Abstract
The nervous systems of cnidarians, pre-bilaterian animals that diverged close to the base of the metazoan radiation, are structurally simple and thus have great potential to inform us about basic structural and functional principles of neural circuits. Unfortunately, cnidarians have thus far been relatively intractable to electrophysiological and genetic techniques and consequently have been largely passed over by neurobiologists. However, recent advances in molecular and imaging methods are fueling a renaissance of interest in and research into cnidarians nervous systems. Here, we review current knowledge on the nervous systems of some cnidarian species and propose that researchers should seize this opportunity and undertake the study of this phylum as strategic experimental systems with great basic and translational relevance for neuroscience.
Keywords: BRAIN Initiative, Imaging, CRISPR, Hydra, Nematostella
The power of a comparative approach to understand neural circuits
Since the time of Cajal, comparative approaches have been central to neuroscience [1]. However, in contrast to Cajal and Sherrington’s “neuron doctrine”, which established that the individual neuron is the functional unit of the nervous system [2], modern neuroscience is now focused on understanding entire neural circuits, as they may be the unit responsible for emergent functional properties [3]. With the advent of innovative methods, researchers expect to record and manipulate entire neural circuits. This could generate a dynamic picture that will reveal, perhaps for the first time in its entirety, how complex neural circuits generate behavior and internal functional states.
The immense complexity of the human brain, consisting of a hundred billion neurons of a yet unknown number of different types, with each neuron able to connect to tens of thousands of other neurons, has made a holistic understanding of how the system works, or how neuronal circuits work on a scale of the entire organism’s nervous system, virtually impossible. Therefore, there is a need to study alternative models with smaller and simpler nervous systems.
Previous examples of the strength of this comparative approach in neuroscience in the 20th century include the use of invertebrate models, such as the marine mollusc Aplysia californica, to elucidate mechanisms of neural function that specifically mediate habituation, sensitization, and forms of associative learning [4]. The large neurons of Aplysia, for example, allowed the detailed examination of neuronal architecture, physiology and control of behaviors at the level of single characterized cells and defined signaling pathways. Other classical examples of breakthroughs that were made possible by using a comparative approach include the understanding of the ionic basis of the action potential (squid) [5], the discovery of adult neurogenesis (canary) [6], conditioned reflexes (dog) [7], and even the earlier discovery of dendritic spines (chicken) [8]. More recently, significant advancements in temporal control of neuronal function through optogenetics have been made thanks to the characterization of channel-rhodopsins in algae [9].
In contrast to the comparative tradition in neuroscience, the power of molecular genetics has driven the exploitation of a selected number of model organisms, particularly: yeast, Caenorhabditis elegans, Drosophila melanogaster, zebrafish and the mouse. Without doubt, these model organisms have revolutionized our understanding of living processes. At the same time, particularly for neuroscience, the emphasis on these model organisms has come at the cost of essentially ignoring the rich biological diversity demonstrated by the structural and functional variety of nervous systems in disparate animal groups. This situation principally arose due to the difficulty of applying developed genetic or molecular methods to most species, thereby leaving them outside the purview of molecular neuroscience.
This outlook has changed dramatically in the last few years due to the introduction of novel methods that can be directly applied to a wide variety of species. In many of these cases, it was the Human Genome Project [10] that opened the way for systematically sequencing genomes of representatives of every phylum. Every area of biology has been transformed by the development of sequencing technologies and transgenics. This, together with gene-editing techniques such as CRISPR-Cas9 [11], have led to a “democratization” of molecular biology, with functional and genomic analyses now possible in a wide range of species. A similar case can be made for the use of calcium imaging of neural circuits [12], which has enabled access to functional information from neurons that were previously too difficult to record from with electrical methods. Taken together, these advances underpin a revitalized interest in applying comparative approaches to nervous system evolution, structure and function, by making use of newly-emerging model animals with “simple” nervous systems. Such investigations are now not only possible, but also appear increasingly necessary for understanding the structural determinants of behavior in more complex animals.
In the following we will briefly review some of the basic features of the neurobiology of different non-bilaterians and then illustrate some examples of how modern techniques are starting to yield significant insights into their nervous systems. We end with a “call to arms”, pointing out the unique opportunity and potential benefits for neuroscience to study these organisms.
Nervous systems were present in the common ancestor of Cnidaria and Bilateria
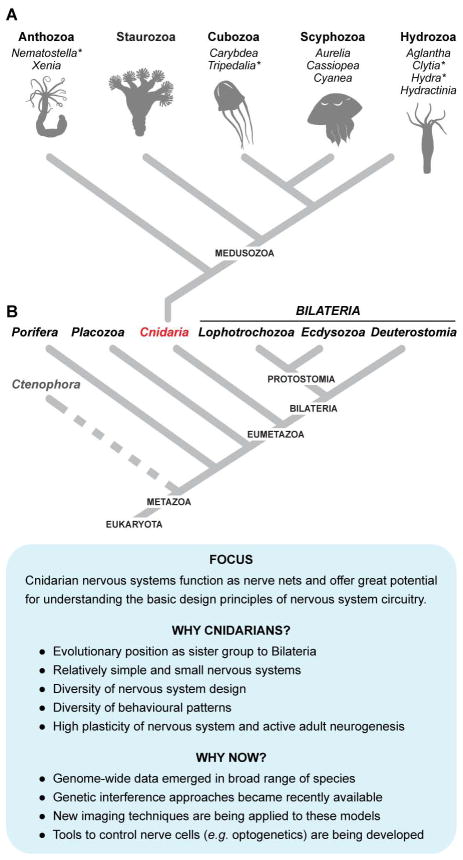
If one aims to understand how nervous systems function by identifying cardinal shared features of neurons and neural circuits via the comparative approach, it is profitable to study the earliest evolutionary examples. Remarkably, nervous systems appeared very early in animal evolution and were certainly in place prior to the origin of bilaterally symmetric animals – also known as Bilateria (Figure 1). Of the lineages that diverged prior to the bilaterian radiation, nervous systems are present only in Ctenophora and Cnidaria. Intriguingly, while cnidarian and bilaterian nervous systems have many characteristics in common, those of the Ctenophora appear to differ fundamentally – for example, glutamate and neuropeptides may be the sole neurotransmitters employed [13]. In addition to the controversial phylogenetic position of the Ctenophora [14,15], a paucity of data and limited amenability to technical approaches make these animals difficult candidates for comparative studies of neurobiology. A more practical choice for such studies are cnidarians, a phylum of approximately 11,000 aquatic animals that occupies a strongly supported position as the sister clade of the Bilateria (Figure 1). Cnidarians, which include jellyfishes and polyps and are distinguished by their nematocysts (stinging cells), have long been utilized in laboratories as experimental organisms to address diverse biological questions [16–20].
Figure 1. Cnidarians as model organisms for studies of nervous systems.
A: Schematic phylogenetic tree showing the relationships of the five classes within the phylum Cnidaria. Species of cnidarians used for research on nerve systems and referred to in this Review are listed. Complete genome sequences are available for several cnidarian species (marked with asterisk) [21]. B: Schematic phylogenetic tree showing main branches of metazoan evolution and position of Cnidaria among the non-bilaterian Metazoa. As the phylogenetic position of comb-jellies (Ctenophora) remains controversial [13– 15], the branch leading to this group is represented by a dashed line.
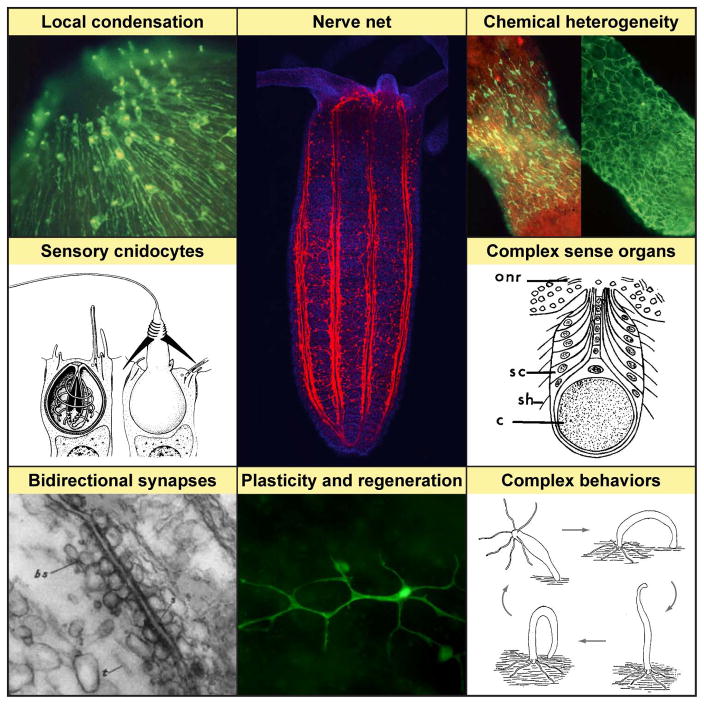
Interestingly, in stark contrast to the centralized nervous systems of bilaterians, cnidarian nervous systems function as diffuse nerve nets that can possess varying degrees of regional condensation and specialization (Figure 2). This difference in neural architecture alone offers great potential for understanding the basic design principles and evolutionary trajectories of nervous system circuitry.
Figure 2. Distinctive features of cnidarian nervous system.
A diffuse nerve net is a basic design of cnidarian nerve systems. Nerve net of the sea anemone Nematostella vectensis revealed by expression of a mCherry protein under nerve-specific elav promotor. Modified from [98]. Local condensation of the nerve net is observed in most cnidarians [69,121,122]. For instance, in Hydra high density of the JRI-positive sensory neurons is observed around the mouth opening (hypostome) of a polyp. Modified from [93]. Chemical heterogeneity of the cnidarian nerve net contrasts its apparent morphological simplicity. A broad range of neurotransmitters present in cnidarian nerve cells are expressed by particular subsets of neurons. For instance, in Hydra RFamide (left) and GLWamide (right) are produced by two apparently not-overlapping populations of neurons. Modified from [93]. Sensory cnidocytes represent a type of mechanosensory cell found exclusively in cnidarians. Mechanical stimulation of a resting cnidocyte (left) triggers rapid discharge of a nematocyst and release of a harpoon to spear and paralyze prey. Modified from [123]. Complex sense organs are found in cnidarians. A statocyst from the umbellar margin of the hydrozoan jellyfish Aglantha contains a concretion (c) surrounded by sensory cells (sc) with sensory cilia (sh), that are connected to the umbrella’s outer nerve ring (onr). Declination of the concretion stimulates cilia, enabling the medusa’s vestibular sense. Modified from [66]. Bidirectional synapses are common for cnidarian. An electron micrograph of a synapse between two axons in the scyphozoan Cyanea represents neurotubules (t) and small vesicles (bs) on both sides of a synaptic cleft (s). Modified from [36]. Plasticity and regeneration are characteristic for nervous systems of cnidarians undergoing constant asexual proliferation. In vivo imaging of transgenic cells (here a ganglion neuron of Hydra, modified from [124]) allows studying how this plasticity is accomplished by integrating stem cell derived migratory neuronal precursor cells into the nervous system. Complex behaviours, such as somersaulting in Hydra, emerge from the activity of simple nerve nets; the cellular and molecular processes behind remain poorly understood. Modified from [92].
In support of the viability of the analysis of the development, structure and function of cnidarian nervous systems, significant advances have been made not only in the application of experimental technologies, but also in the number of cnidarian species that can be interrogated by such means (Figure 1). In recent years, the classical freshwater polyp Hydra has been joined in the laboratory by a sea anemone, Nematostella vectensis, a jellyfish, Clytia hemispherica, and a colonial hydrozoan Hydractinia echinata [21]. For each of these species, extensive genomic and transcriptomic resources now exist, and transgenesis and gene-editing technologies are available. Thus, a suite of pre-bilaterian nervous systems is now open to functional analyses addressing questions that range from the molecular to the behavioral and evolutionary level. In addition, an extensive set of genomic and transcriptomic databases is becoming available for several sponge species -Amphimedon queenslandica, Oscarella carmela and Sycon ciliatum, as well as for a placozoan Trichoplax adhaerens [22–25]. Since sponges and placozoans represent an “outgroup” to cnidarians and bilaterians (Figure 1), and are phyla of metazoans lacking nervous systems, these data are very valuable for comparative genomic approaches.
Most nervous system components have ancient origins
Although phylostratigraphy [26] implies that vertebrate nervous systems have their origins in the common ancestor of Cnidaria and Bilateria (herein referred to as the Eumetazoa), individual components clearly predate this. For example, the processing machinery for neuropeptides, a dual function peptidylglycine alpha-amidating monooxygenase (PAM), is present in early diverging phyla such as sponges (Porifera) which lack a nervous system, supporting the idea that, within the Metazoa, amidated peptides may have originally functioned in communication between epithelial cells [27]. Also, most of the structural components of the post-synaptic density (PSD) are present in the sponge Amphimedon [22,28], and some have clear homologs in choanoflagellates and other unicellular holozoans [29,30]. Other classes of “synaptic” proteins, particularly those involved in vesicle exocytosis, are also represented in unicellular holozoans and homologs of some of the SNARE proteins (e.g. synaptobrevin2, syntaxin1) and their interacting partners (e.g. tomosyn) have been even identified in fungi and plants [31].
Thus, remarkably, some individual neuronal components predate the Eumetazoa, and the cnidarian repertoire of proteins associated with nervous systems is essentially complete. In fact, homologs of some vertebrate synaptic proteins not present in the model invertebrates Drosophila and Caenorhabditis, such as Narp/Pentraxin, are present in cnidarians (though absent from sponges, placozoans and protists). An “ancestral complexity” of cnidarians in terms of synaptic proteins is therefore apparent [28,32]. Moreover, whereas orthologs of synaptic proteins may be present in organisms that lack nervous systems, their roles may be quite different [33], suggesting that the eumetazoan ancestor co-opted these to enable neuronal communication.
In fact, very few synaptic proteins are actually unique to vertebrates; of the 74 protein components of the mammalian nervous system listed by Burkhardt et al. [33], only three (SynCam, Piccolo and Bassoon) lack homologs in other phyla. A caveat here is that generalizations about early nervous system evolution are currently based on relatively sparse taxon sampling – as yet, whole genome assemblies are available for only a few non-bilaterian animals or holozoans. The patchy distribution of some gene families across the Metazoa, perhaps resulting from stochastic gene loss [34] implies that more extensive taxon sampling may eventually identify larger numbers of synaptic proteins in both cnidarians and other groups, mandating the revisiting of evolutionary scenarios based on presently available data.
The hidden complexity of cnidarian nervous systems
The anatomical simplicity of the cnidarian nervous system masks some remarkable neurophysiological specializations [35] including, for example, bidirectional chemical synapses (Figure 2) [36,37], signaling by a diversity of peptide-gated channels [38–40], the rapid discharge of nematocysts (Figure 2) [41] and axons with two kinds of impulse propagation [35,42–45]. In the following section, we will briefly touch on these mechanisms and other distinct neural characteristics to illustrate the functional sophistication of these “primitive” nervous systems and highlight fertile avenues for neurobiological research. These examples could also help put in perspective standard mechanisms of synaptic transmission and neuronal integration found in bilaterians.
Neurotransmitters and receptors
Functional analysis of the cnidarian nerve net requires knowledge of the mode(s) of synaptic transmission between individual neurons. This which could reveal fundamental insights into basic design principles of excitatory and inhibitory transmission, and into how this design for communication is used within a neural circuit. While unidirectional synapses are the norm in both cnidarians and bilaterians, bidirectional synapses can be found, for example, in the mammalian retina and olfactory system [46,47]. Bidirectional synapses in Cnidaria (Figure 2) were first described at the ultrastructural level, containing synaptic vesicles accumulated at both sides of the synapse [36]. Electrophysiological recordings revealed that such bidirectional synapses are excitatory and non-polarized, and conduct equally well in either direction [37].
Ample histochemical, biochemical and functional data have been accumulated, indicating the presence of different small molecule neurotransmitters, such as catecholamines, serotonin, acetylcholine, glutamate and GABA in Cnidaria [48,49]. Their precise functional role, however, remains largely undefined, mainly because receptors for these putative transmitters have not yet been identified.
In Cnidaria, cloning and characterization of transmitter receptors has in the past been relatively slow and mainly performed by researchers focused on individual gene families [38,39], thus leaving the receptor repertoire of Cnidaria as a mostly unknown territory. The sequencing in recent years of the whole genome of different cnidarian species, most notably those of the anthozoan Nematostella vectensis and the hydrozoan Hydra magnipapillata (Figure 1) [50,51], has offered the exciting possibility to quickly gather an overview on the entire complement of receptors in these species. Moreover, the possibility to perform in-situ hybridization (ISH), in principle, allows a quick overview of the expression of a specific receptor in the whole organism.
In particular, the metabotropic and ionotropic receptor repertoire of Nematostella has been inspected by genomic analysis in some detail [52] revealing the presence of several metabotropic glutamate and GABA(B) receptors as well as a large number of G protein-coupled receptors for monoamines and melatonin [52]. Strikingly, specific biogenic amine-synthesizing enzymes are sparse, serotonin is apparently lacking, and it has been concluded that aminergic-like transmitters unique to sea anemones act on these receptors [52]. Interestingly, many transmitter-related protein classes appeared closer to vertebrate than to invertebrate counterparts; for example, octopamine and its receptors, which are common in invertebrates, are missing in Nematostella [52]. While not present in Nematostella [52], two muscarinic acetylcholine receptors have been identified in Hydra [53].
Fast synaptic transmission and synaptic plasticity in Bilateria relies on ion channel receptors. The functional requirements of complex neuronal signaling have driven the evolution of distinct ion channel gene families that differ with respect to ligands, ion selectivity and kinetics (onset of activation and desensitization time course). In addition to metabotropic receptors, the Nematostella genome contains several genes for the major classes of ionotropic receptors (nicotinic acetylcholine receptors, glutamate receptors of the AMPA, kainate and NMDA type, GABA(A), and P2X); only an 5-HT3 receptor homolog is apparently lacking [52]. Thus, the diversity of receptor subtypes in cnidarians is, in principle, comparable to that of chordates and notably includes AMPA and NMDA receptors, which mediate synaptic plasticity in the vertebrate central nervous system [54]. The presence of these receptors suggests that the cnidarian nerve net is chemically complex and uses several small molecule transmitters. It also suggests that cnidarians build their nervous systems using essentially the same building blocks present in bilaterians, so nervous system evolution may result more from the rearrangement of existing molecular components than the invention of novel ones. It should be emphasized, however, that the assignment of cnidarian genes to a receptor class was exclusively based on homology to bilaterian receptors [52]. An unequivocal assignment still requires the detailed functional analysis of ligand specificity and, in the case of ionotropic receptors, ion selectivity. Of further note, the diversity of ion channel receptors in Cnidaria is in stark contrast to the relative paucity of ion channel families present in ctenophores, supporting views of independent neural evolution in the Ctenophora [13].
In addition to small molecule neurotransmitters, the cnidarian nervous system extensively, and perhaps predominately, uses neuropeptides [52,55]. In Nematostella the diversity of neuropeptides is matched by over 80 putative GPCRs for neuropeptides [52]. Strikingly, neuropeptide signaling in Hydra is mediated not only by GPCRs, but also by ion channel receptors [38,40,56]. These channels are directly gated by RFamide neuropeptides, which are also present in vertebrates and invertebrates [57]. The presence of RFamide neuropeptides in large-dense core vesicles at the neuromuscular junction [58] and of the ionotropic RFamide-receptors (iRFa-Rs) in epitheliomuscular cells together with pharmacological evidence suggests that the iRFa-Rs are involved in neuromuscular transmission [39,40]. Homologous channels also exist in Nematostella [56] and in some Protostomia [59], suggesting that neuropeptides have a broad function in fast neuronal communication.
While in most cases it is not clear whether these transmitters and receptors identified in cnidarians have a neuronal location and function, the large repertoire of GPCRs and ion channel receptors suggests that the cnidarian nerve net is chemically complex. The cnidarian nerve net thus appears to be an intriguing object to study how a complex toolbox of transmitters and receptors is used to enable the emergence of behaviors within a morphologically simple nerve net structure. This apparent complexity makes it possible that cnidarians use a “chemical connectome” [60], i.e., a sophisticated chemical agonist-receptor matching space that implements the selectivity necessary for specific behavioral patterns. From this view, a specific muscle contraction, for example, could be achieved not by precisely connecting a motor neuron with a particular muscle fiber, but by expressing a particular receptor combination in that muscle fiber, while the neurotransmitter is widely distributed across all muscle fibers. Thus, instead of implementing computations in its physical wiring, the cnidarian nervous system may primarily operate using a “chemical wiring diagram”. We note that this view supports earlier ideas proposed by Carl Pantin [61].
Specialized cellular composition of cnidarian nervous systems
While the comparably low morphological complexity of their nerve nets is a basic feature of cnidarian nervous systems (Figure 2), they have independently evolved several fascinating neural structures and properties that provide unique opportunities to understand how more complex nervous systems can evolve.
For example, all Cnidaria are endowed with specialized mechanosensory cells (the nematocyst-containing cnidocytes, which give name to the phylum, Figure 2) that enable them to spear and paralyze prey with supersonic harpoons. Indeed, the discharge of nematocysts is one of the fastest processes in biology. This discharge is triggered by mechanical stimulation, and employs a fast-recoiling silk-like elastic protein, cnidoin, that incorporates into the capsule of the nematocyst and stores kinetic energy [41]. There is a striking structural similarity between nematocysts and the ciliary-microvillar sensory apparatus of mechanosensory cells in some cnidarians [62,63], suggesting that nematocysts and mechanosensory structures are evolutionarily related. The ciliated mechanosensory cells in cnidarians are also similar to the hair cells of vertebrates, and may represent descendants of an ancient metazoan sensory cell type [64]. A better understanding of their cell biology could inform us further about the evolution and diversification of animal mechanosensory systems.
Sense organs and navigation
In addition to cnidocytes, some cnidarians have elaborate sense organs and perform intricate behaviors (Figure 2) that suggest advanced neural integration. For example, the statocysts in Scyphozoa (Figure 1) are small tentacle-like organs that hang at the outer side of the bell of medusae and are involved in gravisensation, a sense still poorly understood in bilaterians (Figure 2). Statocysts contain a ‘concretion’ at their distal part, and are surrounded by non-motile mechanosensory cilia [65,66]. Movement deflects cilia, allowing the medusa to sense its orientation in the water. These sense organs, therefore, are intriguing objects to study ancestral feedback control mechanisms and may even be informative for understanding bilaterian vestibular sensing systems..
In terms of vision, an interesting specialization is found in the cubomedusae Tripedalia and Carybdea (Cubozoa, Figure 1), which have four sensory structures, the rhopalia, which contain camera-type eyes. These eyes enable the box jellyfish to avoid obstacles and to navigate in the water based on terrestrial cues [67–69]. Rhopalia are integrated with the rest of the nervous system [70], and visual input modulates the pacemaker system that controls the contraction of the swimming bell [69,71]. The structure and function of such visual systems in non-bilaterians provides further support to the view [72] that camera-type eyes evolved several times. Further investigations may further reveal how complex eyes can function in the absence of a centralized nervous system.
Giant axons
Another fascinating specialization of cnidarians is found in some medusae, which are endowed with larger axonal structures. The giant axon system of a hydrozoan Aglantha digitale (Figure 1) represents an advanced specialization that has been extensively studied due to its amenability to electrophysiological recordings [42,45,73–78]. Interestingly, the giant axons in the jellyfish Aglantha were reported to have two kinds of impulse propagation [79]. During slow swimming, low amplitude Ca2+ spikes drive weak muscle contractions, but during predator avoidance the jellyfish switches to fast escape swimming that relies on strong contractions elicited by rapidly conducted Na+-dependent action. The eight giant motor axons thus can simultaneously mediate slow and fast swimming modes, by directly synapsing on the contractile myoepithelial cells as part of an elaborate neural circuitry. This system arguably represents the best-understood circuitry in any cnidarian [80]. The circuits mediating locomotion, feeding, and tentacle contractions in Aglantha are composed of at least 14 distinct neuron types with dedicated functions, including a relay, a carrier, and a pacemaker system [42,45,73,74]. This complexity and functional sophistication stands in contrast to the default concept of a simple nerve net in cnidarians, and indicates that Aglantha has found ingenious ways to implement different behaviors using the same available “hardware”.
Behavior in cnidarians
Perhaps one of the greatest surprises in cnidarian neuroscience has been the realization that their behavioural repertoire is surprisingly complex, given the apparent structural simplicity of their nerve nets (Figure 2). This is something that was already appreciated in the 18th century, when Abraham Trembley first described somersaulting locomotion in Hydra [81] (Figure 2). As a single footed polyp, Hydra cannot translate its position in the bottom of fresh water ponds and rivers without being carried away by the current. As a solution, Hydra bends its body wall, attaches its tentacles and mouth to the substrate, releases its foot, swings its body over to reattach its foot ahead of the mouth, and then releases the mouth to become erect again. It is currently not at all understood how this complicated behavior, which indeed not all humans can master, is achieved via a diffuse net of neurons.
Spontaneous body column contractions in Hydra is another coordinated behavior, which is based on a subpopulation of nerve cells [44,82,83]. Beyond Hydra, spontaneous rhythmic pulsation of tentacles in soft corals (as Xenia, Anthozoa, Figure 1) [84] were first noted by Lamarck nearly 200 years ago. Among medusae, such as Cassiopea (Scyphozoa, Figure 1) the spontaneous rhythmic pulsations of the medusa bell present a behavior pattern in which the influence of certain sense organs and the overall control of the nervous system has been investigated [85,86]. Moreover, growth pulsations, i.e. successive rhythmic extensions and retractions of the shoot and stolon growing tips, are thought to be essential for growth and morphogenesis of colonial hydroid polyps (Hydrozoa, Figure 1) [87–89]. Another impressive behaviour among cnidarians is the ‘wedding dance’, or courting behaviour described for some cubozoan medusae [90]. In the sexually dimorphic cubomedusa Carybdea sivickisi (Cubozoa, Figure 1), sexually mature females produce conspicuous velar spots and males only court females with these spots. During mating, the male attaches a tentacle to the female and brings their oral openings (manubriums) into direct contact. The male forms a spermatophore, which is then transferred to one of the female’s tentacles. Following release by the male, the female inserts the spermatophore into her manubrium [91]. This behaviour includes elements of recognition of the other sex and the assessment of sexual maturity. Since the conspicuous velar spots of the females are important for mating, recognition is likely mediated by visual cues.
Feeding behaviour in jellyfish and polyps also represents another highly coordinated behaviour, including nematocyte discharge, tentacle flexing, and lip flaring to ingest prey [74,92,93]. In Hydra, for example, this behavior can be effectively elicited in the laboratory by micromolar concentrations of reduced glutathione, therefore allowing one to study and manipulate behavior under controlled conditions. Feeding behavior of the polyps can be easily monitored using a dissection microscope, and video-recorded to register and quantify sequence and timing of the events. Thus, the Hydra nervous system must implement a sustained behavioral plan, composed of many independent modules that are precisely arranged in time. This provides us with an example of an early-evolved fixed action pattern.
Plasticity of the nerve net
Cnidarians have a long history as experimental animals for whole body regeneration beginning with Abraham Trembley’s bisection experiments in 1744 [81] and the classic “developmental organizer” transplant experiments of Ethel Brown in Thomas Morgan’s lab [94]. The spectacular ability to rebuild any missing body part includes the generation of large numbers of new nerve cells that seamlessly connect with the remains of the existing nervous system (Figure 2). In Hydra, this is accomplished by integrating stem cell derived migratory neuronal precursor cells into the nervous system [95,96]. Indeed an increase in nerve cell density is the first change detected in cell distribution upon budding and regeneration [97]. In a frame of 12–24 hours, the local density of neurons may double in the amputation region or emerging bud.
Both normal and regeneration-induced neurogenesis implies fast and effective integration of new neurons into the nerve net, through re-wiring and reestablishment of synaptic contacts. This remarkable neurogenic potential also characterizes the initial formation of the nervous system during embryogenesis. For example, in the sea anemone Nematostella vectensis neurons are generated throughout the ectoderm as well as in the endoderm [98]. The molecular control of neurogenesis is remarkably similar to that in vertebrates: Notch signalling controls the number of neural progenitor cells, soxB and bHLH transcription factor encoding genes regulate their further development and Wnt signaling gradients are involved in the patterning of the nervous system [99–101]. The spatially broad neurogenic potential of cnidarians poses specific challenges for the developmental control of neurogenesis, e.g. how is the balance between neural and non-neural cell types regulated? Which mechanisms determine the number of neural progenitor cells? Given the molecular similarities, understanding these questions in cnidarians can inform research on vertebrate neurogenesis.
Certainly, while the analysis of shared and divergent aspects of cnidarian and bilaterian neural development will help to extract core principles of neurogenesis, understanding in detail how the cnidarian neurogenic program can be promiscuously activated throughout both germ layers of the embryo and during regeneration has the potential to stimulate research to improve the generation of neurons in pathological conditions in human patients.
Modern methods reach Cnidaria
There are several fascinating aspects of the cnidarian nervous system that merit further attention (see Outstanding Questions), from the molecular and subcellular level, through the cellular, neural circuit and organ levels and up to the behavioural level. Studying the neurobiology of these properties in cnidarians permits fundamental questions about the function and evolution of nervous systems to be addressed, such as: how can relatively complex macro-behaviours such as swimming/feeding/reproducing be coordinated by non-centralized neural networks; and what are the core structural and functional features of all metazoan neurons.
Outstanding Questions.
How do simple nerve nets work? Reconstructing entire neural circuits, recording their dynamic maps could lead to understanding organism-wide neural connectivity.
What was the original function of the nervous system? Did it emerge to control motility or monitor the environment and to orchestrate multiple functions, as development, tissue homeostasis, and host-microbiome interactions?
How are constant neurogenesis, nerve net regeneration and plasticity achieved in adult asexually proliferating cnidarians? What are the core principles and the main molecular mechanisms of neurogenesis that stay active throughout the life of a cnidarian?
What were the main trajectories of nervous systems evolution? Reconstructing evolution of nervous systems would clarify the role of invention of novel molecular elements and rearrangement of existing ones. It would reveal ancient and therefore fundamental principles of nerve system’s structure and function.
How is the complex toolbox of transmitters, receptors and ion channels used in cnidarians to enable emergence of complex behaviors with a seemingly simple nerve net structure?
The relevance of cnidarians for understanding the evolution of nervous systems and information processing in “simple” nerve nets, has long been recognized, but now faces a re-ignited interest (Figure 1). Indeed, over the past decade, modern molecular biology technology has been implemented in several cnidarian species allowing many of the questions that have been inaccessible in the past to be finally addressed. Stable transgenesis was first achieved in Hydra [102] and is also now available in both Nematostella and Hydractinia [103,104] (Figure 2). The use of cell type-specific promoters for reporter genes has also improved the visualization of neuronal morphology and tracking of the developmental origin of nerve cells [98,100]. Such transgenic lines could also be used to isolate specific neuronal subpopulations by FACS and determine their transcriptome and proteome profiles. The availability of stable transgenesis and neuron-specific promoters now allows the conditional ablation of neurons in adult animals, opening the possibility to analyze the regenerative capacity of cnidarians in new experimental paradigms. In addition to well-established transient knockdown strategies (dsRNA, morpholinos), genome-editing technologies like TALENs or CRISPR/Cas9 are now available in some cnidarian model systems [105,106].
The expression of pre- and postsynaptic marker proteins will significantly facilitate the description of neural connectivity and how it is established. Indeed, the first experiments using genetically encoded calcium indicators to monitor organism-wide neural activity are now underway [107] and in conjunction with the use of optogenetics tools (e.g. light controlled ion channels), these will be instrumental in understanding how non-centralized nervous systems generate spontaneous behaviour and react in response to environmental cues. Finally, exquisitely detailed maps of the entire nervous system in cnidarian models generated with the aforementioned technologies may become a valuable input for computational models (artificial neural networks), intended to simulate function of a nerve net and provide testable predictions.
Concluding remarks and future perspectives
In summary, we are at the beginning of what could constitute a revolution in the study of the neurobiology of cnidarians and other non-bilaterian metazoans. The systematic use of large scale imaging, optogenetics and molecular engineering methods could permit the elucidations of basic principles of neural circuits and the expansion of synthetic biology to metazoans. The comparative analysis of cnidarian and bilaterian nervous systems may allow the identification of ancient and therefore fundamental principles of nervous system structure and function (Box 1). The great molecular similarity between the cnidarian and bilaterian nervous systems makes tenable the possibility that many of the basic circuit mechanisms could be also conserved. At present, there are large research communities deciphering these principles in the nervous systems of Caenorhabditis and Drosophila. A large body of literature covers the molecular make-up and the wiring of their nervous systems and how specific behaviors emerge from these properties. In this context, a small investment elucidating the cnidarian nervous system could have a major payoff. Although the standard model animals have been very useful in the past and will no doubt continue to deliver important insights in the future, one has to be aware that both Caenorhabditis and Drosophila are highly diverged organisms characterized by high levels of gene loss and sequence divergence, whereas at least some cnidarians have retained much of the ancestral genetic complexity of metazoans [108–110]. Reflecting this ancestral core, cnidarian genes are often more similar to vertebrate genes than vertebrate genes are to those of more diverged animals like Caenorhabditis or Drosophila [50,109]. Thus, in addition to the simple design of the cnidarian nervous system, its comparative analysis might reveal new and unexpected basic features of nervous systems. It seems to us that, armed with a new generation of methods, the time is ripe for a deep exploration of the neurobiology of cnidarians.
Box 1. Potential promise of cnidarian neuroscience.
Cnidaria offer a rich playground to explore in relatively simple and accessible nervous systems many of the classical questions in neuroscience. Using calcium imaging of the complete pattern of neural activity, the neural basis of many behaviours such as feeding, swimming, egestion and somersaulting, for example, could be elucidated, given the relatively small number of neurons that some cnidarian have. These efforts could be aided by the reconstructions of their connectomes, a task that appears very feasible in a relatively sparse nerve net, such as those present in many cnidarians. Similarly, complete access to such simple systems could also enable the rigorous modelling of neural circuits, with simulations that are constrained completely, in terms of the number of neurons, connections between the neurons and activity patterns.
The decoding of the activity patterns involved in behaviour could reveal general principles of neural circuit function used more widely in evolution. For example, it is likely that neural circuits generate emergent states of activity, such as dynamical attractors [111]. These attractors are very difficult to identify and manipulate in most laboratory species, due to the fact that functional recordings of the activity are always incomplete. In the case of a cnidarian, however, one could start by measuring the complete record of neural activity for every neuron for a long period of time. This could enable the rigorous identification of a large portion of the entire dynamical space of the nerve net, and the systematic mapping of attractors to behavioural or functional states. While it is impossible to predict how these dynamical landscapes will appear, it is likely that insights acquired in this research could help illuminate a similar research agenda in Bilateria. The initial results in the “brain-wide” imaging of Hydra vulgaris, which displays robust endogenous and sensory-drive dynamics [107], bodes well for the success of such an experimental program.
Besides offering a comparative approach to classical questions, there is a deep evolutionary question that cnidarian could help answer, namely, the function of the first nervous systems. Distressingly, the original function of the nervous system remains somewhat mysterious [112]. While most ascribe the invention of nervous systems to the evolutionary need for motor control, some animals move without a nervous system, such as sponges and nerve-free Hydra, where skin-muscle tissue forms excitable epithelia. Conversely, there are animals that are sessile and still have a nervous system. Understanding in detail the neurobiology of the cnidarian, which are among the most primitive representatives of the first nervous systems in evolution, can shed light on this issue.
Cnidarians can also help explore non-canonical functions of nervous systems. Current opinion mostly perceives the nervous system as a means of communication and information exchange between the central nervous system, the rest of the body and the environment. However, the spectrum of functions performed by nervous systems is much broader. Non-canonical functions may include a role in regeneration, development of innervated tissues, and tissue homeostasis [113,114] as well as bidirectional communication with the commensal microbiota [115–120].
Trends.
Accumulating genomic data strongly support the position of Cnidaria as the sister clade to Bilateria. The emergence of a simple nerve net together with biological, structural and functional diversity within this taxonomic group make cnidarians highly informative for comparative approaches.
Recently sequenced genomes and transcriptomes provide insights into the molecular complexity of cnidarian nerve nets. The diversity of synaptic proteins, small neurotransmitters, neuropeptides, their processing machinery and receptors, is comparable with that of chordates.
Recent advances in imaging and gene manipulation techniques make cnidarians now amenable to functional analysis addressing molecular, behavioral and evolutionary questions.
Accumulating evidences point to multiple roles of the “simple” nervous systems. Emerging evidences point to functions of nervous systems beyond the sensory and motor coordination.
Acknowledgments
The work in the Bosch lab (T.B., A.K., A.M., K.S.) related to this review was supported in part by grants from the Deutsche Forschungsgemeinschaft (DFG), the CRC 1182 (“Origin and Function of Metaorganisms“) and the Cluster of Excellence “Inflammation at Interfaces”. Support by the Alexander von Humboldt foundation (A.K.) and Max Plank Institute for Evolutionary Biology (A.M.) is gratefully acknowledged. Work in the lab of S.G. was supported by the DFG grant GR1771/7-1. F.R. and G.R. are supported by the Sars Centre core budget and a Marie Curie Incoming Postdoctoral Fellowship, respectively. D.J.M. gratefully acknowledges the support of the Australian Research Council, both directly (DP1095343) and indirectly via the ARC Centre of Excellence for Coral Reef Studies (CE14100020). U.T. is funded by the Austrian Science Fund (FWF P27353). T.D.-L. acknowledges Adris Foundation and City of Zagreb.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cajal SR. In Nobel lectures, physiology or medicine 1901–1921. 1906. [Google Scholar]
- 2.Shepherd GM. Foundations of the neuron doctrine. Oxford Univ Press; 1991. [Google Scholar]
- 3.Yuste R. From the neuron doctrine to neural networks. Nat Rev Neurosci. 2015;16:487–497. doi: 10.1038/nrn3962. [DOI] [PubMed] [Google Scholar]
- 4.Kandel E, Schwartz J. Principles of Neural Science, Fifth Edition. McGraw-Hill Education; 2013. [Google Scholar]
- 5.Hodgkin AL, Huxley AF. The components of membrane conductance in the giant axon of Loligo. J Physiol. 1952;116:473–496. doi: 10.1113/jphysiol.1952.sp004718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nottebohm F, et al. Song learning in birds: the relation between perception and production. Philos Trans R Soc London B Biol Sci. 1990;329:115–124. doi: 10.1098/rstb.1990.0156. [DOI] [PubMed] [Google Scholar]
- 7.Pavlov IP. Conditioned reflex. Feldsher Akush. 1951;10:3–10. [PubMed] [Google Scholar]
- 8.Cajal SR. Estructura de los centros nerviosos de las aves. 1888. [Google Scholar]
- 9.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yager TD, et al. The Human Genome Project: creating an infrastructure for biology and medicine. Trends Biochem Sci. 1991;16:454. doi: 10.1016/0968-0004(91)90177-w. [DOI] [PubMed] [Google Scholar]
- 11.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science (80- ) 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 12.Yuste R, Katz LC. Control of postsynaptic Ca 2+ influx in developing neocortex by excitatory and inhibitory neurotransmitters. Neuron. 1991;6:333–344. doi: 10.1016/0896-6273(91)90243-s. [DOI] [PubMed] [Google Scholar]
- 13.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–14. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jékely G, et al. The phylogenetic position of ctenophores and the origin (s) of nervous systems. Evodevo. 2015;6:1. doi: 10.1186/2041-9139-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pisani D, Liu AG. Animal Evolution: Only Rocks Can Set the Clock. Curr Biol. 2015;25:R1079–R1081. doi: 10.1016/j.cub.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Technau U, Steele RE. Evolutionary crossroads in developmental biology: Cnidaria. Development. 2011;138:1447–1458. doi: 10.1242/dev.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe H, et al. Nodal signalling determines biradial asymmetry in Hydra. Nature. 2014;515:112–115. doi: 10.1038/nature13666. [DOI] [PubMed] [Google Scholar]
- 18.Kelava I, et al. Evolution of eumetazoan nervous systems: insights from cnidarians. Phil Trans R Soc B. 2015;370:20150065. doi: 10.1098/rstb.2015.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosch TCG. Rethinking the role of immunity: lessons from Hydra. Trends Immunol. 2014;35:495–502. doi: 10.1016/j.it.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Bosch TCG, et al. How do environmental factors influence life cycles and development? An experimental framework for early-diverging metazoans. Bio Essays. 2014;36:1185–1194. doi: 10.1002/bies.201400065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Technau U, Schwaiger M. Recent advances in genomics and transcriptomics of cnidarians. Mar Genomics. 2015;24:131–138. doi: 10.1016/j.margen.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 24.Fortunato SAV, et al. Comparative analyses of developmental transcription factor repertoires in sponges reveal unexpected complexity of the earliest animals. Mar Genomics. 2015;24:121–129. doi: 10.1016/j.margen.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Riesgo A, et al. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol. 2014 doi: 10.1093/molbev/msu057. [DOI] [PubMed] [Google Scholar]
- 26.Šestak MS, et al. Phylostratigraphic profiles reveal a deep evolutionary history of the vertebrate head sensory systems. Front Zool. 2013;10:1. doi: 10.1186/1742-9994-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attenborough RMF, et al. A “neural” enzyme in nonbilaterian animals and algae: preneural origins for peptidylglycine α-amidating monooxygenase. Mol Biol Evol. 2012;29:3095–3109. doi: 10.1093/molbev/mss114. [DOI] [PubMed] [Google Scholar]
- 28.Sakarya O, et al. A post-synaptic scaffold at the origin of the animal kingdom. PLoS One. 2007;2:e506. doi: 10.1371/journal.pone.0000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suga H, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 2013;4 doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkhardt P. The origin and evolution of synaptic proteins– choanoflagellates lead the way. J Exp Biol. 2015;218:506–514. doi: 10.1242/jeb.110247. [DOI] [PubMed] [Google Scholar]
- 31.Roshchina VV. Microbial Endocrinology: Interkingdom Signaling in Infectious Disease and Health. Springer; 2016. New trends and perspectives in the evolution of neurotransmitters in microbial, plant, and animal cells; pp. 25–77. [DOI] [PubMed] [Google Scholar]
- 32.Alié A, Manuel M. The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol Biol. 2010;10:1. doi: 10.1186/1471-2148-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkhardt P, et al. Evolutionary insights into premetazoan functions of the neuronal protein homer. Mol Biol Evol. 2014;31:2342–2355. doi: 10.1093/molbev/msu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forêt S, et al. New tricks with old genes: the genetic bases of novel cnidarian traits. Trends Genet. 2010;26:154–158. doi: 10.1016/j.tig.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Meech RW. Electrogenesis in the lower Metazoa and implications for neuronal integration. J Exp Biol. 2015;218:537–550. doi: 10.1242/jeb.111955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horridge GA, Mackay B. Naked axons and symmetrical synapses in coelenterates. J Cell Sci. 1962;3:531–541. doi: 10.1038/193899a0. [DOI] [PubMed] [Google Scholar]
- 37.Anderson PA. Physiology of a bidirectional, excitatory, chemical synapse. J Neurophysiol. 1985;53:821–835. doi: 10.1152/jn.1985.53.3.821. [DOI] [PubMed] [Google Scholar]
- 38.Golubovic A, et al. A peptide-gated ion channel from the freshwater polyp Hydra. J Biol Chem. 2007;282:35098–35103. doi: 10.1074/jbc.M706849200. [DOI] [PubMed] [Google Scholar]
- 39.Dürrnagel S, et al. Three homologous subunits form a high affinity peptide-gated ion channel in Hydra. J Biol Chem. 2010;285:11958– 11965. doi: 10.1074/jbc.M109.059998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Assmann M, et al. The comprehensive analysis of DEG/ENaC subunits in Hydra reveals a large variety of peptide-gated channels, potentially involved in neuromuscular transmission. BMC Biol. 2014;12:1. doi: 10.1186/s12915-014-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckmann A, et al. A fast recoiling silk-like elastomer facilitates nanosecond nematocyst discharge. BMC Biol. 2015;13:1. doi: 10.1186/s12915-014-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackie G, Meech R. Central circuitry in the jellyfish Aglantha. I: The relay system. J Exp Biol. 1995;198:2261–2270. doi: 10.1242/jeb.198.11.2261. [DOI] [PubMed] [Google Scholar]
- 43.Westfall JA, et al. Neuro-epitheliomuscular cell and neuro-neuronal gap junctions inHydra. J Neurocytol. 1980;9:725–732. doi: 10.1007/BF01205015. [DOI] [PubMed] [Google Scholar]
- 44.Takaku Y, et al. Innexin gap junctions in nerve cells coordinate spontaneous contractile behavior in Hydra polyps. Sci Rep. 2014;4 doi: 10.1038/srep03573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackie G, Meech R. Central circuitry in the jellyfish Aglantha. II: The ring giant and carrier systems. J Exp Biol. 1995;198:2271–2278. doi: 10.1242/jeb.198.11.2271. [DOI] [PubMed] [Google Scholar]
- 46.Shepherd GM. The synaptic organization of the brain. Oxford University Press; 1990. [Google Scholar]
- 47.Sterling P. Retina. In: Shepherd GM, editor. The synaptic organization of the brain. Oxford University Press; 1990. [Google Scholar]
- 48.Kass-Simon G, Pierobon P. Cnidarian chemical neurotransmission, an updated overview. Comp Biochem Physiol Part A Mol Integr Physiol. 2007;146:9–25. doi: 10.1016/j.cbpa.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Pierobon P. Coordinated modulation of cellular signaling through ligand-gated ion channels in Hydra vulgaris (Cnidaria, Hydrozoa) Int J Dev Biol. 2012;56:551–565. doi: 10.1387/ijdb.113464pp. [DOI] [PubMed] [Google Scholar]
- 50.Putnam NH, et al. Sea Anemone Genome Reveals Ancestral Eumetazoan Gene Repertoire and Genomic Organization. Sci. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 51.Chapman Ja, et al. The dynamic genome of Hydra. Nature. 2010;464:592–6. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anctil M. Chemical transmission in the sea anemone Nematostella vectensis: A genomic perspective. Comp Biochem Physiol Part D Genomics Proteomics. 2009;4:268–289. doi: 10.1016/j.cbd.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Collin C, et al. Two types of muscarinic acetylcholine receptors in Drosophila and other arthropods. Cell Mol life Sci. 2013;70:3231–3242. doi: 10.1007/s00018-013-1334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan TJ, Grant SGN. The origin and evolution of synapses. Nat Rev Neurosci. 2009;10:701–712. doi: 10.1038/nrn2717. [DOI] [PubMed] [Google Scholar]
- 55.Grimmelikhuijzen CJP, et al. Cell signalling in prokaryotes and lower Metazoa. Springer; 2004. Neuropeptides in cnidarians; pp. 115–139. [Google Scholar]
- 56.Gründer S, Assmann M. Peptide-gated ion channels and the simple nervous system of Hydra. J Exp Biol. 2015;218:551–561. doi: 10.1242/jeb.111666. [DOI] [PubMed] [Google Scholar]
- 57.Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci. 2013;110:8702–8707. doi: 10.1073/pnas.1221833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koizumi O, et al. Ultrastructural localization of RFamide-like peptides in neuronal dense-cored vesicles in the peduncle of Hydra. J Exp Zool. 1989;249:17–22. doi: 10.1002/jez.1402490105. [DOI] [PubMed] [Google Scholar]
- 59.Lingueglia E, et al. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 60.Andrews AM. The BRAIN Initiative: Toward a Chemical Connectome. ACS Chem Neurosci. 2013;4:645. doi: 10.1021/cn4001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pantin CFA. The origin of the nervous system. Pubbl Staz Zool Napoli. 1956;28:171–181. [Google Scholar]
- 62.Mattern CFT, et al. Electron microscope observations on the structure and discharge of the stenotele of Hydra. J Cell Biol. 1965;27:621–638. doi: 10.1083/jcb.27.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tardent P, Schmid V. Ultrastructure of mechanoreceptors of the polyp Coryne pintneri (Hydrozoa, Athecata) Exp Cell Res. 1972;72:265–275. doi: 10.1016/0014-4827(72)90589-7. [DOI] [PubMed] [Google Scholar]
- 64.Arendt D, et al. Gastric pouches and the mucociliary sole: setting the stage for nervous system evolution. Phil Trans R Soc B. 2015;370:20150286. doi: 10.1098/rstb.2015.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horridge GA. Statocysts of medusae and evolution of stereocilia. Tissue Cell. 1969;1:341–353. doi: 10.1016/s0040-8166(69)80029-7. [DOI] [PubMed] [Google Scholar]
- 66.Singla CL. Fine structure of the sensory receptors of Aglantha digitale (Hydromedusae: Trachylina) Cell Tissue Res. 1983;231:415–425. doi: 10.1007/BF00222191. [DOI] [PubMed] [Google Scholar]
- 67.Garm A, et al. Visually guided obstacle avoidance in the box jellyfish Tripedalia cystophora and Chiropsella bronzie. J Exp Biol. 2007;210:3616–3623. doi: 10.1242/jeb.004044. [DOI] [PubMed] [Google Scholar]
- 68.Garm A, et al. Box jellyfish use terrestrial visual cues for navigation. Curr Biol. 2011;21:798–803. doi: 10.1016/j.cub.2011.03.054. [DOI] [PubMed] [Google Scholar]
- 69.Satterlie RA. Multiple conducting systems in the cubomedusa carybdea marsupialis. Biol Bull. 2014;227:274–284. doi: 10.1086/BBLv227n3p274. [DOI] [PubMed] [Google Scholar]
- 70.Garm A, et al. Rhopalia are integrated parts of the central nervous system in box jellyfish. Cell Tissue Res. 2006;325:333–343. doi: 10.1007/s00441-005-0134-8. [DOI] [PubMed] [Google Scholar]
- 71.Garm A, Bielecki J. Swim pacemakers in box jellyfish are modulated by the visual input. J Comp Physiol A. 2008;194:641–651. doi: 10.1007/s00359-008-0336-0. [DOI] [PubMed] [Google Scholar]
- 72.Nilsson DE. Eye evolution and its functional basis. Vis Neurosci. 2013;30:5–20. doi: 10.1017/S0952523813000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mackie GO, Meech RW. Central circuitry in the jellyfish Aglantha digitale. III The rootlet and pacemaker systems. J Exp Biol. 2000;203:1797–1807. doi: 10.1242/jeb.203.12.1797. [DOI] [PubMed] [Google Scholar]
- 74.Mackie GO, et al. Central circuitry in the jellyfish Aglantha digitale IV. Pathways coordinating feeding behaviour. J Exp Biol. 2003;206:2487–2505. doi: 10.1242/jeb.00450. [DOI] [PubMed] [Google Scholar]
- 75.Mackie GO, et al. Giant axons and escape swimming in Euplokamis dunlapae (Ctenophora: Cydippida) Biol Bull. 1992;182:248–256. doi: 10.2307/1542118. [DOI] [PubMed] [Google Scholar]
- 76.Mackie GO. Central neural circuitry in the jellyfish Aglantha. Neurosignals. 2004;13:5–19. doi: 10.1159/000076155. [DOI] [PubMed] [Google Scholar]
- 77.Roberts A, Mackie GO. The giant axon escape system of a hydrozoan medusa, Aglantha digitale. J Exp Biol. 1980;84:303–318. doi: 10.1242/jeb.84.1.303. [DOI] [PubMed] [Google Scholar]
- 78.Kerfoot PA, et al. Neuromuscular transmission in the jellyfish Aglantha digitale. J Exp Biol. 1985;116:1–25. doi: 10.1242/jeb.116.1.1. [DOI] [PubMed] [Google Scholar]
- 79.Mackie GO, Meech RW. Separate sodium and calcium spikes in the same axon. 1985. [DOI] [PubMed] [Google Scholar]
- 80.Katsuki T, Greenspan RJ. Jellyfish nervous systems. Curr Biol. 2013;23:R592–R594. doi: 10.1016/j.cub.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 81.Trembley A. In: Mémoires pour servir à l’histoire d’un genre de polypes d’eau douce, à bras en forme de cornes. Par A. Trembley. Jean chez, Verbeek Herman., editors. 1744. [Google Scholar]
- 82.Anderson PA. Evolution of the first nervous systems. Plenum Press; 1990. [Google Scholar]
- 83.Mackie GO. The elementary nervous system revisited. Am Zool. 1990;30:907–920. [Google Scholar]
- 84.Kremien M, et al. Benefit of pulsation in soft corals. Proc Natl Acad Sci. 2013;110:8978–8983. doi: 10.1073/pnas.1301826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamlet C, et al. A numerical study of the effects of bell pulsation dynamics and oral arms on the exchange currents generated by the upside-down jellyfish Cassiopea xamachana. J Exp Biol. 2011;214:1911– 1921. doi: 10.1242/jeb.052506. [DOI] [PubMed] [Google Scholar]
- 86.Santhanakrishnan A, et al. Flow structure and transport characteristics of feeding and exchange currents generated by upside-down Cassiopea jellyfish. J Exp Biol. 2012;215:2369–2381. doi: 10.1242/jeb.053744. [DOI] [PubMed] [Google Scholar]
- 87.Labas YA, et al. On Pulsating Growth in Multicellular Organisms. Dokl Akad Nauk SSSR. 1981;257:1247–1250. [Google Scholar]
- 88.Beloussov LV, et al. Growth pulsations in hydroid polyps: kinematics, biological role and cytophysiology. Oscil Morphog 1993 [Google Scholar]
- 89.Kosevich IA. Mechanics of growth pulsations as the basis of growth and morphogenesis in colonial hydroids. Russ J Dev Biol. 2006;37:90–101. [PubMed] [Google Scholar]
- 90.Werner B. Spermatozeugmen und paarungsverhalten bei Tripedalia cystophora (Cubomedusae) Mar Biol. 1973;18:212–217. [Google Scholar]
- 91.Lewis C, Long TAF. Courtship and reproduction in Carybdea sivickisi (Cnidaria: Cubozoa) Mar Biol. 2005;147:477–483. [Google Scholar]
- 92.Wagner G. Memoirs: On Some Movements and Reactions of Hydra. J Cell Sci. 1905;2:585–622. [Google Scholar]
- 93.Koizumi O. The Cnidaria, Past, Present and Future. Springer; 2016. Origin and Evolution of the Nervous System Considered from the Diffuse Nervous System of Cnidarians; pp. 73–91. [Google Scholar]
- 94.Browne EN. The production of new hydranths in hydra by the insertion of small grafts. J Exp Zool. 1909;7:1–23. [Google Scholar]
- 95.Hager G, David CN. Pattern of differentiated nerve cells in hydra is determined by precursor migration. Development. 1997;124:569–576. doi: 10.1242/dev.124.2.569. [DOI] [PubMed] [Google Scholar]
- 96.Technau U, Holstein TW. Phenotypic Maturation of Neurons and Continuous Precursor Migration in the Formation of the Peduncle Nerve Net inHydra. Dev Biol. 1996;177:599–615. doi: 10.1006/dbio.1996.0189. [DOI] [PubMed] [Google Scholar]
- 97.Bode H, et al. Quantitative analysis of cell types during growth and morphogenesis in Hydra. Wilhelm Roux’Archiv für Entwicklungsmechanik der Org. 1973;171:269–285. doi: 10.1007/BF00577725. [DOI] [PubMed] [Google Scholar]
- 98.Nakanishi N, et al. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development. 2012;139:347–357. doi: 10.1242/dev.071902. [DOI] [PubMed] [Google Scholar]
- 99.Layden MJ, et al. Nematostella vectensis achaete-scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development. 2012;139:1013–1022. doi: 10.1242/dev.073221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richards GS, Rentzsch F. Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development. 2014;141:4681–4689. doi: 10.1242/dev.112029. [DOI] [PubMed] [Google Scholar]
- 101.Watanabe H, et al. Sequential actions of β-catenin and Bmp pattern the oral nerve net in Nematostella vectensis. Nat Commun. 2014;5 doi: 10.1038/ncomms6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wittlieb J, et al. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci U S A. 2006;103:6208–11. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Renfer E, et al. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc Natl Acad Sci. 2010;107:104– 108. doi: 10.1073/pnas.0909148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Künzel T, et al. Migration and differentiation potential of stem cells in the cnidarian Hydractinia analysed in eGFP-transgenic animals and chimeras. Dev Biol. 2010;348:120–129. doi: 10.1016/j.ydbio.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 105.Ikmi A, et al. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat Commun. 2014;5 doi: 10.1038/ncomms6486. [DOI] [PubMed] [Google Scholar]
- 106.Kraus Y, et al. Pre-bilaterian origin of the blastoporal axial organizer. Nat Commun. 2016;7 doi: 10.1038/ncomms11694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Juliano CE, Hobmayer B. Meeting report on “Animal Evolution: New Perspectives From Early Emerging Metazoans”, Tutzing, September 14–17, 2015. Bio Essays. 2016;38:216–219. doi: 10.1002/bies.201500200. [DOI] [PubMed] [Google Scholar]
- 108.Kortschak RD, et al. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13:2190–2195. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 109.Technau U, et al. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. TRENDS Genet. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 110.Raible F, Arendt D. Metazoan evolution: some animals are more equal than others. Curr Biol. 2004;14:R106–R108. [PubMed] [Google Scholar]
- 111.Hopfield JJ. Neural networks and physical systems with emergent collective computational abilities. Proc Natl Acad Sci. 1982;79:2554–2558. doi: 10.1073/pnas.79.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jekely G, et al. An option space for early neural evolution. Phil Trans R Soc B. 2015;370:20150181. doi: 10.1098/rstb.2015.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ivashkin E, et al. A paradigm shift in neurobiology: peripheral nerves deliver cellular material and control development. Zoology. 2014;117:293–294. doi: 10.1016/j.zool.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 114.Adameyko I, Fried K. The nervous system orchestrates and integrates craniofacial development: a review. Front Physiol. 2016;7 doi: 10.3389/fphys.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Williams BB, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barrett E, et al. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 118.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 119.Forsythe P, Kunze WA. Voices from within: gut microbes and the CNS. Cell Mol life Sci. 2013;70:55–69. doi: 10.1007/s00018-012-1028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mu C, et al. Gut microbiota: the brain peacekeeper. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Satterlie RA, Eichinger JM. Organization of the ectodermal nervous structures in jellyfish: scyphomedusae. Biol Bull. 2014;226:29–40. doi: 10.1086/BBLv226n1p29. [DOI] [PubMed] [Google Scholar]
- 122.Eichinger JM, Satterlie RA. Organization of the ectodermal nervous structures in medusae: cubomedusae. Biol Bull. 2014;226:41–55. doi: 10.1086/BBLv226n1p41. [DOI] [PubMed] [Google Scholar]
- 123.Holstein T. The morphogenesis of nematocytes in Hydra and Forsklia: An ultrastructural study. J Ultrastruct Res. 1981;75:276–290. doi: 10.1016/s0022-5320(81)80085-8. [DOI] [PubMed] [Google Scholar]
- 124.Khalturin K, et al. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev Biol. 2007;309:32–44. doi: 10.1016/j.ydbio.2007.06.013. [DOI] [PubMed] [Google Scholar]