Abstract
Our understanding of gene expression has come far since the “one-gene one-polypeptide” hypothesis proposed by Beadle and Tatum. This review addresses the gradual recognition that a growing number of polycistronic genes, originally discovered in viruses, are being identified within the mammalian genome, and that these may provide new insights into disease mechanisms and treatment. We have carried out a systematic literature review identifying 13 mammalian genes for which there is evidence for polycistronic expression via translation through an Internal Ribosome Entry Site (IRES). Although the canonical mechanism of translation initiation has been studied extensively, this review highlights a process of non-canonical translation, IRES-mediated translation, that is a growing source of understanding complex inheritance, elucidation of disease mechanisms, and discovery of novel therapeutic targets. Identification of additional polycistronic genes may provide new insights into disease therapy and allow for new discoveries of translational and disease mechanisms.
Keywords: Bicistronic, polycistronic, IRES, ITAF, eIF, cap-independent
One messenger RNA, multiple polypeptides
George Beadle and Edward Tatum first proposed the model of gene expression in which individual genes encode single enzyme products, recast in subsequent years as the “one-gene one-polypeptide” hypothesis [1, 2]. Since then, in addition to the characterization of alternative splicing of transcripts to generate protein diversity, studies revealing a number of non-canonical translational mechanisms have shown that one gene may code for more than one functionally distinct polypeptide. One prominent example in viruses, protozoans, and invertebrates is the existence of numerous genes bearing multiple open reading frames (polycistronic) that encode two or more independently regulated proteins. Polycistronic genes in protozoans and invertebrates allow for translation initiation at two or more sites along a single mRNA transcript as an efficient means of coordinated gene expression. This expression strategy was not initially attributed to the vertebrate genome repertoire. Recently, a small but growing number of genes have been identified in vertebrates with either tandem or overlapping cistrons. Initiation of translation at a downstream or overlapping open reading frame is usually achieved by capitalizing on a highly structured stem-loop RNA element called an internal ribosome entry site (IRES) (see Glossary) in the primary transcript where ribosomes may bind downstream of the canonical translation start site. IRES function requires interaction with a distinct set of RNA binding proteins (translation initiation factors and IRES trans-acting factors) which may allow differential, but coordinated, control of gene expression relative to the upstream open reading frame. Prominent mammalian polycistronic cellular genes appear to fall into at least four distinct categories based on mRNA structure and function of gene products: 1. Two subunits of a multi-subunit complex whose expression is coordinated in a single transcript; 2. Functionally similar gene products that are differentially co-expressed; 3. Functionally distinct gene products that have programmatically-related expression; 4. Signaling proteins generated by stimulus-coupled protease cleavage or by cap-independent translation (Figure 1). Here we review our current understanding of mechanisms for bicistronic gene translation, examples of prominent polycistronic genes, and some clinical implications of genetic aberrations of these genes and therapeutic possibilities for treatment of resulting disease symptoms.
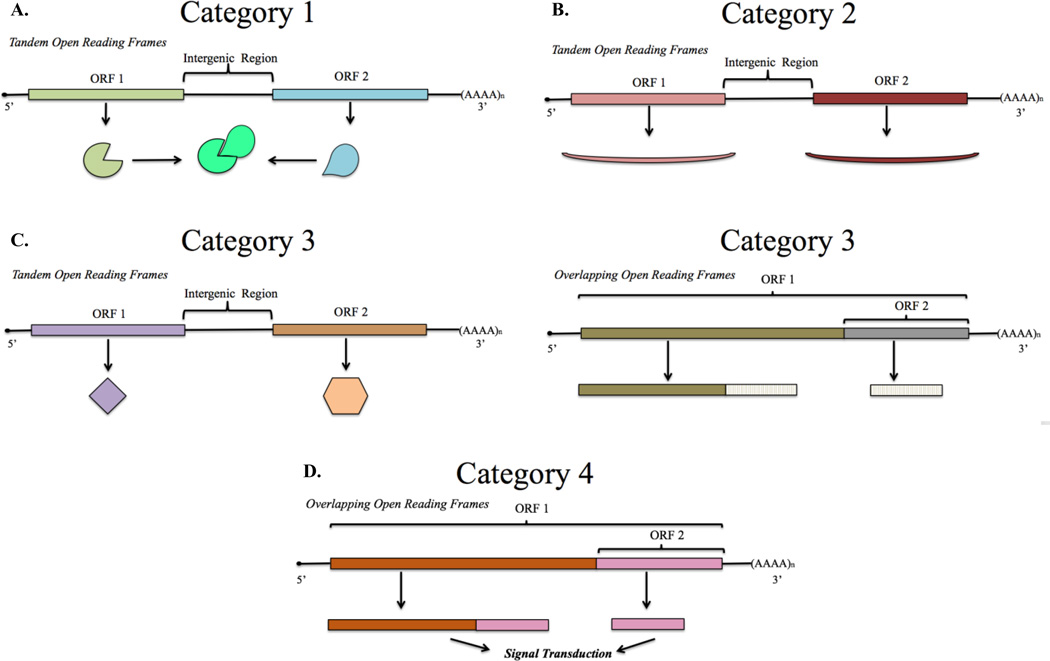
Figure 1. Functional Organization of Bicistronic Genes.
A. Two subunits of a multi-subunit complex whose expression is coordinated in a single transcript: each Open Reading Frame (ORF) codes for a specific subunit of a larger protein complex. B. Functionally similar gene products that are differentially co-expressed: a primary protein is expressed through canonical cap-dependent translation while a secondary and functionally similar protein is differentially expressed through a cap-independent mechanism. C. Functionally distinct gene products that have programmatically-related expression: expression of two differentially functioning proteins is coupled with their operation in a particular biological pathway. Category 3 appears with both tandem and overlapping reading frames. D. Signaling proteins generated by stimulus-coupled protease cleavage or by cap-independent translation: two overlapping ORFs code for necessary products for signal transduction, the primary product is a receptor that initiates signal transduction upon ligand binding while the secondary product is a constitutively active signal.
Canonical protein synthesis in eukaryotes
Translation of mRNA into protein involves three stages - initiation, elongation, and termination. The predominant form of translation initiation in eukaryotes, known as cap-dependent initiation, relies upon recognition of the m7GpppN(7-methylguanosine) “cap” structure at the 5’ end of the mRNA by a complex of canonical initiation factors (eIFs, eukaryotic initiation factors) termed eIF4F. The eIF4F complex, which is composed of eIF4A (a DEAD-box RNA helicase), eIF4E (the cap binding protein), and eIF4G (a multi-domain scaffold protein), recognizes and binds the 5’-cap. The eIF4F complex then recruits the 40S ribosomal subunit along with a second complex, the ternary complex (GTP-bound eIF2 and charged methionine initiator tRNA), as part of the 43S initiation complex, to form the 48S initiation complex. The 48S initiation complex then moves along the 5’-UTR (untranslated region) of the mRNA using the helicase activity of eIF4A to help unwind any potentially inhibitory secondary structure until an AUG in favorable conditions is encountered [3]. GTP is subsequently hydrolyzed to GDP in the presence of eIF5, followed by dissociation of some of the initiation factors. The 60S ribosomal subunit then joins the small subunit, resulting in an elongation-competent 80S ribosome. At this point, translation is initiated (Figure 3). The elongation phase of translation then proceeds until the ribosome encounters a termination codon [4, 5].
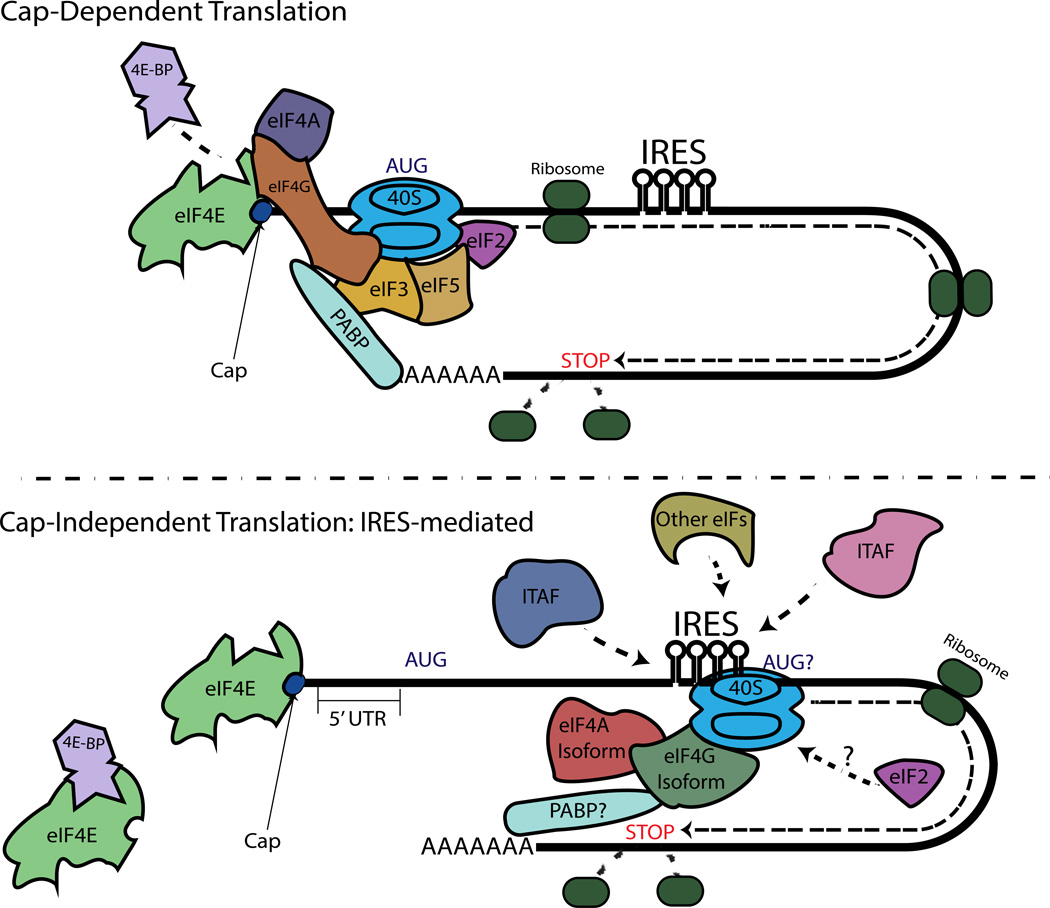
Figure 3. A Brief Overview of Cap-Dependent Translation and IRES-mediated, Cap-Independent Translation.
Mammalian cap-dependent translation functions through the use of all the canonical initiation factors as well as circularization of the mRNA through the interaction of PABP and eIF4G. Key initiation factors are pictured here. Cap-independent IRES-mediated translation is thought to require a subset of ITAFs, an unknown number canonical initiation factors, and may require circularization.
Cap-independent translation in vertebrates
Based on their primary nucleotide sequences, many mRNAs undergo intramolecular base pairing to produce highly ordered secondary structures capable of forming complex stem-loop conformations that facilitate binding of specific proteins with functional consequences. For viral and eukaryotic mRNAs, highly-structured IRES elements can form complexes with ribosomes together with a subset of the canonical eIFs and other RNA binding proteins called IRES transacting factors (ITAFs) to allow for protein synthesis to initiate under conditions not favorable to (or not requiring) cap-dependent initiation (Figure 3) [6–8].The eIF and ITAF requirements differ among IRESs. Interactions of eIFs and ITAFs with ribosomal subunits is thought to allow for correct positioning of the IRES-mediation translation initiation codon at the ribosomal P-site [6]. However, extensive studies on initiation complexes for the IRES region have not been conducted. Once thought to be a property exclusive to viruses, many (~10–15%) vertebrate cellular mRNAs are now believed to be capable of undergoing internal ribosome entry-mediated protein synthesis based on the presence of IRESs located within the 5’ UTR downstream of the cap site [9]. One hundred and fifteen genes containing cellular 5’UTR IRESs have been annotated, and a more recent systematic study provides supporting evidence for known IRESs and suggests potentially novel IRESs using oligonucleotides verified for cap-independent translation activity [10, 11].
Based on the functions of the genes studied to date, it appears that the 5’ IRES provides a means of expression of certain key proteins under conditions of cell stress when cap-dependent translation is inhibited. Alternative translation by cellular 5’UTR IRESs may also occur during phases of angiogenesis, mitosis, and cell proliferation, particularly in genes that have IRESs located in the 5’UTR region such as myc-family genes, cyclin-dependent kinase 11, and vascular endothelial growth factor (VEGF) [6]. The structures, eIFs, and ITAFs have been defined for only a small number of these cellular IRESs. However, in several cases IRES activities appear to be dependent on ITAF expression levels and localization, suggesting a strict regulation of polycistronic genes [6]. Unlike their viral counterparts that comprise distinct families of structurally-related IRESs, cellular IRESs appear to be defined by short motifs and ITAFs [8]. Thus, cellular IRESs have differentiated structures that potentially allow for the diverse functions and localization of the protein products produced under their control.
Polycistronic cellular genes
Nearly all positive-sense RNA viruses have genomic RNAs that encode multiple protein products in the form of precursor polyproteins that are then processed to the functional polypeptides used by the virus during infection. A subset of these viruses (e.g., human rhinovirus, hepatitis C virus, cricket paralysis virus) contain at least one open reading frame (ORF) preceded by IRES structures. However, there is growing evidence that some cellular and vertebrate mRNAs also possess IRES-like structures further downstream of the 5’-UTR, within or after the 5’ proximal ORF, enabling the expression of proteins from tandem or overlapping ORFs. As with prokaryotic polycistronic genes, canonical translation typically initiates near the 5’ends of mRNAs of vertebrate bicistronic genes. However, in a few cases it has been documented that the synthesis of a second protein is initiated via an IRES element located downstream of or within the first open reading frame.
Due to multiple mechanisms of alternative gene expression and translation in eukaryotic cells, the identification of mRNAs harboring bona fide IRES sequences requires a number of stringent criteria that must be satisfied. Cryptic promoters in reporter plasmids and alternative splicing events that can lead to distinct transcripts and secondary protein products need to be ruled out [12, 13]. In addition, multiple mechanisms of secondary protein translation including ribosomal scanning, re-initiation, stop codon read-through, or translational frameshifting may be found in the same gene. Conclusive evidence for the presence of an IRES requires exclusion of these mechanisms and several functional studies (Figure 2).
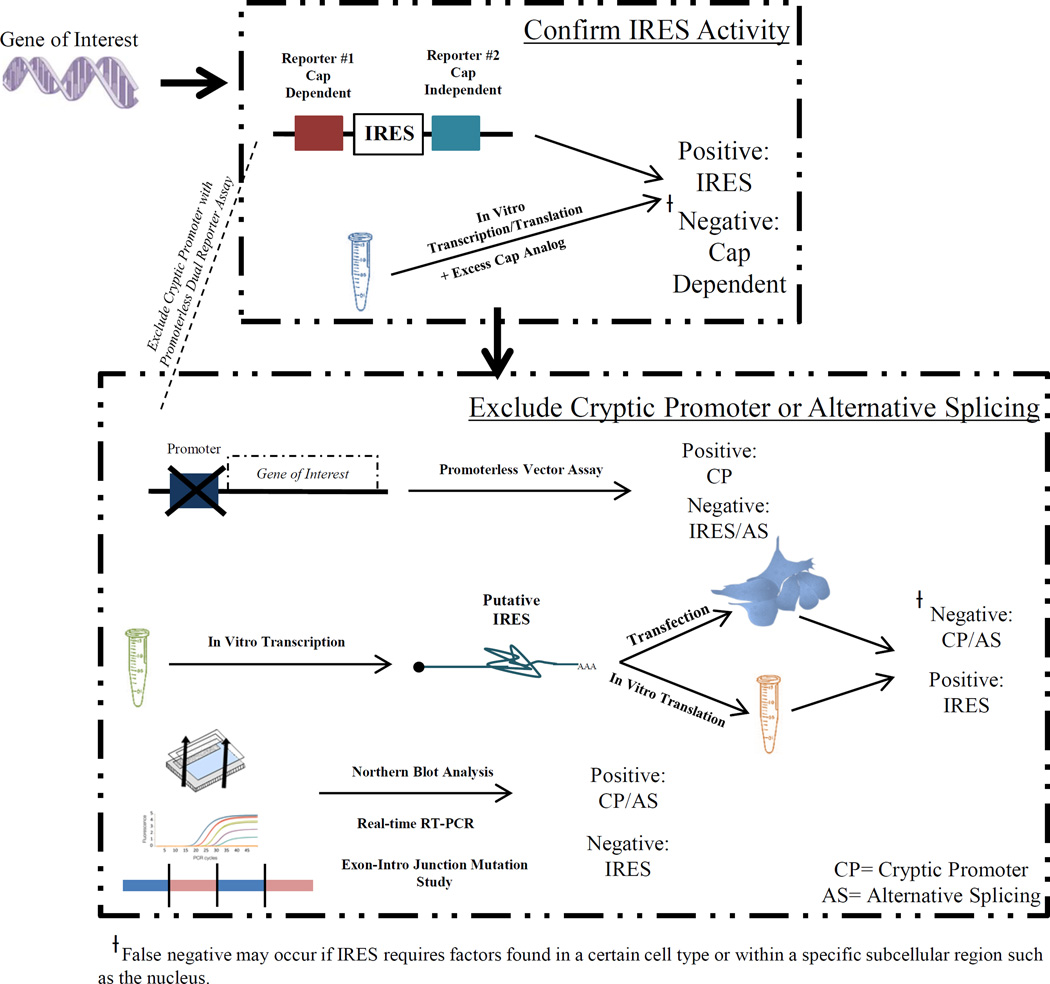
Figure 2. Key Methodology for Identifying IRES-mediated Translation.
Confirmation of IRES activity is done by either: 1) insertion of the putative IRES sequence into a dual reporter vector; or 2) in vitro transcription and translation with excess cap analog. Typical steps for exclusion of cryptic promoter activity or alternative splicing: 1) promoterless vector assay 2) in vitro transcription of a single mRNA, and subsequent in vitro translation or cellular transfection 3) northern blotting and reverse transcription PCR spanning the exons.
Regulation of cellular IRES activity
Although the mechanism of cellular IRES function is not well understood, there is increasing interest in identifying cell-specific expression levels of ITAFs and structural elements that define IRES activity. ITAFs have not been implicated in cap-dependent mechanisms and have been suggested to be specific for cap-independent translation [14]. As a result, ITAFs play an important role in determining the cell- or tissue-specific expression of polycistronic transcripts with IRESs. For example, in meloe, two IRESs code for T-cell antigens that are selectively expressed in melanoma cells but are absent in melanocytes, suggesting that ITAFs are silenced in normal melanocytes [15, 16]. In tissues that do not express or sequester ITAFs critical for IRES function, it would be expected that the polycistronic gene would function in a monocistronic manner, highly reliant on cap-dependent translation. Therefore, polycistronic activity can alter gene expression diversity merely through the availability of these IRES specific factors [6]. However, loss-of and gain-of IRES-mediated translation may contribute to a pathophysiology in cases where disease tissue changes the availability of ITAFs to abnormally promote or eliminate IRES activity. LAMB1, for example, codes for the membrane glycoprotein Laminin B1 that plays a significant role in cancer invasion via a 5’UTR IRES regulated by the ITAF La. La expression, and subsequently IRES-mediated translation of Laminin B1 is decreased in normal hepatocytes compared to hepatocytes after the epithelial-mesenchymal transition (EMT), a critical step in acquisition of invasive properties by carcinoma cells [17].
In addition to expression levels, the subcellular localization of the ITAFs is important for cellular IRES function [6, 14, 18]. A nuclear ITAF may sequester the mRNA until signaling changes allow for translocation to the cytoplasm for translation. Unbound ITAFs may also be compartmentalized until certain cues lead to translocation events necessary for translation of the IRES (e.g. the movement of an inhibitory ITAF from the cytoplasm to the nucleus) [6, 14]. These hypotheses suggest that a separation of ITAFs from mRNA via subcellular compartmentalization could be a critical regulatory mechanism for polycistronic mRNAs.
Lastly, the structural elements of the IRES are important for understanding their function and role in disease. microRNA (miRNA), for example, have been shown to bind to a site in the 3’UTR and act as regulators of IRES-mediated translation for vascular endothelial growth factor A (VEGF) gene that contains two IRESs in its 5’UTR [19]. Moreover, cellular IRESs have been categorized into those that depend on short motifs and those that require an overall complex secondary or tertiary structure [11]. This categorization suggests that mutations that disrupt either short motifs or RNA structure would lead to a disruption in the normal function of the IRES and could lead to disease.
Why polycistronic genes?
Cellular polycistronic genes have several potential advantages for coordinated gene expression, organized here into four distinct categories that indicate expression mechanisms of each polycistronic gene (Figure 1). Furthermore, while polycistronic gene organization allows a unique and selective mechanism for control of protein expression, in the presence of genetic mutations or dysregulation of the IRES this genetic strategy has several potential adverse clinical outcomes. Distinct mutations of polycistronic genes lead to complex and different phenotypes, possibly because of their effects on either ORF or the IRES sequence itself. As a result, polycistronic genes could also open the door to novel therapies.
Table 1 lists thirteen polycistronic genes identified through the literature. Genes were not included that merely expressed truncated forms of the same protein with essentially the same function. Genes for which a cap-independent expression mechanism had not been supported experimentally were also excluded. In the next sections, we provide examples of the four functional classes of polycistronic genes and the biology/gene expression patterns associated with them.
Table 1.
Mammalian Polycistronic Genes
| Gene | Protein(s) | Functional Category |
Primary Protein Function | Secondary Protein Function | Disease Relevance | Mechanism of Expression of Second Cistron | Bicistronic Organization |
Refs |
|---|---|---|---|---|---|---|---|---|
|
TCP-Binding Protein |
PRO1 (8.9kDa) / PRO2 (9.5kDa) |
1 | PRO1: Interacts with PRO2 to make a low-affinity phencyclidine binding entity that is part of a synaptic membrane complex |
PRO2: Interacts with PRO1 to make a low-affinity phencyclidine binding entity that is part of a synaptic membrane complex |
None reported | Putative IRES sequence between PRO1 ORF and PRO2 ORF |
Tandem | [20] |
| FFAR1 | GPR40 (31kDa) / GPR41 (38kDa) |
2 | GPR40: Long-chain fatty acid receptor | GPR41: Short-chain fatty acid receptor |
None reported | IRES in the coding region of FFAR1 encodes GPR41 |
Tandem | [21–24] |
| meloe | MELOE-1 (4.3 kDa) / MELOE-2 (5.1 kDa)/ MELOE-3 (5.9 kDa) |
2 | MELOE-3: No known function; Poor immunogenicity, but is expressed in melanoma cells and normal melanocytes |
MELOE-1: No known function; T cell antigen selectively expressed in melanoma cells MELOE-2: No known function; T cell antigen selectively expressed in melanoma cells |
Potential target for T cell immune therapy for melanoma |
IRES upstream of the ORFs of MELOE-1 and MELOE-2; MELOE-3 is produced by cap- dependent translation |
Tandem | [15, 16, 25] |
| MTPN | Myotrophin (12.9 kDa) / MPD6 (6.4 kDa) |
3 | Myotrophin: Allows dimerization of NFκB in cardiac tissue |
MPD6: No known function; antigen upregulated in response to interferon-α (IFN-α) |
Myotrophin induces cardiac hypertrophy; MPD6 elicits humoral immune response in some cancers |
MPD6 antigen ORF is located in the 3' UTR region of the myotrophin gene and is translated by an IRES located upstream |
Tandem | [30] |
| Connexin 43 | Cx43 (43kDa) / Cx43-CT (20kDa) |
3 | Cx43: Highly abundant connexin with junctional and non-junctional function |
Cx43-CT: Implicated in autoregulating the trafficking of Cx43 |
Hypoplastic left heart syndrome 1, Oculodentodigital dysplasia, Syndactyly type III Craniometaphyseal dysplasia |
Putative IRES or Cap-dependent internal translation |
Overlapping | [45, 46] |
| Connexin 55.5 | Cx55.5 (56kDa) / CT-11 (16kDa) |
3 | Cx55.5: A connexin exclusively expressed in horizontal cells |
CT-11: Has nuclear translocation and may be involve in regulating light- dependent plasticity |
Mutations in human homolog associated with cataracts |
Putative IRES upstream of ATG at nucleotide position 1201 |
Overlapping | [47] |
| pVHL-19 | pVHL-30 (30kDa) / pVHL-19 (19kDa) |
3 | pVHL-30: Tumor suppressor residing in cytosol |
pVHL-19: Comparable tumor suppressor in nucleus and cytoplasm with possibly different molecular substrates |
von Hippel-Lindau disease | Mechanism not investigated | Overlapping | [48, 49] |
|
CdcL1/CdcL2 (PITSLRE genes) |
p110 (PITSLRE) (110 kDa) / p58(PITSLRE) (58kDa) cyclin- dependent kinase isoforms |
3 | p110 (PITSLRE): Regulates transcriptional processing by binding elongation factors and RNA-binding proteins |
p58 (PITSLRE): Binds cyclin D3 in G2/M phases of the cell cycle |
None reported | Increased translation of p58 during G2/M phase due to Unr and phosphorylated eIF-2α binding via IRES near C-terminal end of p110 |
Overlapping | [26] |
| SNRPN/SNURF | SmN (24.6kDa) / SNURF (8.4kDa) |
3 | SmN: Splicing factor important for RNA processing |
SNURF: Potentially implicated in the regulatory mechanism behind gene imprinting |
Prader-Willi syndrome; Angelman syndrome |
Mechanism not investigated | Tandem | [50] |
| GDF1-LASS1 | GDF1 (39kDa) / LASS1 (39kDa) |
3 | GDF1: TGFβ superfamily member implicated in cell growth and differentiation with role in left-right patterning and mesoderm induction |
LASS1 (CerS1) ceramide synthase: TGFβ superfamily member implicated in cell growth and differentiation; catalyzes the synthesis of C18 ceramide in a fumonisin B1- independent manner |
GDF1: cardiovascular malformations; LASS1: progressive myoclonic epilepsy-8 (EPM8) |
Mechanism not Investigated | Tandem | [51] |
| CACNA1A | a1A (220kDa) / a1ACT (75kDa) |
3 | a1A: Voltage-gated calcium channel subunit involved in pre and postsynaptic Ca2+ signaling, gene expression |
a1ACT: Transcription factor important for expression of neural and Purkinje cell development genes |
Spinocerebellar ataxia type 6 (SCA6), Episodic ataxia type 2 (EA-2), Familial hemiplagic migraine type 1 (FHM-1), Epilepsy |
a1ACT is made via an IRES in the C-terminal coding region of CACNA1A |
Overlapping | [27–29] |
| Notch2 | Notch2 (265kDa) / N2ICD (v-Notch2) (70kDa) |
4 | Notch2: Cell surface transmembrane receptor involved in cell fate decision during development by release of N2ICD |
N2ICD: The intracellular domain decoupled and independent of Notch2 surface ligand |
Hajdu-Cheney Syndrome; Alagille syndrome |
Proposed dual mechanism of second cistron initiation: ribosomal reinitiation and IRES both equally contributing to expression of N2ICD |
Overlapping | [31–37] |
| Her2 | HER2 (185 kDa) / HER2 CTF (70 kDa) |
4 | HER2: Tyrosine kinase receptor canonically known to act in secretory and endocytic pathways but also acts within the cell nucleoplasm and nucleus |
HER2 CTFs: C-terminal fragments that encompass the transmembrane and cytoplasmic domains of full length HER2, are associated with nodal metastasis, and localize to the cytoplasm and nucleus of cells |
Hereditary breast cancer, cancer metastasis, tumor development |
Hypothesized IRES within coding region (not confirmed); two methionines at positions 611 and 687 act as initiators of translation for CTFS |
Overlapping | [38] |
1. Two subunits of a multi-subunit complex whose expression is coordinated in a single transcript
TCP-BP
Tenocyclidine [1-(1-(2-Thienyl)cyclohexyl)piperidine, or TCP] binding protein (TCP-BP) was identified as one of a complex of four proteins purified from rat brain synaptic membranes that bind glutamate agonists. TCP-BP is comprised of two subunits, PRO-1 and PRO-2, found to be co-expressed subunits encoded by two tandem ORFs within a single bicistronic transcript. An IRES element preceding the second ORF that leads to translation of the PRO-2 subunit was identified [20]. PRO-1 translation begins at an AUG at the start of ORF1 present 160 base pairs downstream of the mRNA start site. PRO-2 translation occurs from ORF2, 400 base pairs downstream of ORF1 in the intergenic region of the mRNA. Both PRO-1 and PRO-2 are expressed in neurons in brain and show a tendency to form a complex required for TCP-binding. PRO-1 and PRO-2 are thus shown to be formed through coordinately controlled, independent translation of two ORF regions via both a cap-dependent mechanism (PRO-1) and an IRES element (PRO-2). This rat brain gene reveals a potentially rare class of mammalian bicistronic genes in which two subunits of a multiprotein complex are translated by different expression strategies. There have yet to be any clinical associations with this bicistronic gene.
2. Functionally similar gene products that are differentially co-expressed
Two proteins with similar structure and function are differentially expressed in the same biological system.
FFAR1: GPR40 and GPR41
GPR40 and GPR41 are encoded by a bicistronic transcript with GPR40 expression driven by a cap-dependent mechanism whereas GPR41 expression is regulated via an IRES [21]. GPR40, a medium and long-chain fatty acid receptor, causes increased Gαq/11-mediated intracellular calcium levels that stimulate the release of insulin in the presence of long-chain fatty acids [22]. GPR40 is expressed in a highly selective pattern, primarily in pancreatic beta and gastrointestinal tract cells [23]. GPR41 is also coupled to Gαq/11, but is sensitive to short-chain fatty acids. Although its role in the metabolic pathways of beta cells has not been fully characterized, GPR41 has been implicated in energy homeostasis. Both proteins are also co-expressed in adipose tissue and the peripheral nervous system [23, 24]. In beta cells, the expression of both receptors may be necessary to affect metabolic pathways. Dysregulation of either or both of the cistrons may affect the pathology of metabolic disease. It has been hypothesized that co-expression of these fatty acid-activated receptors could sensitize beta cells to different types and levels of fatty acids [21].
meloe: MELOE-1, MELOE-2, and MELOE-3
Cells of the melanocyte lineage express two polypeptides of unknown function, MELOE-1 and MELOE-2, from tandem cistrons in the meloe mRNA. Specific T cell clones recognize melanoma cells but not normal melanocytes based on the expression of one of these melanoma antigens on the surface of the melanoma cells [15, 25]. Therefore, MELOE-1 and MELOE-2 have been studied for potential use in T cell immunotherapy in metastatic melanoma [15]. IRES sequences in the meloe mRNA were found upstream of both ORFs and it was speculated that the different translational mechanisms of 5’ cap-dependent and IRES-mediated translation might be responsible for the selective expression of MELOE-1 and MELOE-2 only in melanoma cells [15]. Differential expression of ITAFs between normal melanocytes and melanoma cells could also account for the activation of these antigens selectively in cancer cells [15]. Furthermore, an additional ORF was identified in the 5’ region of meloe coding for MELOE-3, translated by a cap-dependent pathway [16]. MELOE-3 was found to have poor immunogenicity, suggesting the importance of targeting the IRES-dependent proteins MELOE-1 and MELOE-2 for immunotherapy of melanoma.
3) Functionally distinct gene products that have programmatically-related expression
Two proteins with distinct functions are coupled for co-expression in the same biological pathway
CdcL1/CdcL2 (PITSLRE genes): p110 and p58
PITSLRE/CDK11 duplicate genes CdcL1 and CdcL2 each code for two cyclin-dependent protein kinase isoforms, p110 and p58. The full length CdcL1/CdcL2 transcripts encode p110 proteins. p58 is produced from an open reading frame at the C-terminal end of the main ORF via IRES-mediated translation [26]. p110 is a known transcriptional regulator that regulates RNA-binding proteins and elongation factors. Furthermore, p110 was detected in all phases of the cell cycle [26]. Meanwhile, p58 binds cyclin D3 in the G2/M phases of the cell cycle [26]. Since translation of p58 is up-regulated during G2/M phases, the transcriptional activity of p110 during the cell cycle is likely coupled with the selective expression of p58 in the G2/M phase. Thus, these two PITSLRE genes code for functionally distinct proteins that are programmatically related and may undergo coupled expression. Although no specific disease association has been found with CdcL1 and CdcL2, cyclin-dependent protein kinases carry out critical phosphorylation steps during the cell cycle that are crucial for proper cell function.
CACNA1A: α1A and α1ACT
At least one member of the family of genes encoding voltage-gated Ca2+ channels, CACNA1A, is bicistronic. CACNA1A encodes the pore-forming calcium channel subunit, α1A, through canonical, cap-dependent translation and a novel transcription factor, α1ACT, through IRES-mediated translation from at least one spliced form of the same CACNA1A mRNA [27, 28]. The α1A voltage-gated Ca2+ channel is highly expressed in axon terminals of neurons, particularly in the cerebellum where it mediates calcium entry to facilitate vesicular neurotransmitter release. α1ACT translocates to the nucleus and binds to enhancer sequences containing an AT-rich motif to increase expression of several genes expressed in Purkinje cells. α1ACT expression enhances Purkinje cell dendritic growth and cerebellar cortical development. The α1A mRNA bears a CAG repeat encoding a polyglutamine (polyQ) tract that is expanded in the autosomal dominant disease, spinocerebellar ataxia type 6 (SCA6). SCA6 results from selective degeneration of Purkinje cells. α1ACT bearing an expanded polyQ tract has reduced transcription factor function and is toxic to neuronal cells [27]. Selective suppression of α1ACT by targeting the IRES (e.g. via RNA interference with miRNA) in adults prior to disease onset may be a therapeutic strategy for SCA6 [29].
MTPN: Myotrophin and MPD6
The gene, MTPN, expresses two proteins from a tandem bicistronic transcript. Myotrophin is translated by canonical cap-dependent translation, and MPD6 is produced from a second cistron via IRES-mediated translation. As with CACNA1A, the functions of the two proteins encoded by MTPN appear to be distinct, but presumably programmatically related. The primary protein product, myotrophin, functions in the dimerization of NFκB in cardiac tissue. MPD6 is an antigen upregulated in response to interferon-α (IFN-α) [30]. As a result, MPD6 is implicated in the immune response in a subset of polycythemia vera, chronic myelogenous leukemia, and prostate cancer patients [30]. Due to the possibility of IFN-α selectively increasing IRES translation and the implications for immunotherapy, this presents a striking similarity to selective regulation of meloe IRESs in cancer cells. Different ITAF expression between melanocytes and melanoma cells has been attributed to different cistron expression in meloe. In the case of MPD6, this differentiation appears attributable to a response to the production of IFN-α by host cells. Thus, selective regulation of expression in the bicistronic gene presents a more complex view of the role of MTPN in normal and abnormal cellular function.
4) Signaling proteins generated by stimulus-coupled protease cleavage or by cap-independent translation
The secondary protein normally coupled to expression and ligand binding of the first protein has a constitutively active route for expression in physiological or pathological states.
NOTCH2: Notch2 and N2ICD
Notch2 codes for one of four receptor genes (Notch1-4) involved in the highly conserved ligand-receptor notch-signaling pathway. Interaction of these cell surface receptors with extracellular notch ligand leads to protease cleavage of the C terminal polypeptide known as notch intracellular domain (NICD). NICD translocates to the nucleus and induces transcription of target genes through interactions with co-repressors and co-activators [31]. Apart from the canonical pathway used by Notch1-4, Notch2-ICD (N2ICD) is produced as an independent peptide through a cap-independent process involving an IRES element present within the coding region of the Notch2 mRNA [32]. The IRES-based expression of N2ICD provides an additional mode of notch signaling under different regulation [32]. Additionally, this dual mechanism of expression might have implications in understanding the various conditions associated with notch-signaling dysregulation, especially given that widely different clinical conditions result from mutations in different domains of Notch2 [33–37]
Her2: HER2 and HER2 CTF
Her2 is part of the ErbB/Her family of receptor tyrosine kinases implicated in cancers and neurodegenerative disease. Her2 is a polycistronic gene that codes for a full-length 185 kDa HER2 protein and several independent C-terminal fragments (~ 70kD HER2 CTF) [38]. Two methionines, one at position 611 and the other at 687, act as initiators of translation for CTFs. The existence of these internal initiation sites suggests the presence of an IRES within the coding region of HER2, although canonical IRES identification methods have not been conducted. The HER2 protein is a tyrosine kinase receptor known to act in secretory and endocytic pathways. The full length protein also potentially localizes in the cell nucleoplasm. HER2 CTFs encompass the transmembrane and cytoplasmic domains of HER2 and may be involved in gene expression. Over-activation of Her2 is implicated in hereditary breast cancer and cancer metastasis. Therapeutic administration of antibodies to surface HER2 is a mainstay of immunotherapy for breast cancer. However, HER2 CTFs expressed in the absence of HER2 surface receptors were shown to be unresponsive to therapeutic antibodies. These tumor cells were only sensitive to kinase inhibitors [38]. As a result, revelations in polycistronic regulation of Her2 cancer may allow for improved development and selection of cancer therapies.
Concluding Remarks
The organization and expression of specific proteins from mammalian polycistronic mRNAs appears to provide a similar layer of coordinated expression control to that used constitutively by invertebrates and protozoans, adding to other expression controls believed exclusive to mammals. Among the few examples of human polycistronic genes recognized, the additional cistrons appear to allow for 1) co-expression of multiple subunits of a protein that is part of a complex; 2) co-expression of similarly functioning proteins under slightly different spatial or temporal patterns, 3) or co-production of novel, independently functioning but programmatically related proteins. Although little is understood at present about the cellular machinery involved in the regulation of these IRESs, or whether the upstream cistron product may have a role in the function of the IRES or expression of the second protein, these controls may have important implications in the understanding of health and disease. ITAFs have been shown to both promote and inhibit the translation of cellular IRESs, suggesting their dynamic role in regulating downstream cistron expression [6, 39]. In addition, over-expression or depleted expression of ITAFs could cause a direct dysregulation of the secondary protein that could potentially result in disease. A possible therapeutic strategy could be to use genetic knockdown or drug intervention to restore the balance of ITAF expression [40]. Additionally, a disease state may result in alternative localization or alternative cell-specificity of ITAFs and thereby change regulation of secondary protein expression. This is a critical consideration to take into account when developing therapeutics against diseases involving genes with functional IRESs [41].
In cases where polycistronic activity is compromised through mutations in important regions within the IRES, therapeutics could target the expression of the second protein independent of the IRES (e.g. via adeno-associated virus injection of a cap-dependent form of the secondary protein’s mRNA sequence) or possibly genetic editing to restore IRES function. Additionally, if the secondary protein expression needs to be abrogated, the IRES may be targeted directly through RNAi (i.e. miRNA) or small molecule inhibitors [29, 40]. For example, selective inhibition of the function of an IRES may enable selective suppression of a disease protein, as we have shown in animal model of SCA6, and could, in theory, be the case with persistent expression of HER2 intracellular protein in treatment-resistant breast tumors [29, 38]. Additionally, second cistron expression of an antigen exclusively in cancer cells, such as in the case of meloe, presents an opportunity for immunotherapy [15, 25]. Thus, if the IRES-specific mechanisms governing the expression of the additional cistrons are relatively unique they may be ideal drug targets. Furthermore, elucidating various IRES control mechanisms such as eIFs, ITAFs, microRNAs and other regulatory features may provide insight into the mechanism of poly amino acid expression seen in diseases associated with RAN translation [42, 43]. Finally, if the function of the upstream cistron-encoded protein plays a role in the expression of the downstream protein, genetic aberrations of this gene could in theory have pleiotropic consequences. For example, because depolarization may affect the translocation of the CACNA1C calcium channel transcription factor, CCAT, to the nucleus, gain or loss of function mutations of the calcium channel portion of this gene may affect the entry of CCAT into the nucleus and expression of the array of genes under its control [44]. Thus, a further understanding of the control of polycistronic gene expression in vertebrate tissues should provide new insights into different human genotype-phenotype correlations as well as treatments of human disease.
Outstanding Questions Box.
What additional polycistronic genes can be identified by candidate gene approach or non-biased screens that are regulated by IRES-mediated translation?
What are the molecular and cellular pathways that regulate these novel IRESs?
What off-target effects could arise if small molecule inhibitors or immunotherapy were used to target pathological secondary protein production in polycistronic genes?
Insights into mechanisms of alternative-translation control of these IRESs, such as initiation factors and ITAFs could reveal new sites for clinical treatment, particularly in diseases associated with RAN translation.
Trends Box.
When compared to well-studied systems in viruses, protozoans, and invertebrates, recently identified mammalian polycistronic genes appear to maintain a similar level of coordinated expression control and offer a unique strategy for temporal and spatial control of gene expression.
Studies done on IRES-translation in polycistronic mammalian genes demonstrate multiple possible functions of the resulting proteins.
Mutations that affect the expression of multiple cistrons and therefore different molecular pathways could explain some observations of genetic and clinical complexity.
Newly discovered polycistronic genes demonstrate that targeting of secondary proteins translated by IRES-mediated mechanisms could be a viable method of disease treatment using miRNA targeting or immunotherapy.
Glossary
* See Table 1 for additional explanation of gene and protein function
- 4E-BP
eIF4E-binding protein; binds eIF4E preventing interaction between eIF4E and eIF4G and thereby preventing cap-dependent translation initiation
- 5’ UTR
5’ Untranslated Region; region within the mRNA upstream of start codon of coding sequence often with complex secondary structures that may contain regulatory elements for canonical translation or an IRES
- CACNA1A
calcium voltage-gated channel subunit alpha1 A; encodes the alpha1 A subunit of a calcium channel and the alpha1ACT transcription factor
- CCAT
CACNA1C calcium channel associated transcription regulator; transcription factor produced by the CACNA1C gene
- CdcL1/CdcL2 (PITSLRE)
PITSLRE serine/threonine-protein kinases
- Cx43
Connexin 43
- Cx55.5
Connexin 55.5
- eIF
eukaryotic initiation factor
- FFAR1
Free fatty acid receptor 1; encodes the G-protein coupled receptors GPR40 and GPR41
- GDF1
Growth Differentiation Factor 1
- Her2
human epidermal growth factor receptor 2
- IRES
Internal Ribosome Entry Site; a specialized region within mRNA that can initiate translation independent of a 5’ cap
- ITAF
IRES trans-acting factor; a biomolecule involved in the regulation of the IRES
- LASS1
Ceramide Synthase 1, also Longevity Assurance Gene 1 Protein Homolog 1
- LAMB1
encodes Laminin B1, a membrane glycoprotein associated with cancer invasion
- meloe
melanoma-overexpressed antigen; encodes three antigens expressed in melanocytes and overexpressed in melanomas
- MTPN
Myotrophin; in addition to encoding myotrophin which functions to NFκB in cardiac tissue, the gene expressed the antigen MPD6
- NOTCH2
Neurogenic locus notch homolog protein
- PABP
Poly(A)-binding protein; involved in the circularization of mRNA through its interaction with the poly(A) tail and eIF4G
- pVHL
Von Hippel-Lindau Tumor Suppressor
- RAN translation
Repeat associated non-ATG translation; specialized, novel method of translation that appears to require codon repeats for initiation
- SCA6
spinocerebellar ataxia type 6; ataxia caused by a polyglutamine repeat expansion in the voltage-gated Ca2+ channel, CACNA1A
- SNRPN
Small Nuclear Ribonucleoprotein Polypeptide N
- SNURF
SNRPN Upstream Reading Frame
- TCP-BP
Tenocyclidine [1-(1-(2-Thienyl)cyclohexyl)piperidine binding protein gene; a protein comprised of two subunits that forms a complex of proteins that bind glutamate agonists
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beadle GW, Tatum EL. Genetic Control of Biochemical Reactions in Neurospora. Proceedings of the National Academy of Sciences of the United States of America. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 3.Peri S, Pandey A. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta. 2009;1789:518–528. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson RJ, et al. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komar AA, Hatzoglou M. Cellular IRES-mediated translation. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King HA, et al. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 8.Baird SD, et al. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spriggs KA, et al. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 10.Mokrejs M, et al. IRESite--a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Research. 2010;38:D131–D136. doi: 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weingarten-Gabbay S, et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science (New York, N.Y.) 2016:351. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 12.Van Eden ME, et al. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranick BT, et al. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl. Acad. Sci. U.S A. 2008;105:4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]
- 15.Carbonnelle D, et al. The melanoma antigens MELOE-1 and MELOE-2 are translated from a bona fide polycistronic mRNA containing functional IRES sequences. PloS One. 2013;8:e75233. doi: 10.1371/journal.pone.0075233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charpentier M, et al. IRES-dependent translation of the long non coding RNA meloe in melanoma cells produces the most immunogenic MELOE antigens. Oncotarget. 2016 doi: 10.18632/oncotarget.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petz M, et al. La enhances IRES-mediated translation of laminin B1 during malignant epithelial to mesenchymal transition. Nucleic Acids Res. 2012;40:290–302. doi: 10.1093/nar/gkr717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spriggs KA, et al. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 19.Karaa ZS, et al. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15:249–254. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui D, et al. A rat brain bicistronic gene with an internal ribosome entry site codes for a phencyclidine-binding protein with cytotoxic activity. The Journal of Biological Chemistry. 2009;284:2245–2257. doi: 10.1074/jbc.M807063200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahar Halpern K, et al. GPR41 gene expression is mediated by internal ribosome entry site (IRES)-dependent translation of bicistronic mRNA encoding GPR40 and GPR41 proteins. The Journal of Biological Chemistry. 2012;287:20154–20163. doi: 10.1074/jbc.M112.358887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burant CF. Activation of GPR40 as a therapeutic target for the treatment of type 2 diabetes. Diabetes Care. 2013;36(Suppl 2):S175–S179. doi: 10.2337/dcS13-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edfalk S, et al. Gpr40 is expressed in enteroendocrine cells and mediates free fatty acid stimulation of incretin secretion. Diabetes. 2008;57:2280–2287. doi: 10.2337/db08-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Covington DK, et al. The G-protein-coupled receptor 40 family (GPR40-GPR43) and its role in nutrient sensing. Biochemical Society Transactions. 2006;34:770–773. doi: 10.1042/BST0340770. [DOI] [PubMed] [Google Scholar]
- 25.Godet Y, et al. MELOE-1 is a new antigen overexpressed in melanomas and involved in adoptive T cell transfer efficiency. The Journal of Experimental Medicine. 2008;205:2673–2682. doi: 10.1084/jem.20081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornelis S, et al. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Molecular Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 27.Du X, et al. A second cistron in the CACNA1A gene encodes a transcription factor that mediates cerebellar development and SCA6. Cell. 2013;154:118–133. doi: 10.1016/j.cell.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du X, et al. Revelations from a bicistronic calcium channel gene. Cell Cycle. 2014;13:875–876. doi: 10.4161/cc.28199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazaki Y, et al. An miRNA-mediated therapy for SCA6 blocks IRES-driven translation of the CACNA1A second cistron. Sci Transl Med. 2016;8:347ra394. doi: 10.1126/scitranslmed.aaf5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong Z, et al. An Unconventional Antigen Translated by a Novel Internal Ribosome Entry Site Elicits Antitumor Humoral Immune Reactions. Journal of immunology (Baltimore, Md. : 1950) 2006;177:4907–4916. doi: 10.4049/jimmunol.177.7.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guruharsha KG, et al. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nature reviews. Genetics. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauring AS, Overbaugh J. Evidence that an IRES within the Notch2 coding region can direct expression of a nuclear form of the protein. Molecular Cell. 2000;6:939–945. doi: 10.1016/s1097-2765(05)00084-5. [DOI] [PubMed] [Google Scholar]
- 33.Turnpenny PD, Ellard S. Alagille syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet. 2012;20:251–257. doi: 10.1038/ejhg.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(Spec No 1):R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 35.Penton AL, et al. Notch signaling in human development and disease. Semin Cell Dev Biol. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Descartes M, et al. Hajdu-Cheney syndrome: phenotypical progression with de-novo NOTCH2 mutation. Clin Dysmorphol. 2014;23:88–94. doi: 10.1097/MCD.0000000000000034. [DOI] [PubMed] [Google Scholar]
- 37.Stathopoulos IP, et al. Severe osteoporosis and mutation in NOTCH2 gene in a woman with Hajdu-Cheney syndrome. Bone. 2013;52:366–371. doi: 10.1016/j.bone.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 38.Anido J, et al. Biosynthesis of tumorigenic HER2 C-terminal fragments by alternative initiation of translation. The EMBO journal. 2006;25:3234–3244. doi: 10.1038/sj.emboj.7601191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kullmann M, et al. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komar AA, Hatzoglou M. Exploring Internal Ribosome Entry Sites as Therapeutic Targets. Front Oncol. 2015;5:233. doi: 10.3389/fonc.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters B, Thompson SR. Cap-Independent Translational Control of Carcinogenesis. Front Oncol. 2016;6:128. doi: 10.3389/fonc.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013;22:45–51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cleary JD, Ranum LP. Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr. Opin. Genet. Dev. 2014;26:6–15. doi: 10.1016/j.gde.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez-Ospina N, et al. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salat-Canela C, et al. Internal translation of the connexin 43 transcript. Cell Commun Signal. 2014;12:31. doi: 10.1186/1478-811X-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ul-Hussain M, et al. Internal ribosomal entry site (IRES) activity generates endogenous carboxyl-terminal domains of Cx43 and is responsive to hypoxic conditions. J Biol Chem. 2014;289:20979–20990. doi: 10.1074/jbc.M113.540187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ul-Hussain M, et al. IRES-mediated translation of the carboxy-terminal domain of the horizontal cell specific connexin Cx55.5 in vivo and in vitro. BMC Mol Biol. 2008;9:52. doi: 10.1186/1471-2199-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iliopoulos O, et al. pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc Natl Acad Sci U S A. 1998;95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minervini G, et al. Isoform-specific interactions of the von Hippel-Lindau tumor suppressor protein. Sci Rep. 2015;5:12605. doi: 10.1038/srep12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray TA, et al. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang B, et al. Cloning and characterization of a LASS1-GDF1 transcript in rat cerebral cortex: conservation of a bicistronic structure. DNA sequence: the journal of DNA sequencing and mapping. 2007;18:92–103. doi: 10.1080/10425170601060947. [DOI] [PubMed] [Google Scholar]