25: 397–407, 2011. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J Pediatr Gastroenterol Nutr 38: 414–421, 2004. doi: 10.1097/00005176-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Balsari A, Ceccarelli A, Dubini F, Fesce E, Poli G. The fecal microbial population in the irritable bowel syndrome. Microbiologica 5: 185–194, 1982. [PubMed] [Google Scholar]
- 9.Bangsgaard Bendtsen KM, Krych L, Sørensen DB, Pang W, Nielsen DS, Josefsen K, Hansen LH, Sørensen SJ, Hansen AK. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One 7: e46231, 2012. doi: 10.1371/journal.pone.0046231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbara G, Feinle-Bisset C, Ghoshal UC, Quigley EM, Santos J, Vanner S, Vergnolle N, Zoetendal EG. The intestinal microenvironment and functional gastrointestinal disorders. Gastroenterology 150: 1305–1318.e8, 2016. doi: 10.1053/j.gastro.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, Pasquinelli G, Morselli-Labate AM, Grady EF, Bunnett NW, Collins SM, Corinaldesi R. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology 126: 693–702, 2004. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 12.Bassotti G, Villanacci V, Antonelli E, Morelli A, Salerni B. Enteric glial cells: new players in gastrointestinal motility? Lab Invest 87: 628–632, 2007. doi: 10.1038/labinvest.3700564. [DOI] [PubMed] [Google Scholar]
- 13.Belmonte L, Beutheu Youmba S, Bertiaux-Vandaële N, Antonietti M, Lecleire S, Zalar A, Gourcerol G, Leroi AM, Déchelotte P, Coëffier M, Ducrotté P. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS One 7: e42777, 2012. doi: 10.1371/journal.pone.0042777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107: 18933–18938, 2010. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17: 478–482, 2011. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Böhn L, Störsrud S, Liljebo T, Collin L, Lindfors P, Törnblom H, Simrén M. Diet low in FODMAPs reduces symptoms of irritable bowel syndrome as well as traditional dietary advice: a randomized controlled trial. Gastroenterology 149: 1399–1407.e2, 2015. doi: 10.1053/j.gastro.2015.07.054. [DOI] [PubMed] [Google Scholar]
- 17.Bradford K, Shih W, Videlock EJ, Presson AP, Naliboff BD, Mayer EA, Chang L. Association between early adverse life events and irritable bowel syndrome. Clin Gastroenterol Hepatol 10: 385–390.e1–3, 2012. doi: 10.1016/j.cgh.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol 106: 329–336, 2011. doi: 10.1038/ajg.2010.438. [DOI] [PubMed] [Google Scholar]
- 19.Camilleri M, Gorman H. Intestinal permeability and irritable bowel syndrome. Neurogastroenterol Motil 19: 545–552, 2007. doi: 10.1111/j.1365-2982.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- 20.Canavan C, West J, Card T. The epidemiology of irritable bowel syndrome. Clin Epidemiol 6: 71–80, 2014. doi: 10.2147/CLEP.S40245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cenac N. Protease-activated receptors as therapeutic targets in visceral pain. Curr Neuropharmacol 11: 598–605, 2013. doi: 10.2174/1570159X113119990039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chadwick VS, Chen W, Shu D, Paulus B, Bethwaite P, Tie A, Wilson I. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology 122: 1778–1783, 2002. doi: 10.1053/gast.2002.33579. [DOI] [PubMed] [Google Scholar]
- 23.Chan CL, Facer P, Davis JB, Smith GD, Egerton J, Bountra C, Williams NS, Anand P. Sensory fibres expressing capsaicin receptor TRPV1 in patients with rectal hypersensitivity and faecal urgency. Lancet 361: 385–391, 2003. doi: 10.1016/S0140-6736(03)12392-6. [DOI] [PubMed] [Google Scholar]
- 24.Chang FY, Lu CL, Chen TS. The current prevalence of irritable bowel syndrome in Asia. J Neurogastroenterol Motil 16: 389–400, 2010. doi: 10.5056/jnm.2010.16.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang L, Toner BB, Fukudo S, Guthrie E, Locke GR, Norton NJ, Sperber AD. Gender, age, society, culture, and the patient’s perspective in the functional gastrointestinal disorders. Gastroenterology 130: 1435–1446, 2006. doi: 10.1053/j.gastro.2005.09.071. [DOI] [PubMed] [Google Scholar]
- 26.Creed F, Ratcliffe J, Fernandes L, Palmer S, Rigby C, Tomenson B, Guthrie E, Read N, Thompson DG; North of England IBS Research Group . Outcome in severe irritable bowel syndrome with and without accompanying depressive, panic and neurasthenic disorders. Br J Psychiatry 186: 507–515, 2005. doi: 10.1192/bjp.186.6.507. [DOI] [PubMed] [Google Scholar]
- 27.Crouzet L, Gaultier E, Del’Homme C, Cartier C, Delmas E, Dapoigny M, Fioramonti J, Bernalier-Donadille A. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol Motil 25: e272–e282, 2013. doi: 10.1111/nmo.12103. [DOI] [PubMed] [Google Scholar]
- 28.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13: 701–712, 2012. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 29.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563, 2014. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107: 14691–14696, 2010. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dey N, Soergel DA, Repo S, Brenner SE. Association of gut microbiota with post-operative clinical course in Crohn’s disease. BMC Gastroenterol 13: 131, 2013. doi: 10.1186/1471-230X-13-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dey N, Wagner VE, Blanton LV, Cheng J, Fontana L, Haque R, Ahmed T, Gordon JI. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell 163: 95–107, 2015. doi: 10.1016/j.cell.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinan TG, Stilling RM, Stanton C, Cryan JF. Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res 63: 1–9, 2015. doi: 10.1016/j.jpsychires.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 34.Dlugosz A, Nowak P, D’Amato M, Mohammadian Kermani G, Nyström J, Abdurahman S, Lindberg G. Increased serum levels of lipopolysaccharide and antiflagellin antibodies in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 27: 1747–1754, 2015. doi: 10.1111/nmo.12670. [DOI] [PubMed] [Google Scholar]
- 35.Dohrman A, Miyata S, Gallup M, Li JD, Chapelin C, Coste A, Escudier E, Nadel J, Basbaum C. Mucin gene (MUC 2 and MUC 5AC) upregulation by Gram-positive and Gram-negative bacteria. Biochim Biophys Acta 1406: 251–259, 1998. doi: 10.1016/S0925-4439(98)00010-6. [DOI] [PubMed] [Google Scholar]
- 36.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107: 11971–11975, 2010. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duboc H, Rainteau D, Rajca S, Humbert L, Farabos D, Maubert M, Grondin V, Jouet P, Bouhassira D, Seksik P, Sokol H, Coffin B, Sabaté JM. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 24: 513–520, 2012. doi: 10.1111/j.1365-2982.2012.01893.x. [DOI] [PubMed] [Google Scholar]
- 38.Dunlop SP, Hebden J, Campbell E, Naesdal J, Olbe L, Perkins AC, Spiller RC. Abnormal intestinal permeability in subgroups of diarrhea-predominant irritable bowel syndromes. Am J Gastroenterol 101: 1288–1294, 2006. doi: 10.1111/j.1572-0241.2006.00672.x. [DOI] [PubMed] [Google Scholar]
- 40.Ferrer M, Martins dos Santos VA, Ott SJ, Moya A. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut Microbes 5: 64–70, 2014. doi: 10.4161/gmic.27128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19: 731–743, 2016. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gwee KA, Lu CL, Ghoshal UC. Epidemiology of irritable bowel syndrome in Asia: something old, something new, something borrowed. J Gastroenterol Hepatol 24: 1601–1607, 2009. doi: 10.1111/j.1440-1746.2009.05984.x. [DOI] [PubMed] [Google Scholar]
- 43.Halmos EP, Christophersen CT, Bird AR, Shepherd SJ, Gibson PR, Muir JG. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut 64: 93–100, 2015. doi: 10.1136/gutjnl-2014-307264. [DOI] [PubMed] [Google Scholar]
- 44.Halmos EP, Power VA, Shepherd SJ, Gibson PR, Muir JG. A diet low in FODMAPs reduces symptoms of irritable bowel syndrome. Gastroenterology 146: 67–75.e5, 2014. doi: 10.1053/j.gastro.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 45.Hong SN, Rhee PL. Unraveling the ties between irritable bowel syndrome and intestinal microbiota. World J Gastroenterol 20: 2470–2481, 2014. doi: 10.3748/wjg.v20.i10.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes PA, Harrington AM, Castro J, Liebregts T, Adam B, Grasby DJ, Isaacs NJ, Maldeniya L, Martin CM, Persson J, Andrews JM, Holtmann G, Blackshaw LA, Brierley SM. Sensory neuro-immune interactions differ between irritable bowel syndrome subtypes. Gut 62: 1456–1465, 2013. doi: 10.1136/gutjnl-2011-301856. [DOI] [PubMed] [Google Scholar]
- 47.Hughes PA, Zola H, Penttila IA, Blackshaw LA, Andrews JM, Krumbiegel D. Immune activation in irritable bowel syndrome: can neuroimmune interactions explain symptoms? Am J Gastroenterol 108: 1066–1074, 2013. doi: 10.1038/ajg.2013.120. [DOI] [PubMed] [Google Scholar]
- 48.Husain N, Chaudhry IB, Jafri F, Niaz SK, Tomenson B, Creed F. A population-based study of irritable bowel syndrome in a non-Western population. Neurogastroenterol Motil 20: 1022–1029, 2008. doi: 10.1111/j.1365-2982.2008.01143.x. [DOI] [PubMed] [Google Scholar]
- 49.Jalanka J, Salonen A, Fuentes S, de Vos WM. Microbial signatures in post-infectious irritable bowel syndrome–toward patient stratification for improved diagnostics and treatment. Gut Microbes 6: 364–369, 2015. doi: 10.1080/19490976.2015.1096486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeffery IB, O’Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EM, Simrén M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 61: 997–1006, 2012. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 51.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156: 3216–3223, 2010. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 52.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, Li L, Ruan B. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 48: 186–194, 2015. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 53.Jones RC III, Xu L, Gebhart GF. The mechanosensitivity of mouse colon afferent fibers and their sensitization by inflammatory mediators require transient receptor potential vanilloid 1 and acid-sensing ion channel 3. J Neurosci 25: 10981–10989, 2005. doi: 10.1523/JNEUROSCI.0703-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, Pettersson S, Pachnis V. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 85: 289–295, 2015. doi: 10.1016/j.neuron.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kajander K, Hatakka K, Poussa T, Färkkilä M, Korpela R. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6-month intervention. Aliment Pharmacol Ther 22: 387–394, 2005. doi: 10.1111/j.1365-2036.2005.02579.x. [DOI] [PubMed] [Google Scholar]
- 56.Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, Tache Y, Pasricha PJ, Knight R, Farrugia G, Sonnenburg JL. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 144: 967–977, 2013. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 133: 24–33, 2007. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci 9: 392, 2015. doi: 10.3389/fncel.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, Knol J, Akkermans LM. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol 60: 236–245, 2011. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 60.Kerckhoffs AP, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, Akkermans LM. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol 15: 2887–2892, 2009. doi: 10.3748/wjg.15.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One 3: e3064, 2008. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ki Cha B, Mun Jung S, Hwan Choi C, Song ID, Woong Lee H, Joon Kim H, Hyuk J, Kyung Chang S, Kim K, Chung WS, Seo JG. The effect of a multispecies probiotic mixture on the symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J Clin Gastroenterol 46: 220–227, 2012. doi: 10.1097/MCG.0b013e31823712b1. [DOI] [PubMed] [Google Scholar]
- 63.Knowles SR, Nelson EA, Palombo EA. Investigating the role of perceived stress on bacterial flora activity and salivary cortisol secretion: a possible mechanism underlying susceptibility to illness. Biol Psychol 77: 132–137, 2008. doi: 10.1016/j.biopsycho.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Koloski NA, Jones M, Weltman M, Kalantar J, Bone C, Gowryshankar A, Walker MM, Talley NJ. Identification of early environmental risk factors for irritable bowel syndrome and dyspepsia. Neurogastroenterol Motil 27: 1317–1325, 2015. doi: 10.1111/nmo.12626. [DOI] [PubMed] [Google Scholar]
- 65.Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Mäkivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol 9: 95, 2009. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, Sokol H. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22: 598–605, 2016. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lembo A, Zaman M, Jones M, Talley NJ. Influence of genetics on irritable bowel syndrome, gastro-oesophageal reflux and dyspepsia: a twin study. Aliment Pharmacol Ther 25: 1343–1350, 2007. doi: 10.1111/j.1365-2036.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 68.Levy RL, Jones KR, Whitehead WE, Feld SI, Talley NJ, Corey LA. Irritable bowel syndrome in twins: heredity and social learning both contribute to etiology. Gastroenterology 121: 799–804, 2001. doi: 10.1053/gast.2001.27995. [DOI] [PubMed] [Google Scholar]
- 69.Levy RL, Whitehead WE, Von Korff MR, Feld AD. Intergenerational transmission of gastrointestinal illness behavior. Am J Gastroenterol 95: 451–456, 2000. doi: 10.1111/j.1572-0241.2000.01766.x. [DOI] [PubMed] [Google Scholar]
- 70.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol 1: 183–197, 2008. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Locke GR III, Zinsmeister AR, Talley NJ, Fett SL, Melton LJ III. Familial association in adults with functional gastrointestinal disorders. Mayo Clin Proc 75: 907–912, 2000. doi: 10.4065/75.9.907. [DOI] [PubMed] [Google Scholar]
- 72.Locke GR III, Zinsmeister AR, Talley NJ, Fett SL, Melton LJ. Risk factors for irritable bowel syndrome: role of analgesics and food sensitivities. Am J Gastroenterol 95: 157–165, 2000. doi: 10.1111/j.1572-0241.2000.01678.x. [DOI] [PubMed] [Google Scholar]
- 73.Lutgendorff F, Akkermans LM, Söderholm JD. The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr Mol Med 8: 282–298, 2008. doi: 10.2174/156652408784533779. [DOI] [PubMed] [Google Scholar]
- 74.Lyra A, Krogius-Kurikka L, Nikkilä J, Malinen E, Kajander K, Kurikka K, Korpela R, Palva A. Effect of a multispecies probiotic supplement on quantity of irritable bowel syndrome-related intestinal microbial phylotypes. BMC Gastroenterol 10: 110, 2010. doi: 10.1186/1471-230X-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lyra A, Rinttilä T, Nikkilä J, Krogius-Kurikka L, Kajander K, Malinen E, Mättö J, Mäkelä L, Palva A. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol 15: 5936–5945, 2009. doi: 10.3748/wjg.15.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52: 827–833, 2003. doi: 10.1136/gut.52.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol Gastronintest Physiol 276: G941–G950, 1999. [DOI] [PubMed] [Google Scholar]
- 78.Malinen E, Krogius-Kurikka L, Lyra A, Nikkilä J, Jääskeläinen A, Rinttilä T, Vilpponen-Salmela T, von Wright AJ, Palva A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol 16: 4532–4540, 2010. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol 100: 373–382, 2005. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 80.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55: 205–211, 2006. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9: e1001221, 2011. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maxwell PR, Rink E, Kumar D, Mendall MA. Antibiotics increase functional abdominal symptoms. Am J Gastroenterol 97: 104–108, 2002. doi: 10.1111/j.1572-0241.2002.05428.x. [DOI] [PubMed] [Google Scholar]
- 83.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118, 2005. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 84.McKernan DP, Gaszner G, Quigley EM, Cryan JF, Dinan TG. Altered peripheral toll-like receptor responses in the irritable bowel syndrome. Aliment Pharmacol Ther 33: 1045–1052, 2011. doi: 10.1111/j.1365-2036.2011.04624.x. [DOI] [PubMed] [Google Scholar]
- 85.McMurtry VE, Gupta RW, Tran L, Blanchard EE IV, Penn D, Taylor CM, Ferris MJ. Bacterial diversity and Clostridia abundance decrease with increasing severity of necrotizing enterocolitis. Microbiome 3: 11, 2015. doi: 10.1186/s40168-015-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mendall MA, Kumar D. Antibiotic use, childhood affluence and irritable bowel syndrome (IBS). Eur J Gastroenterol Hepatol 10: 59–62, 1998. doi: 10.1097/00042737-199801000-00011. [DOI] [PubMed] [Google Scholar]
- 87.Michail S, Durbin M, Turner D, Griffiths AM, Mack DR, Hyams J, Leleiko N, Kenche H, Stolfi A, Wine E. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis 18: 1799–1808, 2012. doi: 10.1002/ibd.22860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Michl T, Jocic M, Heinemann A, Schuligoi R, Holzer P. Vagal afferent signaling of a gastric mucosal acid insult to medullary, pontine, thalamic, hypothalamic and limbic, but not cortical, nuclei of the rat brain. Pain 92: 19–27, 2001. doi: 10.1016/S0304-3959(00)00467-X. [DOI] [PubMed] [Google Scholar]
- 89.Mohammed I, Cherkas LF, Riley SA, Spector TD, Trudgill NJ. Genetic influences in irritable bowel syndrome: a twin study. Am J Gastroenterol 100: 1340–1344, 2005. doi: 10.1111/j.1572-0241.2005.41700.x. [DOI] [PubMed] [Google Scholar]
- 90.Monsbakken KW, Vandvik PO, Farup PG. Perceived food intolerance in subjects with irritable bowel syndrome–etiology, prevalence and consequences. Eur J Clin Nutr 60: 667–672, 2006. doi: 10.1038/sj.ejcn.1602367. [DOI] [PubMed] [Google Scholar]
- 91.Morris-Yates A, Talley NJ, Boyce PM, Nandurkar S, Andrews G. Evidence of a genetic contribution to functional bowel disorder. Am J Gastroenterol 93: 1311–1317, 1998. doi: 10.1111/j.1572-0241.1998.440_j.x. [DOI] [PubMed] [Google Scholar]
- 92.Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, Mortha A, Leboeuf M, Li XM, Mucida D, Stanley ER, Dahan S, Margolis KG, Gershon MD, Merad M, Bogunovic M. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 158: 300–313, 2014. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nanda R, James R, Smith H, Dudley CR, Jewell DP. Food intolerance and the irritable bowel syndrome. Gut 30: 1099–1104, 1989. doi: 10.1136/gut.30.8.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol 298: G807–G819, 2010. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- 95.O’Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O’Sullivan GC, Kiely B, Collins JK, Shanahan F, Quigley EM. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 128: 541–551, 2005. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 96.O’Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EM, Cryan JF, Dinan TG. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry 65: 263–267, 2009. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 97.O’Sullivan A, Farver M, Smilowitz JT. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights 8, Suppl 1: 1–9, 2015. doi: 10.4137/NMI.S29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pace F, Zuin G, Di Giacomo S, Molteni P, Casini V, Fontana M, Porro GB. Family history of irritable bowel syndrome is the major determinant of persistent abdominal complaints in young adults with a history of pediatric recurrent abdominal pain. World J Gastroenterol 12: 3874–3877, 2006. doi: 10.3748/wjg.v12.i24.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Patel RM, Myers LS, Kurundkar AR, Maheshwari A, Nusrat A, Lin PW. Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am J Pathol 180: 626–635, 2012. doi: 10.1016/j.ajpath.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr Res 61: 37–41, 2007. doi: 10.1203/01.pdr.0000250014.92242.f3. [DOI] [PubMed] [Google Scholar]
- 101.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr 139: 1619–1625, 2009. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Perez-Burgos A, Wang L, McVey Neufeld KA, Mao YK, Ahmadzai M, Janssen LJ, Stanisz AM, Bienenstock J, Kunze WA. The TRPV1 channel in rodents is a major target for antinociceptive effect of the probiotic Lactobacillus reuteri DSM 17938. J Physiol 593: 3943–3957, 2015. doi: 10.1113/JP270229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, Neulinger SC, Däumer C, Heinsen FA, Latorre A, Barbas C, Seifert J, dos Santos VM, Ott SJ, Ferrer M, Moya A. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 62: 1591–1601, 2013. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pimentel M, Lembo A, Chey WD, Zakko S, Ringel Y, Yu J, Mareya SM, Shaw AL, Bortey E, Forbes WP, Group TS; TARGET Study Group . Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 364: 22–32, 2011. doi: 10.1056/NEJMoa1004409. [DOI] [PubMed] [Google Scholar]
- 105.Plöger S, Stumpff F, Penner GB, Schulzke JD, Gäbel G, Martens H, Shen Z, Günzel D, Aschenbach JR. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci 1258: 52–59, 2012. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- 106.Pozuelo M, Panda S, Santiago A, Mendez S, Accarino A, Santos J, Guarner F, Azpiroz F, Manichanh C. Reduction of butyrate- and methane-producing microorganisms in patients with irritable bowel syndrome. Sci Rep 5: 12693, 2015. doi: 10.1038/srep12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141: 1792–1801, 2011. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 108.Rajilić-Stojanović M, Jonkers DM, Salonen A, Hanevik K, Raes J, Jalanka J, de Vos WM, Manichanh C, Golic N, Enck P, Philippou E, Iraqi FA, Clarke G, Spiller RC, Penders J. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am J Gastroenterol 110: 278–287, 2015. doi: 10.1038/ajg.2014.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rodes L, Khan A, Paul A, Coussa-Charley M, Marinescu D, Tomaro-Duchesneau C, Shao W, Kahouli I, Prakash S. Effect of probiotics Lactobacillus and Bifidobacterium on gut-derived lipopolysaccharides and inflammatory cytokines: an in vitro study using a human colonic microbiota model. J Microbiol Biotechnol 23: 518–526, 2013. doi: 10.4014/jmb.1205.05018. [DOI] [PubMed] [Google Scholar]
- 110.Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, Alvarez JI, Kébir H, Anandasabapathy N, Izquierdo G, Jung S, Obholzer N, Pochet N, Clish CB, Prinz M, Prat A, Antel J, Quintana FJ. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 22: 586–597, 2016. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323, 2009. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rousseaux C, Thuru X, Gelot A, Barnich N, Neut C, Dubuquoy L, Dubuquoy C, Merour E, Geboes K, Chamaillard M, Ouwehand A, Leyer G, Carcano D, Colombel JF, Ardid D, Desreumaux P. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat Med 13: 35–37, 2007. doi: 10.1038/nm1521. [DOI] [PubMed] [Google Scholar]
- 113.Saito YA. The role of genetics in IBS. Gastroenterol Clin North Am 40: 45–67, 2011. doi: 10.1016/j.gtc.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saito YA, Schoenfeld P, Locke GR III. The epidemiology of irritable bowel syndrome in North America: a systematic review. Am J Gastroenterol 97: 1910–1915, 2002. [DOI] [PubMed] [Google Scholar]
- 115.Sansone RA, Sansone LA. Irritable bowel syndrome: relationships with abuse in childhood. Innov Clin Neurosci 12: 34–37, 2015. [PMC free article] [PubMed] [Google Scholar]
- 116.Santos J, Yang PC, Söderholm JD, Benjamin M, Perdue MH. Role of mast cells in chronic stress induced colonic epithelial barrier dysfunction in the rat. Gut 48: 630–636, 2001. doi: 10.1136/gut.48.5.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141: 1782–1791, 2011. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schmulson M, Pulido-London D, Rodriguez O, Morales-Rochlin N, Martinez-García R, Gutierrez-Ruiz MC, López-Alvarenga JC, Robles-Díaz G, Gutiérrez-Reyes G. Lower serum IL-10 is an independent predictor of IBS among volunteers in Mexico. Am J Gastroenterol 107: 747–753, 2012. doi: 10.1038/ajg.2011.484. [DOI] [PubMed] [Google Scholar]
- 119.Schoepfer AM, Schaffer T, Seibold-Schmid B, Müller S, Seibold F. Antibodies to flagellin indicate reactivity to bacterial antigens in IBS patients. Neurogastroenterol Motil 20: 1110–1118, 2008. doi: 10.1111/j.1365-2982.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 120.Schreiber J, Chapman KA, Summar ML, Ah Mew N, Sutton VR, MacLeod E, Stagni K, Ueda K, Franks J, Island E, Matern D, Peña L, Smith B, Urv T, Venditti C, Chakarapani A, Gropman AL. Neurologic considerations in propionic acidemia. Mol Genet Metab 105: 10–15, 2012. doi: 10.1016/j.ymgme.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 121.Scully P, McKernan DP, Keohane J, Groeger D, Shanahan F, Dinan TG, Quigley EM. Plasma cytokine profiles in females with irritable bowel syndrome and extra-intestinal co-morbidity. Am J Gastroenterol 105: 2235–2243, 2010. doi: 10.1038/ajg.2010.159. [DOI] [PubMed] [Google Scholar]
- 122.Si JM, Yu YC, Fan YJ, Chen SJ. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol 10: 1802–1805, 2004. doi: 10.3748/wjg.v10.i12.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smirnova MG, Guo L, Birchall JP, Pearson JP. LPS up-regulates mucin and cytokine mRNA expression and stimulates mucin and cytokine secretion in goblet cells. Cell Immunol 221: 42–49, 2003. doi: 10.1016/S0008-8749(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 124.Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol 11: 227–238, 2013. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 125.Soret R, Chevalier J, De Coppet P, Poupeau G, Derkinderen P, Segain JP, Neunlist M. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 138: 1772–1782, 2010. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 126.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290, 2011. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 127.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol 558: 263–275, 2004. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sullivan A, Edlund C, Nord CE. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis 1: 101–114, 2001. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 129.Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr 100: 297–305, 2008. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- 130.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet 6: 81, 2015. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil 22: 512–519, 2010. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 132.Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré J, Störsrud S, Le Nevé B, Öhman L, Simrén M. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology 152: 111–123.e8, 2016. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 133.Taverniti V, Guglielmetti S. Methodological issues in the study of intestinal microbiota in irritable bowel syndrome. World J Gastroenterol 20: 8821–8836, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thabane M, Marshall JK. Post-infectious irritable bowel syndrome. World J Gastroenterol 15: 3591–3596, 2009. doi: 10.3748/wjg.15.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, Guyonnet D, Legrain-Raspaud S, Trotin B, Naliboff B, Mayer EA. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 144: 1394–1401, 2013. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tlaskalová-Hogenová H, Stěpánková R, Kozáková H, Hudcovic T, Vannucci L, Tučková L, Rossmann P, Hrnčí T, Kverka M, Zákostelsk Z, Klimešová K, Pibylová J, Bártová J, Sanchez D, Fundová P, Borovská D, Srtková D, Zídek Z, Schwarzer M, Drastich P, Funda DP. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol 8: 110–120, 2011. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature 457: 480–484, 2009. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Urbanska AM, Paul A, Bhathena J, Prakash S. Suppression of tumorigenesis: modulation of inflammatory cytokines by oral administration of microencapsulated probiotic yogurt formulation. Int J Inflamm 2010: 894972, 2010. doi: 10.4061/2010/894972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Uusijärvi A, Bergström A, Simrén M, Ludvigsson JF, Kull I, Wickman M, Alm J, Olén O. Use of antibiotics in infancy and childhood and risk of recurrent abdominal pain–a Swedish birth cohort study. Neurogastroenterol Motil 26: 841–850, 2014. doi: 10.1111/nmo.12340. [DOI] [PubMed] [Google Scholar]
- 140.Van Oudenhove L, McKie S, Lassman D, Uddin B, Paine P, Coen S, Gregory L, Tack J, Aziz Q. Fatty acid-induced gut-brain signaling attenuates neural and behavioral effects of sad emotion in humans. J Clin Invest 121: 3094–3099, 2011. doi: 10.1172/JCI46380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vázquez-Frias R, Gutiérrez-Reyes G, Urbán-Reyes M, Velázquez-Guadarrama N, Fortoul-van der Goes TI, Reyes-López A, Consuelo-Sánchez A. Proinflammatory and anti-inflammatory cytokine profile in pediatric patients with irritable bowel syndrome. Rev Gastroenterol Mex 80: 6–12, 2015. [DOI] [PubMed] [Google Scholar]
- 142.Verdú EF, Bercik P, Verma-Gandhu M, Huang XX, Blennerhassett P, Jackson W, Mao Y, Wang L, Rochat F, Collins SM. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut 55: 182–190, 2006. doi: 10.1136/gut.2005.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Villarreal AA, Aberger FJ, Benrud R, Gundrum JD. Use of broad-spectrum antibiotics and the development of irritable bowel syndrome. WMJ 111: 17–20, 2012. [PubMed] [Google Scholar]
- 144.Weston AP, Biddle WL, Bhatia PS, Miner PB Jr. Terminal ileal mucosal mast cells in irritable bowel syndrome. Dig Dis Sci 38: 1590–1595, 1993. doi: 10.1007/BF01303164. [DOI] [PubMed] [Google Scholar]
- 145.Whitehead WE, Crowell MD, Robinson JC, Heller BR, Schuster MM. Effects of stressful life events on bowel symptoms: subjects with irritable bowel syndrome compared with subjects without bowel dysfunction. Gut 33: 825–830, 1992. doi: 10.1136/gut.33.6.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Winston J, Shenoy M, Medley D, Naniwadekar A, Pasricha PJ. The vanilloid receptor initiates and maintains colonic hypersensitivity induced by neonatal colon irritation in rats. Gastroenterology 132: 615–627, 2007. doi: 10.1053/j.gastro.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 147.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108, 2011. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhou Q, Zhang B, Verne GN. Intestinal membrane permeability and hypersensitivity in the irritable bowel syndrome. Pain 146: 41–46, 2009. doi: 10.1016/j.pain.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148a.Zoetendal EG, Akkermans AD, Akkermans-van Vliet WM, Visser JA, de Vos WM. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis 13: 129–134, 2001. doi: 10.3402/mehd.v13i3.8013. [DOI] [Google Scholar]
- 149.Zuckerman MJ, Nguyen G, Ho H, Nguyen L, Gregory GG. A survey of irritable bowel syndrome in Vietnam using the Rome criteria. Dig Dis Sci 51: 946–951, 2006. doi: 10.1007/s10620-005-9005-0. [DOI] [PubMed] [Google Scholar]