Abstract
CRISPR/Cas9 has been widely used for genomic editing in many organisms. Many human diseases are caused by multiple mutations. The CRISPR/Cas9 system provides a potential tool to introduce multiple mutations in a genome. To mimic complicated genomic variants in human diseases, such as multiple gene deletions or mutations, two or more small guide RNAs (sgRNAs) need to be introduced all together. This can be achieved by separate Pol III promoters in a construct. However, limited enzyme sites and increased insertion size lower the efficiency to make a construct. Here, we report a strategy to quickly assembly multiple sgRNAs in one construct using a polycistronic-tRNA-gRNA (PTG) strategy. Taking advantage of the endogenous tRNA processing system in mammalian cells, we efficiently express multiple sgRNAs driven using only one Pol III promoter. Using an all-in-one construct carrying PTG, we disrupt the deacetylase domain in multiple histone deacetylases (HDACs) in human cells simultaneously. We demonstrate that multiple HDAC deletions significantly affect the activation of the Wnt-signaling pathway. Thus, this method enables to efficiently target multiple genes and provide a useful tool to establish mutated cells mimicking human diseases.
Keywords: genome editing, polycistronic-tRNA-gRNA, HDAC, Wnt
INTRODUCTION
During the past three years, the clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated protein nuclease have emerged as the most powerful tool for genome editing in many organisms [1–4]. The most commonly used endonuclease for genome editing is Streptococcus pyogenes Cas9 (referred as Cas9 hereafter). The Cas9 protein is guided by an artificial short guide RNA (gRNA) to cleave the DNA whose sequence is complement to the 5′-end of gRNA preceded by a protospacer-adjacent motif (PAM, 5′-NGG-3′) [5–7]. This simple RNA-guided DNA targeting system fundamentally changed our ability to access specific genomic sites for genetic manipulation. The CRISPR-Cas9 system has been engineered for targeted mutagenesis, site-specific integration of DNA fragments, and precise manipulation of chromosomes, such as large segment deletion and translocations. The nuclease defective Cas9 (dCas9) and gRNA were also engineered to control expression of targeted genes [8–11] and labeling specific loci of the chromosomes [12] in vivo. These Cas9 based tools greatly facilitate basic research and the practice of biotechnology in various medical and agricultural fields.
The robust and efficient Cas9-mediated genome engineering requires a sophisticate regulation to control the expression of Cas9 and gRNA in vivo. The expression of Cas9 could be readily and precisely controlled by the RNA Polymerase II promoter, but the efficiency of gRNA expression remains a bottleneck. Particularly, many Cas9 based applications require simultaneous expression of multiple gRNAs. For examples, a pair of gRNAs are needed to edit one gene in the Cas9 nickase [13, 14] and dCas9-FokI [15, 16] mediated genome editing, which helps reduce off-targeting risk associated with the original CRISPR-Cas9 system. In dCas9 mediated transcriptional regulation, multiple gRNAs are required for robust gene activation or suppression, or generating sophisticated molecular device of transcriptional circuit [10, 17, 18]. Generally, the gRNA expression is driven by polymerase III promoters, such as the widely-used snoRNA U3 and U6 gene promoters. However, these Pol III promoters transcribed gRNAs from specific nucleotides. For example, U3 promoter transcribed from “A” and U6 promoter starts a transcript with “G”. This restrained the targeting spacer of Cas9- gRNA. Therefore, a more robust gRNA expression strategy is needed to boost the capability of CRISPR-Cas9 tools.
Recently, we demonstrated that the endogenous transfer RNA (tRNA) processing system could be engineered to boost Cas9 multiplex targeting capability [19]. Multiple gRNAs were expressed in tandem from an artificial polycistronic-tRNA-gRNA (PTG) gene. The endogenous RNases P and Z recognize tRNA and precisely cleave the PTG to release tRNAs and gRNAs. This system not only enables simultaneous expression of multiple gRNAs, but also potentially enhances the Pol III transcription since the tRNA also acts as an internal enhancer or promoter. The PTG system was implemented in plants, but we speculated that it would be functional in other organisms because the tRNA processing system is highly conserved in all living organisms. In this study, we applied the Cas9-PTG technology for human genome editing. The PTG technique simultaneously expresses 2 to 6 gRNAs for precise genome editing of one to three histone deacetylase (HDAC) genes from Class I and Class II. Our study demonstrates that the PTG system can be engineered as a versatile tool for efficient multiplex genome editing in human and animal systems. In addition, the Cas9-PTG technology helps reveal the function of HDACs through multi-targeted strategy.
MATERIALS AND METHODS
Plasmid vector construction
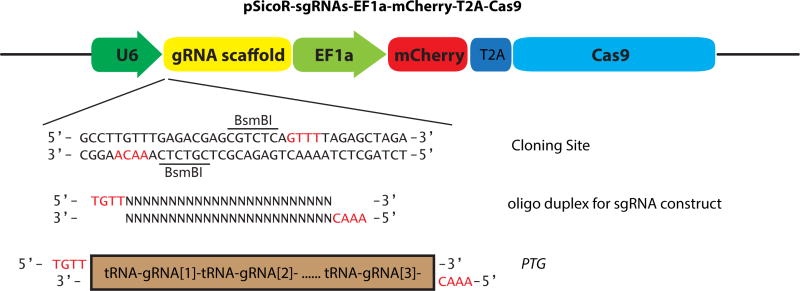
To construct pSicoR-sgRNAs-mCherry-Cas9 vector, the puromycin fragment in pSico-EF1-mCh-Puro (Addgene Plasmid 31845) [20] was replaced with the NLS-Cas9-NLS fragment from pX260 (Addgene Plasmid 42229)[7]. Then the gRNA scaffold with the cloning site containing two BsmBI sites was inserted downstream of the U6 promoter. The schematic of pSicoR-sgRNA-mCherry-Cas9 vector was shown in Figure 1.
Figure 1. Schematic map of the pSicoR-sgRNAs-EF1a-mCherry-T2A-Cas9 vector.
The guide sequence or PTG genes could be expressed after inserting into the BsmbI sites of this vector. The red letters indicated the overhangs in the cloning site.
Assembly of PTG genes
To edit HDAC genes (HDAC1/2/3/4/6) in human cells, a pair of target sites separated by 250–500 bp was selected for each gene based on the presence of PAM and protospacer sequence specificity (Table 1). A total of seven PTG genes (hPTG1 to hPTG7, Table 2) were assembled in vitro using synthesized oligonucleotides (Supplemental Table S1). In this study, the rice glycine-tRNA was used to fuse with gRNA and the in vitro synthesis of hPTG genes was performed as we described previously [19].
Table 1.
Target Sequences for genome editing
| sgRNA | Guide sequence (5′->3′) | Targets | Deletion |
|---|---|---|---|
| HDAC1-sg1 | GTAAGACCACCGCACTAGGC | HDAC1 | 298bp |
| HDAC1-sg2 | GCCCTGCAGCTATTACCATT | ||
| HDAC2-sg1 | GTATTTTAGGATATTGGTGC | HDAC2 | 269bp |
| HDAC2-sg2 | TGTTTCAATCTAACAGTCAA | ||
| HDAC3-sg1 | GAGCAGAACTCAAAGAGCCC | HDAC3 | 437bp |
| HDAC3-sg2 | GCTCAAGTAAGTAGCCCAGG | ||
| HDAC4-sg1 | GAGCTCCTGAATACGTCGCA | HDAC4 | 275bp |
| HDAC4-sg2 | GCTCCTTTGCCGTCCCCGAG | ||
| HDAC6-sg1 | CGACTGGCAGCAGGACGTGC | HDAC6 | 331bp |
| HDAC6-sg2 | CAAGCTGATCCTGTCTCTGG |
Table 2.
Structure of hPTG genes
| Gene Name | Target Genes | # of gRNAs | Encoding gRNAs |
|---|---|---|---|
| hPTG1 | HDAC1 | 2 | HDAC1-sg1, HDAC1-sg2 |
| hPTG2 | HDAC2 | 2 | HDAC2-sg1, HDAC2-sg2 |
| hPTG3 | HDAC3 | 2 | HDAC2-sg1, HDAC2-sg2 |
| hPTG4 | HDAC4 | 2 | HDAC4-sg1, HDAC4-sg2 |
| hPTG5 | HDAC6 | 2 | HDAC6-sg1, HDAC6-sg2 |
| hPTG6 | HDAC1, HDAC2, HDAC3 | 6 | HDAC1-sg1, HDAC1-sg2, HDAC2-sg1, HDAC2-sg2, HDAC2-sg1, HDAC2-sg2 |
| hPTG7 | HDAC4, HDAC6 | 4 | HDAC1-sg1, HDAC1-sg2, HDAC6-sg1, HDAC6-sg2 |
Cell cultures and transfection
Human Embryonic Kidney 293 (HEK293) cells were cultured in DMEM (Sigma) with 10% fetal bovine serum (Atlanta Biologicals), 100 units/mL penicillin and 100 μg/mL streptomycin (Gibco). The culture medium was replaced every 2 days and was sub-cultured when cells reach to 80% to 90% confluency.
For transfection, HEK293 cells were grown in six-well plates at 3×105 cells per well. After reaching 70% confluency, cells were transfected with plasmids and polyethylenimine (PEI). Briefly, cells were fed with fresh culture medium one hour prior to transfection. And then, 3 μg of plasmid DNA and 12 μg of PEI were added to each well. Transfected cells were incubated for 5 hours in a CO2 incubator and cultured for another 48 hours with fresh culture medium.
Circularized RT-PCR (cRT-PCR)
The total RNAs were extracted from HEK293 cells with Trizol Reagent (Life Technologies) and treated with DNase I (New England Labs). To amplify the gRNA, cRT-PCR was performed as we described previously. Briefly, the total RNA was self-ligated with T4 RNA ligase (New England Labs) and then purified with TRIzol Reagent. The ligated total RNA was reverse transcribed using gRNA specific primers and MMLV reverse transcriptase (New England Labs). The cDNA was amplified using specific primers (See Table S1 for sequences) and cloned into pGEM-T easy (Promega) for Sanger sequencing.
DNA extraction and PCR
Transfected cells were washed with phosphate buffered saline and lysed in Lysis buffer (10 mM Tris pH 8.0, 100 mM NaCl and 10 mM EDTA) and genomic DNA were then precipitated with isopropanol. To detect fragment deletion between two target sites within each gene, PCR primers encompass the targeting sites were designed for each HDAC gene (Table S2). PCR amplifications were performed in reactions containing 800 ng genomic DNA, 0.2 mM dNTP, 0.4 μM primers, 1X PCR buffer and 1 unit DreamTaq DNA polymerase (Thermo Fisher Scientific). The PCR product was separated in 1.5% agarose gel containing ethidium bromide to detect chromosomal fragment deletion. The fragment deletion efficiency was estimated with Image J (http://rsb.info.nih.gov/ij/). In addition, the PCR fragment was cloned into pGEM-T easy (Promega) for Sanger sequencing.
Luciferase assay
hPTG constructs were co-transfected with pBARL(β-catenin activated reporter luciferase) [21] firefly luciferase reporter and CTNNB1 to measure activated the Wnt-signaling pathway in HEK293 cells. A pRL-TK renilla luciferase reporter was co-transfected to normalize the transfection efficiency. After 72 hours of the transfection, cells were lysed with Passive Lysis Buffer (Promega). Luminescence was detected in BioTek Synergy 2 using the Dual-Luciferase Reporter Assay System (Promega). Fifteen microliters Luciferase Assay Substrate and Stop & Glo Buffer were mixed in each well with 5 s integration after 5 s delay time. Firefly signals were normalized to the renilla control.
5–Ethynyl–2′–deoxyuridine (EdU) labeling and immunofluorescent staining
Transfected cells were grown with 10 μM EdU for 1 hour. Cells were permeabilized and fixed with 4% formaldehyde. After washing with TBS, cells were incubated with fresh prepared 0.1 M Tris-Cl (pH 8.5), 1 mM CuSO4, 5 μM 6-FAM-Azide and 50 mM Ascorbic Acid for 30 min at room temperature. Cells were washed with TBS with 0.5% Triton X-100 to remove the fluorescent background. After EdU labeling, samples were immunostained with Ki67 (Abcam, 15580) as descried [22]. Nuclear DNA was labeled with DAPI.
RESULTS
PTG enables efficient production of multiple gRNAs in human cells
We demonstrated previously that a PTG with tandem-arrayed tRNA-gRNAs not only allowed precise processing and efficient production of multiple gRNAs, but also enabled Cas9 to simultaneously target multiple genomic sites in plants [19]. We hypothesize that the PTG strategy will boost CRISPR-Cas9 targeting efficiency in animals because tRNA and its processing system is highly conserved in all living organisms. To demonstrate the ability of PTG for genome editing in mammals, we first examined the gRNA excision accuracy from hPTG1 and hPTG2 transgenes in human cells. Each of the artificial PTG constructs encodes two gRNAs targeting the same gene, in which hPTG1 expresses HDAC1-sg1 and HDAC1-sg2 and hPTG2 expresses HDAC2-sg1 and HDAC2-sg2 (Table 2).
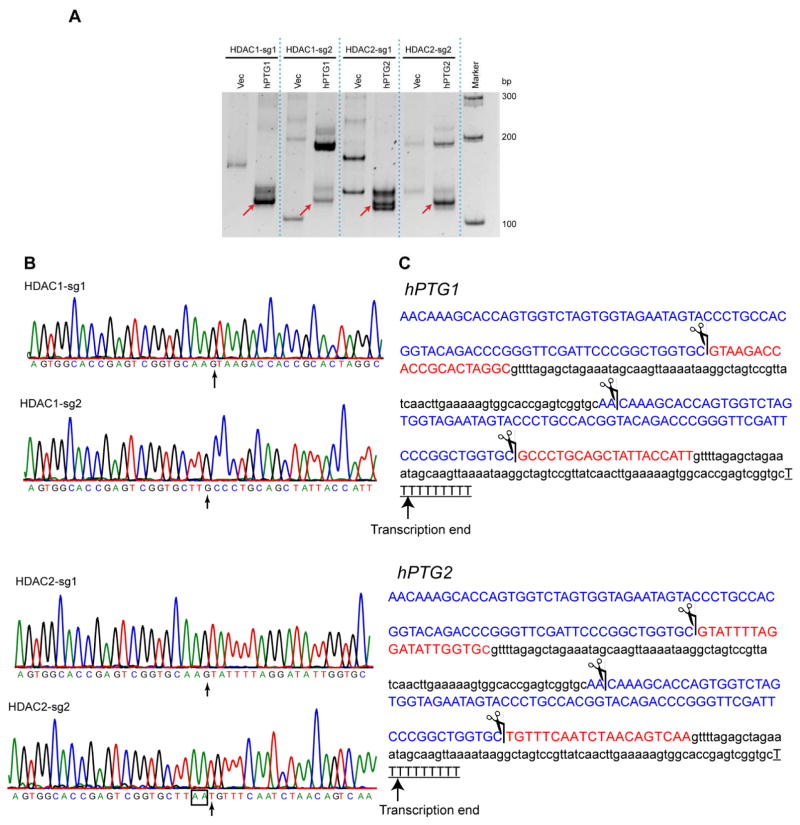
After transfection of hPTG1 and hPTG2 constructs into HEK293 cells, the cRT-PCR was used to amplify the 50 and 30 end of mature gRNAs. Predicted cDNA products with the expected size of single gRNA (~96 nt) were detected by cRT-PCR, despite the presence of some nonspecifically amplified products (Fig. 2A). In Fig. 2B and C, DNA sequencing of these cDNA products indicated that mature gRNAs were precisely processed from hPTG genes with desired 50-end which contains targeting guide sequences. As we observed previously in plants, the gRNA derived from PTG has two additional nucleotides (50-AA-30) at 30-ends if it proceeds tRNA or have two additional T if it precedes a Pol III terminator. We also detected a putative polyadenylation in the 30-end of mature HDAC2-sg2 (Fig. 2C). These results suggest that PTG system could be recognized by the human tRNA processing machinery to produce multiple gRNAs.
Figure 2. Mature gRNAs with desired guide sequences were precisely produced from hPTG strategy.
(A) Electrophoresis of cRT-PCR products in an acrylamide gel. Red arrow indicates the mature gRNAs produced from hPTG1 and hPTG2. Vec, empty vector control. (B) Chromatography of mature gRNA sequences were revealed by cRT-PCR and DNA sequencing. The arrow indicates the first nucleotide at the 5′-end of mature gRNAs. (C) The mapped cleavage site in hPTG1 and hPTG2 according to cRT-PCR results. The scissor indicates the cleaved site of the tRNA processing system. Blue letter, tRNA; red letter, gRNA guide sequence; lowercase letter, gRNA scaffold sequence; underlined letter, Pol III terminator.
Cas9-PTG enables multiplex genome editing in human cells
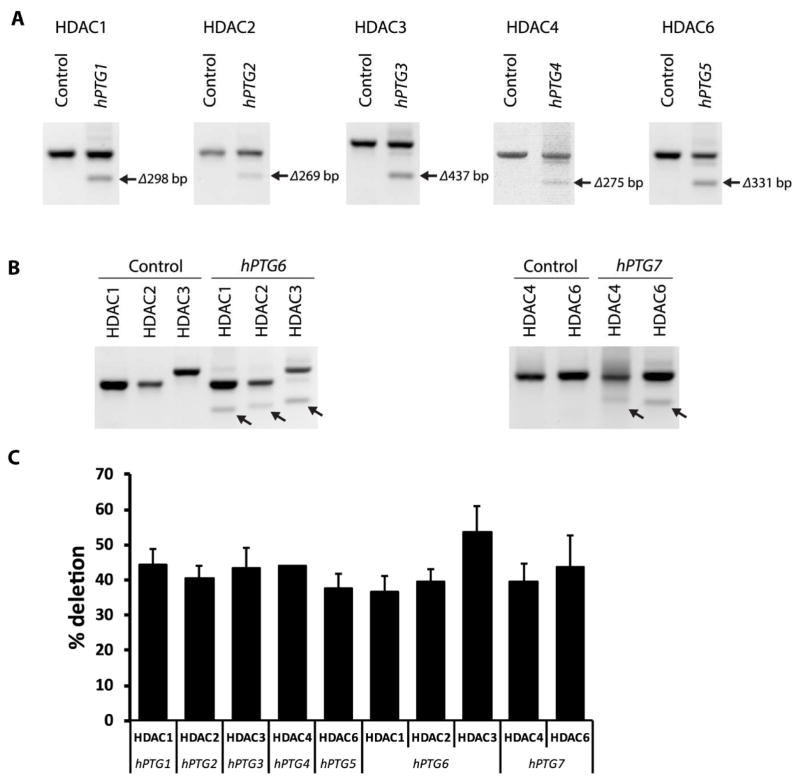
To examine the efficiency of the PTG method for multiplex genome editing, we transfected human cells with seven plasmid constructs expressing different hPTGs and Cas9. These hPTGs encode multiple gRNAs targeting five HDAC genes located in different chromosomes. hPTG1 to hPTG5 were used to target two genomic sites of each gene within HDAC1, 2, 3, 4, and 6; and the hPTG6 and hPTG7 were designed to simultaneously target HDAC3 and HDAC2, respectively (Table 2). Because we used two gRNAs to edit one gene, the efficiency of Cas9-PTG could be measured by the chromosomal fragment deletion frequency within each targeting gene. As predicted, truncated PCR amplicons were detected in all samples (Figure 3A–B). DNA sequence analysis further confirmed these chromosomal deletions was introduced by Cas9-PTGs in transfected cells (Supplemental Figure S1). The deletion frequencies between two gRNA targeting sites were 40%–50% based on the calculation of DNA band intensity (Figure 3C). Interestingly, the efficiency of different hPTG genes with variable number of targets were comparable. For example, hPTG2 (2 gRNAs) and hPTG6 (6 gRNAs) resulted a fragment deletion from HDAC2 locus at ~40% frequency despite their difference in gRNA numbers. PTG with Cas9 in one construct provides an efficiency of genomic deletion that is comparable to previous report [23]. Our results demonstrate that the PTG method can efficiently express multiple gRNAs and simultaneous editing of multiple sites not only in plants, but also in human cells. Because the PTG strategy does not use multiple promoters and terminators, it drastically reduces the size of gRNA construct. As a result, it is more suitable and effective for various genome editing purposes such as the virus-mediated delivery of multiple gRNAs for human gene therapy.
Figure 3. Targeted deletion of chromosomal fragment at HDAC loci.
(A and B) chromosomal fragment deletion were revealed by truncated PCR products (indicated by arrows) at individual HDAC loci in human cells expressing the Cas9-hPTG constructs. (C) Deletion efficiency of hPTG constructs expressing different number of gRNAs.
Cas9-PTG facilitates functional studies for gene family
Histone acetylation change the confirmation of chromatin to allow activating gene transcription. Epigenetic modification alters multiple signaling pathways and controls biological readouts. HDAC inhibitors have been demonstrated to block cell proliferation [24, 25]. We used EdU incorporation to determine the cell proliferation when HDACs were knocked out in human cells [26]. EdU labeling indicated a downregulation of cell proliferation when HDACs were deleted (Fig. 4AeB). Moreover, Ki67 labels all active phases of the cell cycle. We found that all HDAC deletions except for HDAC3 significantly increased cells that are Ki67 negative. This result supports that HDAC loss-of-function results in decreased cell proliferation.
Figure 4. HDAC genomic deletions affect the Wnt signaling pathway and cell proliferation.
(A–B) EdU labeling and immunofluorescence of Ki67 indicate that deletion of HDACs affect the cell proliferation. (C) Luciferase assay indicates the downregulation of the Wnt signaling pathway with single or multiple HDAC deletions.
The Wnt-signaling pathway plays a key role in regulating cell proliferation and differentiation. HDAC inhibitors have been used to suppress the Wnt-signaling pathway [27]. To evaluate Cas9-PTGs, we measure the Wnt-signaling activity using luciferase assay. After normalization to empty vector control without Wnt activation, we found that genomic deletion of HDACs downregulated the Wnt-signaling activity. Interestingly, hPTG1 to hPTG2 showed a mild suppression effect (Figure 4C). However, hPTG5, which deletes HDAC6, showed a stronger inhibition effect on the Wnt-signaling pathway. Our result is consistent with a previous report showing that inhibition of HDAC6 depletes the transcription factor, TCF7L2 [27]. Our data clearly demonstrate that PTG system can efficiently probe functional differences of members in a big gene family.
DISCUSSTION
Multiplex genome editing is one of the fascinating advantage of CRISPR-Cas9 technology, which could be achieved by expressing multiple gRNAs simultaneously. Different strategies of the CRISPR-Cas9 system have been used to modify multiple genes in various organisms, such as the multiplex system that combines several cassettes of Pol III promotors and sgRNAs [28–31]. Besides, ribozyme and Csy4 ribonuclease was also used to express multiple gRNAs from a single transcript [32]. Comparing to these methods, PTG uses the endogenous tRNA processing system without introducing additional components that may be toxic to the cells. The tRNA (~70bp) is smaller than the ribozyme (~200bp) and Pol III promoters (usually 200–300bp). Because the increased size of a viral plasmid can significantly reduce the titer of viral production, PTG has less restraining in the application of the CRISPR-Cas9 system in gene therapy than other methods. Additionally, Pol III promoters obligate to transcribe from specific nucleotide that potentially restrain the targeting range of CRISPR-Cas9. The PTG could be used to express gRNA started with any ribonucleotide as we demonstrated by cRT-PCR (Figure 2). Taken together, the Cas9-PTG would be an efficient strategy for multiplex genome editing.
The feasibility of PTG to simultaneously express multiple gRNAs would enable to design sophisticated CRISPR-Cas9 device. Besides genomic deletion as we showed in this study (Figure 3), the PTG technology can also be used to activate gene transcription or modify epigenetic loci using dCas9-effector system. In addition, using T7 promoter to generate PTG in vitro transcription, equal amount of each desired gRNA will be ensured to present in transfected cells. Because the tRNA system is conserved in living organisms, the applications of Cas9-PTG could be readily expanded to other organisms in addition to rice [19] and human cells (this study).
Members of multi-gene superfamily often carry similar protein structures but exhibit dissimilar biological functions. HDACs superfamily can be classified into four families (class I, IIa, IIb and IV), which differ in enzymatic activity, subcellular localization and expression patterns[33]. Histone deacetylases are often elevated in tumorigenesis [34]. Chemical HDAC inhibitors have been used to suppress the tumor growth [35]. However, it is difficult to identify highly selective chemical inhibitor to each HDAC. Many HDAC inhibitors block activity of multiple HDACs. Comparing to global interference of HDAC activity, single or multiple HDACs manipulation help differentiate the mechanism of epigenetic regulation by individual HDAC. Members in four HDAC classes may have variant mechanisms with similar outcome to control the cell proliferation [35–37].
HDAC1, HDAC2 and HDAC3 can carry similar enzyme activity, but may regulate different biological functions[38]. As HDAC inhibitors unselectively reduce the activity of HDACs, targeting specific HDAC class is important to understand their functions. Homologous deletion of both HDAC1 and HDAC2 in mouse embryonic kidney decreased cytosolic β-catenin and suppress the Wnt signaling [39]. Genomic deletion of either HDAC1, HDAC2 or HDAC3 leads to mild Wnt signaling reduction (Figure 4). The hPTG6 construct, which deletes genomic fragments from the coding region of all HDAC1/2/3, show the highest inhibition of the Wnt signaling. The hPTG7 construct that affects both HDAC4 and HDAC6, also showed a greater suppression of the Wnt activity. Thus, multiplex genomic editing provides an ideal tool to manipulate multiple genes and examine their functional relationships.
Supplementary Material
Highlights.
An efficient strategy to produce multiple gRNAs via the intrinsic tRNA-processing system.
Success in knockout of multiple HDACs in mammalian cells using PTG system.
Demonstration of differential effect of HDACs on the Wnt signaling pathway.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Project 31571374) and the Genetically Modified Breeding Major Projects (2016ZX08010-002-002-003) to K. X and an award from American Heart Association and NIMH 1 R21 MH108983-01 (to Yingwei Mao). We thank Drs. Bernhard Luscher, Timothy Jegla, and Gong Chen for the technical support. The authors have no conflict of interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-Guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 15.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, Clark TA, Williams A, Blume JE, Samal E, Mercola M, Merrill BJ, Conklin BR. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
- 22.Riascos-Bernal DF, Chinnasamy P, Cao LL, Dunaway CM, Valenta T, Basler K, Sibinga NE. beta-Catenin C-terminal signals suppress p53 and are essential for artery formation. Nat Commun. 2016;7:12389. doi: 10.1038/ncomms12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi A, Horiuchi A, Kikuchi N, Hayashi T, Fuseya C, Suzuki A, Konishi I, Shiozawa T. Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int J Cancer. 2010;127:1332–1346. doi: 10.1002/ijc.25151. [DOI] [PubMed] [Google Scholar]
- 26.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gotze S, Coersmeyer M, Muller O, Sievers S. Histone deacetylase inhibitors induce attenuation of Wnt signaling and TCF7L2 depletion in colorectal carcinoma cells. Int J Oncol. 2014;45:1715–1723. doi: 10.3892/ijo.2014.2550. [DOI] [PubMed] [Google Scholar]
- 28.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A. 2014;111:E2967–2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature reviews Genetics. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stojanovic N, Hassan Z, Wirth M, Wenzel P, Beyer M, Schafer C, Brand P, Kroemer A, Stauber RH, Schmid RM, Arlt A, Sellmer A, Mahboobi S, Rad R, Reichert M, Saur D, Kramer OH, Schneider G. HDAC1 and HDAC2 integrate the expression of p53 mutants in pancreatic cancer. Oncogene. 2016 doi: 10.1038/onc.2016.344. [DOI] [PubMed] [Google Scholar]
- 36.Godman CA, Joshi R, Tierney BR, Greenspan E, Rasmussen TP, Wang HW, Shin DG, Rosenberg DW, Giardina C. HDAC3 impacts multiple oncogenic pathways in colon cancer cells with effects on Wnt and vitamin D signaling. Cancer Biol Ther. 2008;7:1570–1580. doi: 10.4161/cbt.7.10.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mak AB, Nixon AM, Kittanakom S, Stewart JM, Chen GI, Curak J, Gingras AC, Mazitschek R, Neel BG, Stagljar I, Moffat J. Regulation of CD133 by HDAC6 promotes beta-catenin signaling to suppress cancer cell differentiation. Cell Rep. 2012;2:951–963. doi: 10.1016/j.celrep.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Yao X, Li Y, Saifudeen Z, Bachvarov D, El-Dahr SS. Histone deacetylase 1 and 2 regulate Wnt and p53 pathways in the ureteric bud epithelium. Development. 2015;142:1180–1192. doi: 10.1242/dev.113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.