Abstract
Renal cell carcinoma (RCC) is the most lethal urological malignancy with high risk of recurrence; thus, new prognostic biomarkers are needed. In this study, a new RCC antigen, PTPL1 associated RhoGAP1 (PARG1), was identified by using serological identification of recombinant cDNA expression cloning with sera from RCC patients. PARG1 protein was found to be differentially expressed in RCC cells among patients. High PARG1 expression is significantly correlated with various clinicopathological factors relating to cancer cell proliferation and invasion, including G3 percentage (P = .0046), Ki-67 score (p expression is also correlated with high recurrence of N0M0 patients (P = .0084) and poor prognosis in RCC patients (P = .0345). Multivariate analysis has revealed that high PARG1 expression is an independent factor for recurrence (P = .0149) of N0M0 RCC patients. In in vitro studies, depletion of PARG1by siRNA in human RCC cell lines inhibited their proliferation through inducing G1 cell cycle arrest via upregulation of p53 and subsequent p21Cip1/Waf1, which are mediated by increased RhoA-ROCK activities. Similarly, PARG1 depletion cells inhibited invasion ability via increasing RhoA-ROCK activities in the RCC cell lines. Conversely, overexpression of PARG1 on human embryonic kidney cell line HEK293T promotes its cell proliferation and invasion. These results indicate that PARG1 plays crucial roles in progression of human RCC in increasing cell proliferation and invasion ability via inhibition of the RhoA-ROCK axis, and PARG1 is a poor prognostic marker, particularly for high recurrence of N0M0 RCC patients.
Introduction
Renal cell carcinoma (RCC) accounts for more than 2% of all adult malignancies [1] and has continuously increased in the world [2]. Almost a third of RCC patients have distant metastases at initial diagnosis [3], and a third of the patients undergoing potentially curative nephrectomy will eventually develop metastases. The prognosis for patients with metastatic RCC is generally poor. The 2-year survival rate for patients with advanced disease is less than 20%. Thus, good biomarkers for predicting prognosis and therapeutic targets for metastatic RCC are needed.
Recently, cell proliferation and invasion ability have become important for cancer progression [4]. In cell proliferation, p53 is one of the key tumor suppressor proteins; inactive p53 function promotes cell growth in cancer cells. Otherwise, RhoA is a key molecule for cell invasion in various cancer cells. However, in RCC, RhoA acts as promoter [5] or suppressor [6] for cell invasion; these different biological characteristics remain controversial. Therefore, it is important for understanding RCC progression to evaluate the function of RhoA associating with cell proliferation and invasion.
In this study, we identified PARG1, a PTPL1-associated RhoGAP1, as a new RCC antigen using serological identification of recombinant cDNA expression cloning (SEREX) for development of a diagnostic method for RCC patients. We clarified the functional role of PARG1 in RCC including relationship between PARG1 expression level and effect of RhoA signaling. PARG1 expression was found to vary among patients, and high PARG1 expression was correlated with clinicopathological features related to cell proliferation and invasion of RCC cells. Moreover, high PARG1 expression was also correlated with higher recurrence and poor survival of RCC patients. As a molecular mechanism, PARG1 was found to increase RCC cell proliferation and invasion through inhibition of RhoA-ROCK pathway. Therefore, PARG1 is involved in malignant characteristics of human RCC and may be an attractive target for development of new diagnostic strategy for patients with RCC.
Material and Methods
RCC Cell Lines and Patient Samples
RCC cell lines used in the study were ACHN (American Type Culture Collection), A498, SW839, 769-p, 786-o, RCCS1 (Department of Urology, Keio University Tokyo, Japan), RCC6, RCC8, and RCC10 (Surgery Branch, National Cancer Institute). These RCC cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS). Human embryonic kidney cell HEK293T (American Type Culture Collection) was cultured in Dulbecco’s modified Eagle’s medium with 10% FBS. Tumor tissues were obtained from patients who had undergone surgical resection at Keio University Hospital (Tokyo, Japan) and National Defense Medical College Hospital (Saitama, Japan) with informed consent according to the institutional guidelines and were stored at −80°C until use. Sera obtained from cancer patients and healthy volunteers were stores at −80°C.
SEREX cDNA Cloning
SEREX cDNA cloning was described in a previous report [7]. cDNA library was constructed using 4 RCC cell lines (RCCS1, RCC6, RCC8, and RCC10) and synthesized by using λZAP-cDNA synthesis kit (Stratagene, La Jolla, CA). Screening was examined by using sera form one RCC patient.
Evaluation of Anti-PARG1 IgG Ab in Sera
Anti-PARG1 IgG Ab from RCC patient sera was detected by ELISA or Western blot using recombinant His-tagged PARG1 protein (amino acid sequence: 558 aa to 891 aa). We performed these experiments according to previous report [7], and the detailed detection method was described.
RT-PCR and quantitative PCR analyses
Total RNA was extracted from RCC cell lines using the RNase mini kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. cDNAs were synthesized with an oligo(dT) 12-18 primer (Invitorogen, Carlsbad, CA) from total RNAs. PARG1 expression was determined by PCR with PARG1-specific primers 3′-GGGCATCAGGTCAACTCTCTAC-5′ and 3′-CCAAGTAGAGGCTGCACAAA-5′and GAPDH as control. qPCR analysis of PARG1 expression in RCC cell lines was performed using 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). The quantitative expression of PARG1 in RCC cells transfected with stealth RNA (siRNA) was evaluated with qPCR using TaqMan Gene Expression Assays with the PARG1-specific probe (Hs00191351) according to the manufacturer's protocol (Applied Biosystems). Human GAPDH (Applied Biosystems) was used as an internal control.
Immunohistochemical Analysis
Immunohistochemical analysis of PARG1 was performed on 74 RCC specimens. These sections contained both tumor and surrounding kidney tissue. Briefly, formalin-fixed, paraffin-embedded tumor sections (4 μm) were deparaffinized in xylene and rehydrated through graded ethanols. Slides were placed in Dako Target Retrieval Solution High pH (Dako Corp., Carpinteria, CA) and heated at 95°C for 50 minutes for antigen retrieval. Endogenous peroxidase activity was quenched with Dako Peroxidase Blocking Reagent (Dako Corp.) for 10 minutes. Sections were incubated in 10% normal goat serum in phosphate-buffered saline (PBS) for 60 minutes at room temperature and subsequently incubated overnight at 4°C with IgY chicken polyclonal anti-PARG1 antibody (GenWay) at 1:200 dilution in PBS. Secondary antibody was anti-chicken IgY (IgG) peroxidase conjugate (Sigma, Saint Louis, MO). Staining was done by a Simple Stain Max PO kit (Nichirei Corp., Tokyo, Japan) according to the manufacturer's instructions. Reaction products were immersed in diaminobenzidine tetrahydrochloride for 3 minutes and counterstained with hematoxylin to visualize. Tubular epithelial cells in surrounding normal kidney tissues were served as a positive internal control. Samples incubated without primary antibody were also stained by the same steps and were used for baseline staining. PARG1 expression levels of each RCC tissue were classified into three levels: level 1: negative or almost negative PARG1 staining, level 2: PARG1 staining in RCC was clearly less than that of normal proximal tubules in the same tissue sections, and level 3: PARG1 staining in RCC was the same as normal proximal tubules in the same tissue sections. Two independent investigators blinded to the patients' clinical information evaluated all specimens. In the same 74 sections, immunostaining of the Ki-67 was also performed basically in the same method as that of PARG1 with the primary antibody for Ki-67 (anti–Ki-67 antibody, mouse monoclonal antibody; ZYMED Laboratories, South San Francisco, CA). RCC cells were cultured in eight-well chamber glass slides, fixed, and blocked. The primary antibodies used were as follows: F-actin/Texas Red(r)-X phalloidin from Molecular Probes (Invitrogen) (1:50 dilution), DNA/DAPI (Invitrogen) (1:100 dilution), and PARG1/ARHGAP29 MaxPab polyclonal antibody (Abnova) (1:50 dilution). The secondary antibody for PARG1 was polyclonal rabbit anti-mouse IgG-FITC (Dako) (1:100 dilution). Confocal microscopy was performed using an LSM 700 (Carl Zeiss MicroImaging, Co., Ltd.), and images were analyzed with the instrument's software.
Knockdown and Overexpression Studies
Knockdown of PARG1 was performed by Stealth RNAi (MediumGC Duplex, Invitrogen Carlsbad, CA). Stealth RNAi Negative Control duplexes were used as a scramble siRNA control (Invitrogen). BlOCK-iT Fluorescent Oligo (Invitrogen) was used to evaluate transfection efficiency. The target sequences of the siRNAs for PARG1 were as follows: si#2:5′-ACGCCTTCCTGACACTTCTAATAAA-3′ and si#3:5′-GCACGCTTGGTAGAGTTTCTCATTA-3′. Cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen). pcDNA3.1-PARG1 plasmid was constructed by using pcDNA3.1(−) (Invitrogen) and PARG1/pcR4-TOPO (Open Biosystems, Huntsville, AL). pcDNA3.1-PARG1 plasmid (1 μg/μl per well) was transfected to HEK293T using Lipofectamine 2000. pcDNA3.1 was used as a control. The WST-1 Cell Proliferation System (Takara, Kyoto, Japan) was used to evaluate cell proliferation. Premix WST-1 solution was added to each well containing cells in 0.1 ml of medium at the second or third day after transfection. Cell cycle distribution was determined by Hoechst 33342 staining with 10 μg/ml of Hoechst 33342 for 90 minutes at 37°C.
Invasion and Migration Assay
In invasion assay, cells were plated in Biocoat Matrigel invasion chambers (BD Biosciences, San Jose, CA) at a cell density of 2.5 × 104 per chamber in serum-free medium according to the manufacturer's instruction. Invasion cells through the membrane were stained with Diff-Quik (Sysmex, Kobe, Japan) and counted using a microscope. The xCELLigence system was used according to the instructions of the supplier (Roche Applied Science and ACEA Biosciences) [8]. Cells were seeded at a density of 20,000 cells/well with RPMI1640 on CIM-plate 16 with 8-μm pores (Roche Diagnostics GmbH). In migration assay, SW839 and 769-p (1.5 × 105 cells/well) or HEK293T (5.0 × 105 cells/well) cells were first plated onto a 24-well culture plate and cultured for 24 hours at 37°C. After rinsing with PBS, experimental wounds were made by dragging a plastic pipette tip across the central line of the culture plate, and the cultured medium was replaced by RPMI or Dulbecco’s modified Eagle’s medium with 10% FBS. Before incubation, a digital image of a space that included the scraped edge on both sides of the wound was taken. After incubation of 10 to 24 hours, a digital image of the same space was taken, and migration of cells was evaluated by counting the average number of migrated cells in the space between the scraped edges.
Rho Activity Assay and Western Blot Analysis
Cells were lysed with a RIPA buffer [20 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.1% Na-deoxycholate, 3 mM EDTA, 0.1% SDS, 1 mM Na3VO4, phosphatase inhibitor cocktail (Sigma), and protease inhibitor cocktail (Roche, Germany)]. Cell lysates were loaded on 7.5% to 15% SDS-PAGE gel. After transfer onto a nitrocellulose membrane (Hybond Extra C, Amersham Biosciences), the membrane was incubated overnight at 4°C with antibody. Anti-GAPDH Ab [rabbit polyclonal IgG (Santa Cruz Biotechnology)] diluted 1:1000 was used for detection of GAPDH for control. Primary antibodies were used as below. PARG1 (ARHGAP29 MaxPab polyclonal antibody) Ab was from Abnova (Taiwan, Taipei), p21Cip1/Waf1 Ab was from Carbioche (Germany), p53 (DO-1) Ab was from Santa Cruz Biotechnology (Delaware Avenue, CA), and phosphor-p53 (ser15) Ab was from Cell Signaling Technology. GAPDH was used as control. The activity of RhoA was determined in SW839, 769-p, and HEK293T cells using specific pull-down assay kit for activated forms of Rho proteins. Rhotekin-RBD was used to precipitate GTP-bound RhoA from cell lysates. Active GTP-bound RhoA and total RhoA were visualized by SDS-PAGE and Western blotting using a RhoA-specific moAB (26C4; Santa Cruz Biotechnology).
Statistical Analyses
Results are presented as the mean ± standard error. Variables of different groups were compared using the Mann-Whitney U test. The independence of fit of categorical data was analyzed by the χ2 test. Survival curves were constructed by the Kaplan-Meier method, and the differences between them were assessed using the log-rank test. Cox's proportional hazard regression model was used for univariate and multivariate analyses. In all tests, a P value less than .05 was considered to indicate statistical significance.
Results
Identification of PARG1 as a New RCC Antigen
To identify RCC antigens which are possibly useful for diagnosis and treatment of RCC patients, we applied SEREX technology with prognostic markers, and potential therapeutic targets have been previously identified [7]. Screening of a cDNA library generated from 4 RCC cell lines with serum from 1 RCC patient resulted in the isolation of a total of 182 positive clones. Among them, 176 were found to be PTPL1-associated RhoGAP1 PARG1. Other positive clones were ferritin, fibronectin, GHRHR, ribosomal protein L23a, ribosomal protein L34, and actin gamma-1. PARG1 was reported to be a negative regulator of RhoA signaling via its RhoGAP activity [9], [10] and involved in malignant phenotypes of some cancers including glioma [11]. However, the roles of PARG1 expressed in RCC have not yet been evaluated. Thus, we have performed further studies on PARG1 expressed in RCC cells.
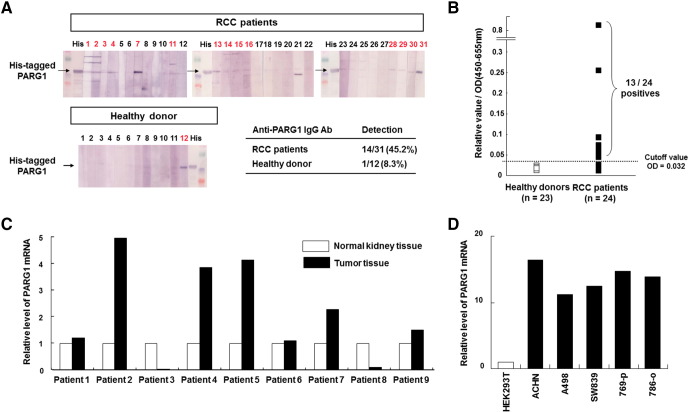
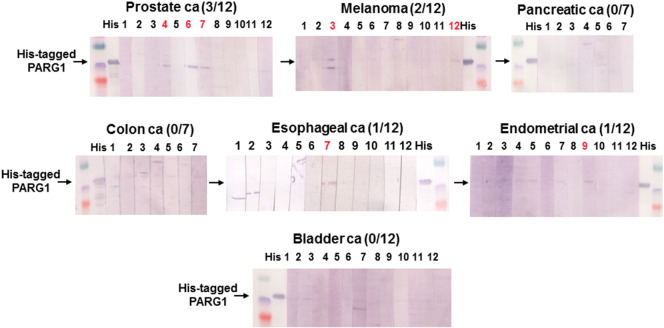
At first, we evaluated the presence of anti-PARG1 IgG using sera from RCC patients and healthy donors. Serum IgG specific for PARG1 was detected in 14 of 31 patients with RCC (45.2%) but detected in only 1 of 12 (8.3%) healthy donors when tested by Western blot analysis using a recombinant His-tagged PARG1 protein (558-891aa) (Figure 1A). Higher titers of anti-PARG1 IgG were detected in sera of RCC patients than those of healthy donors when evaluated by ELISA analysis (Figure 1B). In addition, PARG1-specific IgG was less frequently detected in sera of patients with other cancers (Supplementary Figure 1). Therefore, these results indicated that PARG1 is an immunogenic antigen particularly in RCC patients.
Figure 1.
Identification of RCC antigen PARG1 by SEREX and expression of PARG1 mRNA in normal kidney, RCC tissues, and RCC cell lines.
(A) Presence of anti-PARG1 IgG from sera of RCC was detected by Western blot analysis with recombinant His-tagged PARG1 protein. Detection samples are shown in red. (B) Frequent detection of anti-PARG1 IgG in sera from patients with RCC was evaluated by ELISA. ELISA was done with the recombinant PARG1 protein. The horizontal line indicates the cutoff value for positivity (OD = 0.032: the average absorbance of the healthy individuals plus 2 SD). Positive sera were found in 13 of 24 (54.2%) patients with RCC but not in healthy donors. (C) Expression of PARG1 mRNA in normal kidney and RCC tissue in the same RCC patient sample was detected by qPCR analysis. GAPDH mRNA expression was used as an internal control. (D) Expression of PARG1 in human RCC cell lines was detected by qPCR analysis. GAPDH was used as an internal control. HEK293T was used as control sample for this assay.
Supplementary Figure 1.
Presence of anti-PARG1 IgG from sera of various cancer patients detected by Western blot analysis with recombinant His-tagged PARG1 protein.We checked sera of other cancer patients including prostate cancer, bladder cancer, melanoma, endometrial cancer, esophageal cancer, colon cancer, and pancreatic cancer, but high level of anti-PARG1 IgG Ab was specifically observed in RCC patients. The detailed information was described in Materials and Methods. Detection samples were described by red.
Next, we evaluated the expression level of PARG1 mRNA using various human normal tissues, RCC patient samples, and RCC cell lines. The expression of PARG1 in various human normal tissues was examined by RT-PCR (Supplementary Figure 2). The PARG1 showed relatively high expression in some of the normal tissues including heart, spleen, testis, placenta, stomach, and colon. PARG1 was also expressed in normal kidney. These results were similar with a previous report [9]. Next, we examined the expression level of PARG1 in RCC patient samples including both normal and tumor tissue by qPCR. As shown in Figure 1C, PARG1 expression levels in RCC tissue were higher than normal tissue from several patients (nos. 2, 4, 5, and 7). However, others had the same expression levels of PARG1 (nos. 1, 6, and 9) or lower (nos. 3 and 8). The expression of PARG1 in human RCC cell lines was also evaluated by qPCR. All RCC cell lines expressed PARG1 higher than HEK293T (Figure 1D). These results suggested that PARG mRNA expression level in RCC tissue was different among patients when compared with normal kidney tissue. There was no relationship between presence of anti-PARG1 IgG and PARG1 mRNA expression in RCC patient samples.
Supplementary Figure 2.
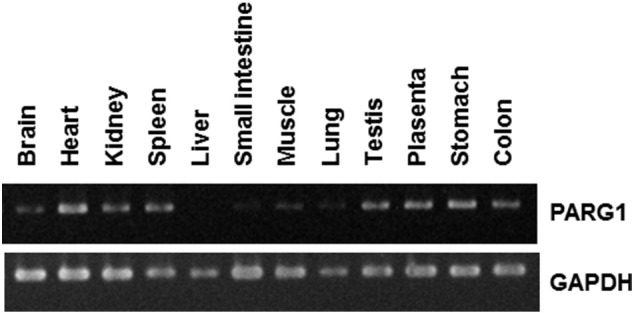
Expression level of PARG1 in normal tissues by RT-PCR.We evaluated the expression level of PARG1 in various human normal tissues by RT-PCR. The expression level was high in heart, spleen, testis, placenta, stomach, and colon. Low expression levels were observed in brain, kidney, small intestine, muscle, and lung. PARG1 was not expressed in liver. Thirty cycles of RT-PCR were done with the PARG1-specific primers as described in Materials and Methods. GAPDH mRNA expression was used as an internal control.
Differential expression of PARG1 in RCC
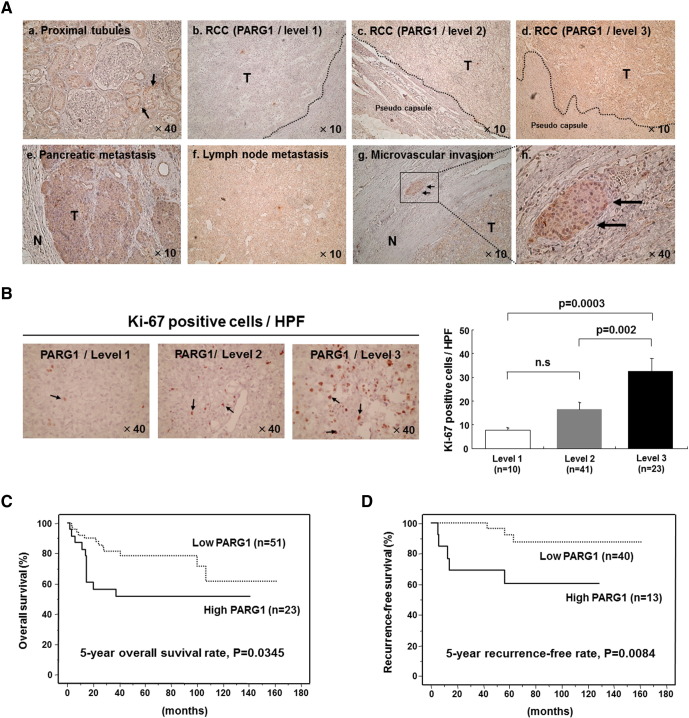
To investigate of expression level of PARG1 in patients with RCC in detail, we performed immunohistochemical analysis using paraffin-embedded RCC tissues. Representative immunostainings were shown in Figure 2A. PARG1 was expressed in proximal tubules of noncancer regions of kidneys (Figure 2Aa), and PARG1 expressions varied among RCC tissues (Figure 2A). The staining intensity of each RCC tissue was determined semiquantitatively by comparing the intensity of normal proximal tubules in the same tissue sections. PARG1 expression levels of each RCC tissue were classified into three levels: level 1: negative or almost negative PARG1 staining (n = 10), level 2: PARG1 staining in RCC was clearly less than that of normal proximal tubules in the same tissue sections (n = 42), and level 3: PARG1 staining in RCC was the same as normal proximal tubules in the same tissue sections (n = 23). The representative results were shown in Figure 2A (b: level 1, c: level 2, d: level 3). The expression of PARG1 protein in some RCC tissues was also evaluated by Western blot analysis using two PARG1 antibodies (mouse and chicken), and similar quantitative results to those of the immunostaining were obtained (data not shown). The distant metastasis and microvascular invasion of RCC showed high PARG1 staining (Figure 2A, e: metastasis in pancreas, f, metastasis in lymph nodes, g and h: microvascular invasion). These results suggested that PARG1 expression level was different among RCC patients but highly expressed in metastatic regions. Therefore, PARG1 was involved in invasive and metastatic activity of RCC cells.
Figure 2.
Expression of PARG1 and survival analysis in patients with RCC.
(A) Representative immunohistochemical analysis of PARG1 protein in paraffin-embedded tissues. (a) Normal proximal tubules (magnification, ×40), (b) RCC tissues (level 1), (c) RCC tissues (level 2), (d) RCC tissues (level 3), (e) pancreatic metastasis region, (f) lymph node metastasis legion, (g) microvascular invasion (magnification, ×10), and (h) microvascular invasion (high magnification, ×40). (B) RCC tissues were stained with anti–Ki-67 Ab, and Ki-67–positive cells (indicated by arrows) in high-powered field (HPF; magnification, ×40) were counted. The right graph shows the number of Ki-67–positive cells in each PARG1 expression level. (C) Kaplan-Meier overall survival curve with respect to low expression level (n = 51) and high expression level (n = 23) of PARG1. Five-year survival rate; P = .035. (D) Kaplan-Meier recurrence-free survival curve with respect to low expression level (n = 40) and high expression level (n = 13) of PARG1 in N0M0 patients with RCC. Five-year recurrence-free survival rate; P = .0084.
Correlation of PARG1 Expression with Poor Prognosis and Recurrence in RCC Patients
We then attempted to investigate the possible roles of PARG1 in malignant characteristics of RCC and clinicopathological features of RCC patients. Seventy-four RCC patients were divided into groups of low PARG1 expression (level 1 and 2, n = 51) or high PARG1 expression (level 3, n = 23). As shown in Table 1A, high PARG1 expression was significantly correlated with higher pT stages (P = .0366), lymph node metastasis (P = .0003), presence of G3 component (P = .0046), microvascular invasion (P = .0168), and Ki-67 score (P < .001) compared with low PARG1 expression (Table 1). Representative Ki-67 staining results are shown in Figure 2B, and positive correlations were observed between the presence of Ki-67–positive cells and the expression levels of PARG1 (Figure 2B). These results indicated that PARG1 might be involved in the malignant features of RCC because microvascular invasion and lymph node metastasis are related to invasion ability of RCC cells, and Ki-67 score is related to proliferation ability of RCC cells.
Table 1.
Correlation between PARG1 Expression and Clinicopathological Features in RCC Patients
| Clinical Characteristic | Level 1-2 (n = 51) PARG1 Low Group | Level 3 (n = 23) PARG1 High Group | P Value |
|---|---|---|---|
| Age (years) | 61.4 ± 2.1 | 59.2 ± 5.1 | .4170⁎ |
| Gender (F/M) | 15 / 36 | 8 / 15 | .6441† |
| Side (R/L) | 21 / 30 | 5 / 18 | .1050† |
| Size (cm) | 5.4 ± 0.4 | 7.7 ± 1.1 | .0926⁎ |
| pT1 or 2/pT3 or 4 | 39 / 12 | 12 / 11 | .0366† |
| Lymph node metastasis + | 1 (2.0%) | 7 (30.4%) | .0003† |
| Distant metastasis + | 10 (19.6%) | 6 (26.1%) | .5309† |
| Percentage of grade 3 component | 10 (19.6%) | 12 (52.2%) | .0046† |
| Microvascular invasion + | 16 (31.4%) | 14 (60.9%) | .0168† |
| Infiltrative growth | 21 (41.2%) | 15 (65.2%) | .0653† |
| CRP level (mg/dl) | 1.7 ± 0.4 | 3.2 ± 1.2 | .0525⁎ |
| Ki-67 score (positive cells/HPF) | 15.0 ± 2.0 | 39.5 ± 5.4 | <.001⁎ |
Analyzed by Mann-Whitney U test.
Analyzed by χ2 test.
We then examined the relationship between PARG1 expression and overall survival in 74 RCC patients evaluated by Kaplan-Meier analysis. RCC patients with high PARG1 expression had significantly lower survival rates than those with low PARG1 expression (P = .0345) (Figure 2C). Five-year overall survival rate of the patients with high PARG1expression was 51.5%, whereas that of patients with low PARG1 expression was 78.7%. In univariate analysis about the correlation between clinicopathological features and survival in patients with RCC, high PARG1 expression was significantly correlated with poor survival (Table 2). However, in multivariate analysis, high PARG1 was not an independent predictor for survival in RCC patients, and distant metastasis was the only independent predictor (P = .0009) (Table 2). We also examined the relationship between PARG1 expression and recurrence-free survival of 53 RCC patients without distant metastasis or lymph node metastasis (N0M0 patients). Patients with high PARG1 expression had significantly lower recurrence-free survival rates than those with low PARG1 expression (P = .0084) (Figure 2D). Five-year recurrence-free rate of patients with high PARG1 expression was 60.6%, and that of patients with low PARG1 expression was 92.7%. The correlation between various clinicopathological factors including PARG1 expression and recurrence was evaluated in N0M0 RCC patients by univariate and multivariate statistical analyses (Table 3). In the univariate analysis, tumor size (P = .001), grade 3 (P = .0005), microvascular invasion (P = .0058), CRP ≥1 mg/dl (P = .0039), and high PARG1 expression (P = .0192) were significantly correlated with the recurrence. In multivariate analyses, high PARG1 expression was the only independent predictor for recurrence (P = .0149). These results suggested that PARG1 might be a predictive marker of recurrence in N0M0 patients as well as a poor prognostic marker in RCC patients.
Table 2.
Correlation between Clinicopathological Features and Overall Survival in 74 RCC Patients
| Clinical Characteristic | P Value (Univariate) | P Value (Multivariate) | Odds Ratio | Relative Risk Ratio 95% CI |
|---|---|---|---|---|
| Gender | .9035 | |||
| Age | .8462 | |||
| Side of tumor | .5833 | |||
| Tumor size | <.0001 | .9092 | ||
| Presence of grade 3 component | <.0001 | .2166 | ||
| pT3 or 4 | <.0001 | .9286 | ||
| Microvascular invasion + | <.0001 | .0679 | ||
| CRP ≧ 1 mg/dl | <.0001 | .1012 | ||
| Ki-67 score (positive cells/HPF) | .0008 | .1454 | ||
| Thrombocytosis | .0486 | .1042 | ||
| High PARG1 | .0402 | .0711 | ||
| Distant metastasis | <.0001 | .0009 | 14.925 | 3.003-76.923 |
Table 3.
High PARG1 Expression Is an Independent Factor Correlating with 53 RCC Recurrence in N0M0 Patients
| Clinical Characteristic | P Value (Univariate) | P Value (Multivariate) | Odds Ratio | Relative Risk Ratio 95% CI |
|---|---|---|---|---|
| Gender | .6992 | |||
| Age | .5297 | |||
| Side of tumor | .3548 | |||
| Tumor size | .0010 | .0682 | ||
| Presence of grad 3 component | .0005 | .7363 | ||
| pT3 or 4 | .0634 | .0878 | ||
| Microvascular invasion + | .0058 | .1096 | ||
| CRP ≧ 1 mg/dl | .0039 | .3370 | ||
| High PARG1 | .0192 | .0149 | 5.524 | 1.321-23.256 |
Enhancement of RCC Cell Proliferation by PARG1 through Downregulation of p53 and p21
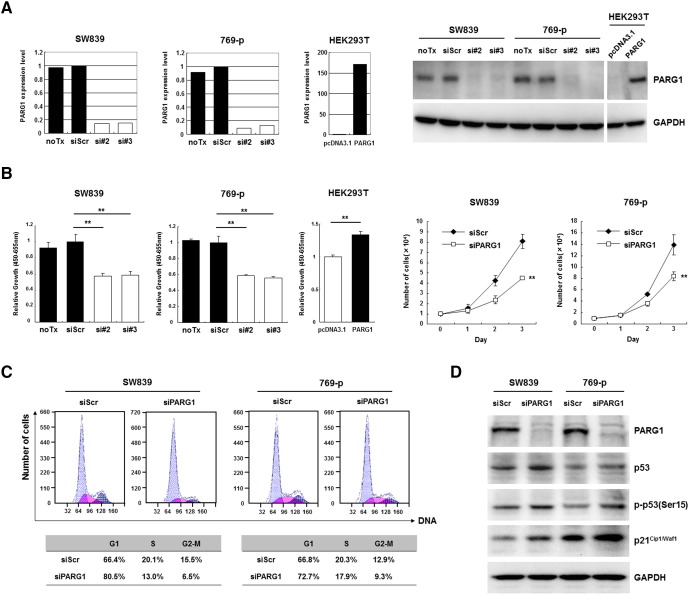
These clinicopathological features of RCC with high PARG1 expression led us to investigate the functional roles of PARG1 in biological characteristics of RCC cells. In in vitro studies, we used two human RCC cell lines (SW839 and 769-p) and HEK2393T (human embryonic kidney cell line) which regulated expression level of PARG1 by siRNA or overexpression vector to investigate cell proliferation and invasion ability. At first, we evaluated the effect of siRNA and overexpression vector by qPCR and Western blotting analysis. SW839 and 769-p significantly downregulated PARG1 expression level of both mRNA and protein by transfected PARG1-specific siRNAs (si#2 and si#3) compared with those with no transfection (noTx) or transfection with control siScr RNA (Figure 3A). HEK293T transfected with overexpression vector (pcDNA3.1-PARG1) significantly upregulated PARG1 expression level of both mRNA and protein compared with pcDNA3.1 control vector (Figure 3A). Knockdown of PARG1 resulted in decreased cell proliferation of SW839 and 769-p RCC cell lines significantly compared with controls, and overexpression of PARG1 resulted in increased cell proliferation of HEK293T cells significantly compared with control when evaluated by WST-1 assay (Figure 3B, left). Similar results were obtained when cell growth was evaluated by counting cell numbers (Figure 3B, right). Cell cycle analysis on SW839 and 769-p with PARG1 siRNA treatment showed that knockdown of PARG1 by siRNAs resulted in G1 cell cycle arrest (Figure 3C). We also performed caspase 3/7 assay with the PARG1 siRNA-treated SW839, and it indicated no apoptosis induction on RCC cells by PARG1 downregulation (Supplementary Figure 3). Western blot analysis of cell cycle–related molecules including p53 and p21Cip1/Waf1 in RCC cell lines (SW839 and 769-p) revealed that downregulation of PARG1 increased p53 protein and phosphorylation of p53 at Ser-15, as well as increased p53 downstream CDK inhibitor p21Cip1/Waf1 which inhibited the cell cycle at G1-S phase (Figure 3D). These results indicated that PARG1 is involved in RCC cell proliferation through downregulation of phosphorylated p53 and p21Cip1/Waf1 expressions but without induction of apoptosis.
Figure 3.
PARG1 was involved in cell proliferation and cell cycle progression through regulation of p53 and p21Cip1/Waf1 in RCC cell lines.
(A) Decrease of PARG1 mRNA and protein was observed after 2 days of incubation with two PARG1-specific siRNAs (si#2 and si#3) in SW839 and 769-p, whereas increase of PARG1 mRNA and protein was observed after 2 days of incubation with pcDNA3.1-PARG1 vector in HEK293T. (B) The inhibition of cell proliferation of SW839 and 769-p after 3 days of incubation with PARG1 siRNAs was observed in WST-1 assay (left graph) or trypan blue cell count (right graph); however, cell proliferation of HEK293T was increased by transfection with pcDNA3.1-PARG1. **P < .01; data are presented as the mean ± SD of three independent experiments. (C) Cell cycle analysis confirmed that treating SW839 and 769-p cells with PARG1 siRNA blocked the cell cycle in G1 phase at day 3 after transfection. (D) PARG1 siRNA upregulated p53, p-p53(Ser15), and p21Cip1/Waf1 protein expression by Western blotting in SW839 and 769-p. GAPDH was used as control. Representative results from three independent experiments (C, D).
Supplementary Figure 3.
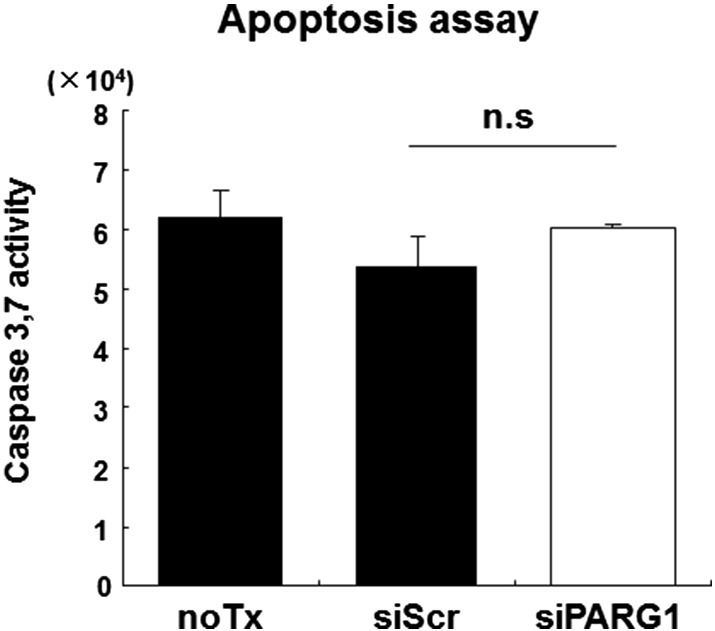
PARG1 was not involved in apoptosis through regulation of p53 and p21 in RCC cell line.We performed apoptosis analysis in SW839 RCC cells. Caspase 3/7 activity in siRNA-transfected cells was measured using the Caspase-Glo 3/7 assay kit (Promega, Madison, WI) according to the manufacturer’s instructions. SW839 cells were plated in 96-well plates at a seeding density of 3 ×103 cells/well, and caspase activity was assayed 72 hours after siRNA transfection. Downregulation of PARG1 by siRNA does not induce cell death and activation of apoptosis pathway. Data are presented as the mean ± SD of three independent experiments.
Enhancement of RCC Cell Invasion Ability by PARG1 through Inhibition of RhoA Activity
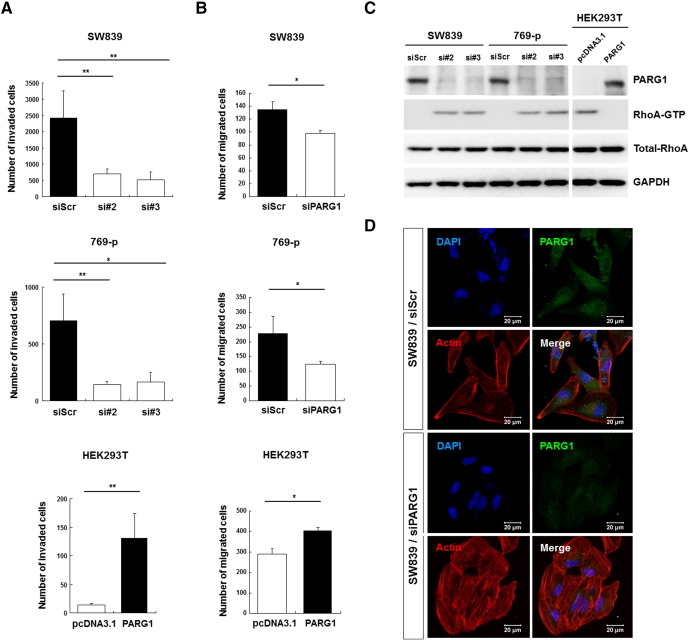
Next, the roles of PARG1 in invasion ability of RCC cells were examined. Knockdown of PARG1 by siRNA significantly inhibited invasion ability of SW839 and 769-p RCC cell lines, and overexpression of PARG1 significantly increased invasion ability of HEK293T compared with corresponding controls when evaluated by Matrigel invasion assay (Figure 4A). Similarly, when cell motilities of these transfected cells were evaluated by wound healing analysis, downregulation of PARG1 significantly inhibited cell motility of SW839 and 769-p, and overexpression of PARG1 significantly increased cell motility of HEK293T cells compared with controls (Figure 4B). Because PARG1 has a RhoGAP activity, we examined its activity by measuring RhoA-GTP pulled down with GST-RBD using Western blot analysis and anti-RhoA Abs. Knockdown of PARG1 by siRNAs increased RhoA-GTP in SW839 and 769-p RCC cells, and overexpression of PARG1 by PARG1-cDNA vector transfection decreased RhoA-GTP in HEK293T cells (Figure 4C). RhoA is the key regulator of the actin cytoskeleton by stimulating actin stress fiber formation [12], [13]. We evaluated actin stress fiber formation of SW839 RCC cells by immunofluorescent staining. As shown in Figure 4D, SW839 treated with PARG1 siRNA showed well-spread–type morphology with constructed actin stress fiber formation (Figure 4D). It means that RhoA activity induced stress fiber formation that showed no invasion type for RCC [14]. MMP2 activity was also decreased in the siRNA-treated SW839, although 769-p did not produce MMP2 (data not shown). These results demonstrated that PARG1 promotes invasion ability of RCC cells mainly through increased cell motility via inhibition of RhoA activity.
Figure 4.
The role of PARG1 involved in cell invasion and migration through inhibition of RhoA activity.
(A) Cell invasion ability was evaluated by Matrigel invasion assay in RCC cell lines and HEK293T. Invasion ability was decreased in PARG1 siRNA-transfected SW839 and 769-p cells, but invasion ability was increased in PARG1 expression vector–transfected HEK293T cells. *P < .05, **P < .01; data are presented as the mean ± SD of three independent experiments. (B) Cell migration ability was performed by wound healing assay. Migration ability was decreased in PARG1 siRNA-transfected SW839 and 769-p cells at 10 hours of incubation; however, migration ability was increased in PARG1 expression vector–transfected HEK293T cells at 24 hours of incubation. *P < .05; data are presented as the mean ± SD of three independent experiments. (C) Effect of PARG1 on RhoA activity was determined using RhoA activation kit (pull-down assay and Western blotting). PARG1 siRNAs induced RhoA-GTP in RCC cell lines, but PARG1 expression vector reduced RhoA-GTP in HEK293T cells. (D) Scramble and PARG1 siRNAs-transfected SW839 cells were stained with PARG1 (FITC), F-actin (Texas red), and DAPI. Downregulation of PARG1 by siRNA induced actin stress fiber formation. Representative results from three independent experiments (C, D).
Enhancement of RCC Cell Proliferation and Invasion by PARG1 through Inhibition of RhoA-ROCK Signaling
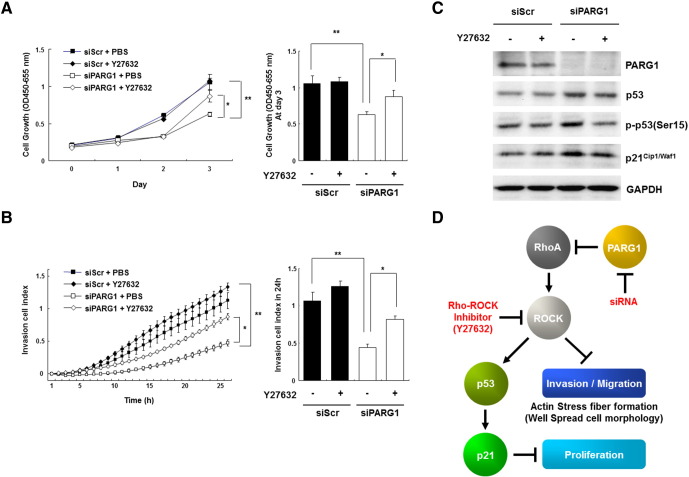
To further investigate RhoA-dependent mechanism of PARG1 on cell proliferation and invasion of RCC cells, we performed rescue experiments using a ROCK inhibitor, Y27632, because a serine/threonine kinase ROCK is one of the major effectors of RhoA, and the RhoA-ROCK axis is involved in functions of various cancer cells [15]. ROCK regulates the phosphorylation of multiple downstream targets and induces actin stress fiber [15]. Cell proliferation of SW839 transfected with PARG1 siRNA was decreased when evaluated by WST-1 assay; however, addition of Y27632 at day 2 in the cell cultures recovered cell proliferation compared with PBD treatment as control (Figure 5A). Similarly, invasion ability of SW839 transfected with PARG1 siRNA was decreased when evaluated using xCELLigence assay; however, addition of Y27632 recovered its invasion ability compared with control (Figure 5B). When we tested the effects of Y27632 on the expression of p53 and p21Cip1/Waf1 in SW839 cells treated with the PARG1 siRNA by Western blot analysis, Y27632 treatment inhibited the increase of p53, p-p53(Ser15), and p21Cip1/Waf1 expressions in PARG1-knockdown SW839 cells compared with control PBS treatment (Figure 5C). These results indicate that PARG1 expressed in human RCC cells promotes cell proliferation via inhibition of RhoA-ROCK-p53-p21 pathway and invasion ability via inhibition of RhoA-ROCK-actin fiber formation (Figure 5D).
Figure 5.
PARG1 promoted cell proliferation and invasion through inhibition of RhoA-ROCK signaling.
(A) Dependency of cell proliferation ability on RhoA-ROCK signaling was evaluated by WST-1 assay with Rho-ROCK inhibitor Y27632 (1 μM) on PARG1-silenced SW839 RCC cells. Cell proliferation was restored in PARG1 siRNA transfected SW839 cell line by addition of Y27632 at day 2, compared with PBS treatment. *P < .05, **P < .01; data are presented as the mean ± SD of three independent experiments. (B) Dependency of cell invasion ability on RhoA-ROCK signaling was evaluated by xCELLigence system analysis as described in Materials and Methods with Y27632 (1 μM) on PARG1-silenced SW839 cells by siRNA. PBS was used as control. Cell invasion ability was rescued by treatment with Y27632 in PARG1 siRNA transfected. SW839 cells transfected with scrambled or PARG1 siRNAs were treated with PBS or Y27632. *P < .05, **P < .01; data are presented as the mean ± SD of three independent experiments. (C) The impact of Rho-ROCK inhibition on expressions of p53, p-p53 (Ser15), and p21Cip1/Waf1 was evaluated by Western blotting in SW839 cells treated with scrambled or PARG1 siRNAs. Representative results from three independent experiments. PBS was used as control. (D) Schematic representation of the functional role of PARG1 involved in cell proliferation, migration, and invasion through regulation of RhoA-ROCK signaling pathway.
Discussion
In this study, we show that PARG1 is expressed in proximal tubule cells of normal kidney and varies among RCC patients. In some RCC patients, PARG1 expression level is lower than normal proximal tubule, although clear cell type of RCC is thought to be derived from proximal tubule cells. RCC cells may originate from proximal tubule stem cells [16] and may change the expression level of PARG1 during malignancy. To reveal the mechanism of differential expression of PARG1 in RCC, identification of PARG1 regulator is needed. In previous reports, Rasip1 (Ras interacting protein 1) capable of binding to PARG1 was shown to suppress RhoA signaling in developing blood vessels, leading to upregulation of C-Raf, pC-Raf, and pErk in endothelial tubulogenesis [17]. Also, IRF6 was shown to induce PARG1 which maintains RhoA inactive state and induces epithelial to mesenchymal transition (EMT) of mouse embryo cells during craniofacial development [18]. These previous reports suggested that Rasip1 and IRF6 may be two of the regulators of PARG1 involving development and EMT, and also it may be possibly related with RCC development. Clarifying the relationship between the mechanism of differential PARG1 expression in RCC and effect of these regulator molecules will be required in future studies.
PARG1, one of the RhoGAPs, contains the GAP domain and functions as a negative regulator of RhoA signaling [9], [10]. The role of PARG1 in cancer is not well known, although it was suggested to be associated with invasion in glioma [11]. Previous reports suggested that RhoA promotes cell invasion and migration in various human cancers [19]. On the other hand, RhoA was reported to inhibit cell motility and invasion in some types of cancers including colon cancer [20] and glioblastoma [21]. Therefore, in RhoA, these different biological roles remain controversial. Our studies show that PARG1 is involved in RCC invasion and migration via inhibition of RhoA-ROCK-actin fiber formation, and PARG1 also stimulated mesenchymal morphology in RCC cell lines. In E-cadherin–deficient human RCC cell lines, a cadherin-associated protein, p120 catenin isoform-1, was reported to promote cell invasion and migration through inhibition of RhoA activity [6]. SW839 and 769-p that we used in in vitro studies are also E-cadherin–deficient cell lines, so PARG1 seems to have the same role with p120 catenin in RCC cells. Future study to reveal the relationship between the regulation of RhoA activity by PARG1 and p120 catenin is required in RCC cells. In addition, downregulation of RhoA was reported to be involved in induction EMT of renal proximal tubular cells through upregulation of snail [22]. However, PARG1 did not affect the expression of EMT-related genes including snail, E-cadherin, and vimentin in RCC cell lines in our study (data not shown). Therefore, PARG1 may not be involved in the typical EMT in RCC cells. Other have shown that low expression of RhoA evaluated by immunohistochemistry was associated with poor outcome in patients with colon cancer [23], and a weak intensity of cytoplasmic RhoA was associated with short overall survival in patients with pancreatic ductal cancer [24]. Our data show that PARG1 expression is related to poor outcome in RCC patients; however, the relationship between RhoA expression level and clinicopathological outcome is not still clear. Further work is needed to reveal impact of PARG1 to RhoA activity in RCC patient samples.
RhoA is also reported to affect cell proliferation as below. The possible dual role of RhoA was previously proposed from the observations that low RhoA activity led to stress fiber formation and high RhoA activity resulted in cell cycle arrest via disruption of the actin cytoskeleton and microtubules in cervical cells [25]. Furthermore, RhoA activates downstream serine threonine kinase ROCK; RhoA-ROCK signaling increased expression of p21 and induced apoptosis in prostate cancer [26], [27] and breast cancer [28]. RhoA-ROCK pathway also induces p53-mediated upregulation of Bax to activate a mitochondrial apoptosis in cardiomyocytes [29]. However, we showed that PARG1 was not involved in the apoptosis of RCC cell lines and only affected cell proliferation ability in our data. We also have clarified the mechanisms of PARG1-RhoA-ROCK-p53-P21 signaling pathway in malignant progression of human RCC cells.
Despite advances in the treatment of patients with RCC, approximately a third of patients who undergo surgery for clinically localized RCC will relapse [30]. Effective molecular markers for prognosis, metastasis, and recurrence of RCC need to be identified. Conventional prognostic factors such as nucleolar grade, TMN stage, performance status, anemia, inflammatory reaction, and thrombocytosis are prognosis predictors in RCC [31], [32], [33]. With regard to the prediction for relapse, clinical factors such as SSIGN (the tumor stage, size, grade, and necrosis) score and preoperative CRP are predictors for recurrence [33]. Molecular markers previously reported to have prognostic significance in RCC patients are CAIX, Ki-67 [34], [35], and growth factors [36], [37]. Low CAIX immunostaining [38] and high proliferative index assessed by Ki-67 [35] were associated with poor survival. As shown in this study, PARG1 expression was found to be associated with cell proliferation–related factors such as Ki-67–positive cells, invasion-related factors such as microvascular invasion and lymph node metastasis, and poor prognosis with high recurrence in RCC patients. Multivariate analysis showed that high PARG1 expression is an independent factor for recurrence in N0M0 RCC patients. Therefore, PARG1 expression in RCC is an attractive biomarker for prediction of recurrence and poor prognosis of RCC patients. In addition to PARG1 itself, serum IgG specific for PARG1 was observed in about half of RCC patients tested but not in healthy donors. Based on the data, PARG1 IgG may also be a biomarker for RCC patients for following up after standard treatments such as surgery, molecular targeted therapy, and immunotherapy. Continuation of high-titer PARG1-specific IgG may suggest the residual RCC in the patients. Presence of PARG1-specific IgG may also suggest the high PARG1 expression in RCC cells, meaning that the patients with high PARG1-specific IgG may have high recurrence and poor prognosis. However, we were not able to evaluate the correlation between presence of serum anti-PARG1 IgG and expression of PARG1 in RCC tissues because of the unavailability of paired samples of serum and tumor tissues. These studies remain to be evaluated. One of the tumor antigens, NY-ESO-1, has recently been reported to be a marker for early recurrence of liver cancer after surgery [39], and serum anti-NY-ESO-1 IgG was also utilized as an early diagnosis of breast cancer [40]. Therefore, PARG1 and specific serum IgG may be useful biomarker for diagnosis in patients with RCC.
Conclusions
Taken together, we revealed that PARG1 is involved in malignant phenotypes of RCC by increasing cancer cell proliferation and invasion ability through inhibition of RhoA-ROCK axis to promote metastasis, recurrence, and poor survival of RCC patients. These findings may provide a new insight in RCC diagnostic and treatment development.
The following are the supplementary data related to this article.
Acknowledgements
We thank Misako Sakamoto and Ryoko Suzuki for preparation of manuscript and Prof. Mototsugu Oya in Department of Urology, Keio University school of Medicine for supports the study.
Footnotes
This work was supported by Grants-in-Aid for Scientific Research (JP14104013, JP17016070, JP23240128, and JP26221005), from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Japan Society for Promotion of Science(KAKENHI), and a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor, Welfare (15-10, 15-17), the P-DIRECT, P-CREATE from Japan Agency for Medical Research and development (AMED), and Keio University Grant-in-Aid for Encouragement of Young Medical Scientists.
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hock LM, Lynch J, Balaji KC. Increasing incidence of all stages of kidney cancer in the last 2 decades in the United States: an analysis of surveillance, epidemiology and end results program data. J Urol. 2002;167:57–60. [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Feng X, Lu X, Man X, Zhou W, Jiang LQ, Knyazev P, Lei L, Huang Q, Ullrich A, Zhang Z. Overexpression of Csk-binding protein contributes to renal cell carcinogenesis. Oncogene. 2009;28:3320–3331. doi: 10.1038/onc.2009.185. [DOI] [PubMed] [Google Scholar]
- 6.Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem. 2008;283:18344–18354. doi: 10.1074/jbc.M801192200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa T, Fujita T, Suzuki Y, Okabe S, Yuasa Y, Iwai T, Kawakami Y. Tumor-specific immunological recognition of frameshift-mutated peptides in colon cancer with microsatellite instability. Cancer Res. 2003;63:5564–5572. [PubMed] [Google Scholar]
- 8.Eisenberg MC, Kim Y, Li R, Ackerman WE, Kniss DA, Friedman A. Mechanistic modeling of the effects of myoferlin on tumor cell invasion. Proc Natl Acad Sci U S A. 2011;108:20078–20083. doi: 10.1073/pnas.1116327108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saras J, Franzén P, Aspenström P, Hellman U, Gonez LJ, Heldin CH. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J Biol Chem. 1997;272:24333–24338. doi: 10.1074/jbc.272.39.24333. [DOI] [PubMed] [Google Scholar]
- 10.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 11.Mariani L, Beaudry C, McDonough WS, Hoelzinger DB, Demuth T, Ross KR, Berens T, Coons SW, Watts G, Trent JM, Wei JS, Giese A, Berens ME. Glioma cell motility is associated with reduced transcription of proapoptotic and proliferation genes: a cDNA microarray analysis. J Neurooncol. 2001;53:161–176. doi: 10.1023/a:1012253317934. [DOI] [PubMed] [Google Scholar]
- 12.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 14.Wacker I, Behrens J. Activin B antagonizes RhoA signaling to stimulate mesenchymal morphology and invasiveness of clear cell renal cell carcinomas. Plos One. 2014;9(10):e111276. doi: 10.1371/journal.pone.0111276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 16.Axelson H, Johansson ME. Renal stem cells and their implications for kidney cancer. Semin Cancer Biol. 2013;1:56–61. doi: 10.1016/j.semcancer.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Xu K, Sacharidou A, Fu S, Chong DC, Skaug B, Chen ZJ, Davis GE, Cleaver O. Blood vessel tubulogenesis requires Rasip1 regulation of GTPase signaling. Dev Cell. 2011;20:526–539. doi: 10.1016/j.devcel.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie EJ, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, Dunnwald M, Lidral AC, Marazita ML, Beaty TH. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol. 2012;94(11):934–942. doi: 10.1002/bdra.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahai E, Maeshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 20.Vial E, Sahai E, Marshall CJ. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 2003;4:67–79. doi: 10.1016/s1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg L, Kloog Y. A Ras inhibitor tilts the balance between Rac and Rho and blocks phosphatidylinositol 3-kinase–dependent glioblastoma cell migration. Cancer Res. 2006;66:11709–11717. doi: 10.1158/0008-5472.CAN-06-1878. [DOI] [PubMed] [Google Scholar]
- 22.Hutchison N, Hendry BM, Sharpe CC. Rho isoforms have distinct and specific functions in the process of epithelial to mesenchymal transition in renal proximal tubular cells. Cell Signal. 2009;10:1522–1531. doi: 10.1016/j.cellsig.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Arango D, Laiho P, Kokko A, Alhopuro P, Sammalkorpi H, Salovaara R, Nicorici D, Hautaniemi S, Alazzouzi H, Mecklin JP, Järvinen H, Hemminki A, Astola J, Schwartz S, Jr, Aaltonen LA. Gene-expression profiling predicts recurrence in Dukes' C colorectal cancer. Gastroenterology. 2005;129:874–884. doi: 10.1053/j.gastro.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 24.Dittert DD, Kielisch C, Alldinger I, Zietz C, Meyer W, Dobrowolski F, Saeger HD, Baretton GB. Prognostic significance of immunohistochemical RhoA expression on survival in pancreatic ductal adenocarcinoma: a high-throughput analysis. Hum Pathol. 2008;39:1002–1010. doi: 10.1016/j.humpath.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Song Y, Wong C, Chang DD. Overexpression of wild-type RhoA produces growth arrest by disrupting actin cytoskeleton and microtubules. J Cell Biochem. 2000;80:229–240. doi: 10.1002/1097-4644(20010201)80:2<229::aid-jcb120>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Xiao L, Eto M, Kazanietz MG. ROCK mediates phorbol ester-induced apoptosis in prostate cancer cells via p21Cip1 up-regulation and JNK. J Biol Chem. 2009;284:29365–29375. doi: 10.1074/jbc.M109.007971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papadopoulou N, Charalampopoulos I, Alevizopoulos K, Gravanis A, Stournaras C. Rho/ROCK/actin signaling regulates membrane androgen receptor induced apoptosis in prostate cancer cells. Exp Cell Res. 2008;314:3162–3174. doi: 10.1016/j.yexcr.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- 29.Del Re DP, Miyamoto S, Brown JH. RhoA/Rho kinase up-regulate Bax to activate a mitochondrial death pathway and induce cardiomyocyte apoptosis. J Biol Chem. 2007;282:8069–8078. doi: 10.1074/jbc.M604298200. [DOI] [PubMed] [Google Scholar]
- 30.Ficarra V, Galfano A, Novara G, Iafrate M, Brunelli M, Secco S, Cavalleri S, Martignoni G, Artibani W. Risk stratification and prognostication of renal cell carcinoma. World J Urol. 2008;2:115–125. doi: 10.1007/s00345-008-0259-y. [DOI] [PubMed] [Google Scholar]
- 31.Scheuring UJ, Cella D, Shah S, Bukowski RM. Randomized phase II trial of first-line treatment with sorafenib versus interferon alfa-2a in patients with metastatic renal cell carcinoma. J Clin Oncol. 2009;27:1280–1289. doi: 10.1200/JCO.2008.19.3342. [DOI] [PubMed] [Google Scholar]
- 32.Suppiah R, Shaheen PE, Elson P, Misbah SA, Wood L, Motzer RJ, Negrier S, Andresen SW, Bukowski RM. Thrombocytosis as a prognostic factor for survival in patients with metastatic renal cell carcinoma. Cancer. 2006;107:1793–1800. doi: 10.1002/cncr.22237. [DOI] [PubMed] [Google Scholar]
- 33.Johnson TV, Abbasi A, Owen-Smith A, Young A, Ogan K, Pattaras J, Nieh P, Marshall FF, Master VA. Absolute preoperative C-reactive protein predicts metastasis and mortality in the first year following potentially curative nephrectomy for clear cell renal cell carcinoma. J Urol. 2010;183:480–485. doi: 10.1016/j.juro.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Bui MH, Visapaa H, Seligson D, Kim H, Han KR, Huang Y, Horvath S, Stanbridge EJ, Palotie A, Figlin RA. Prognostic value of carbonic anhydrase IX and KI67 as predictors of survival for renal clear cell carcinoma. J Urol. 2004;171:2461–2466. doi: 10.1097/01.ju.0000116444.08690.e2. [DOI] [PubMed] [Google Scholar]
- 35.Dudderidge TJ, Stoeber K, Loddo M, Atkinson G, Fanshawe T, Griffiths DF, Williams GH. Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clin Cancer Res. 2005;11:2510–2517. doi: 10.1158/1078-0432.CCR-04-1776. [DOI] [PubMed] [Google Scholar]
- 36.Mizutani Y, Wada H, Yoshida O, Fukushima M, Kawauchi A, Nakao M, Miki T. The significance of thymidine phosphorylase/platelet-derived endothelial cell growth factor activity in renal cell carcinoma. Cancer. 2003;98:730–736. doi: 10.1002/cncr.11570. [DOI] [PubMed] [Google Scholar]
- 37.Rasmuson T, Grankvist K, Jacobsen J, Ljungberg B. Impact of serum basic fibroblast growth factor on prognosis in human renal cell carcinoma. Eur J Cancer. 2001;37:2199–2203. doi: 10.1016/s0959-8049(01)00290-8. [DOI] [PubMed] [Google Scholar]
- 38.Stillebroer AB, Mulders PF, Boerman OC, Oyen WJ, Oosterwijk E. Carbonic anhydrase IX in renal cell carcinoma: implications for prognosis, diagnosis, and therapy. Eur Urol. 2010;58:75–83. doi: 10.1016/j.eururo.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, Gu N, Liu ZB, Zheng M, Xiong F, Wang SY, Li N, Lu J. NY-ESO-1 expression in hepatocellular carcinoma: a potential new marker for early recurrence after surgery. Oncol Lett. 2012;3:39–44. doi: 10.3892/ol.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman C, Murray A, Chakrabarti J, Thorpe A, Woolston C, Sahin U, Barnes A, Robertson J. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol. 2007;18:868–873. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]