Abstract
Stem cell differentiation can be highly sensitive to mechanical inputs from the extracellular matrix (ECM)1–3. Identifying temporal windows during which lineage commitment responds to ECM stiffness, and the signals that mediate these decisions, would advance both mechanistic insights and translational efforts. To address these questions, we investigate adult neural stem cell (NSC) fate commitment using an oligonucleotide-crosslinked ECM platform that for the first time offers dynamic and reversible control of stiffness. “Stiffness pulse” studies in which the ECM was transiently or permanently softened or stiffened at specified initiation times and durations pinpoint a 24-hour window in which ECM stiffness maximally impacts neurogenic commitment. Overexpression of the transcriptional co-activator YAP within this window suppressed neurogenesis, and silencing YAP enhanced it. Moreover, ablating YAP-β-catenin interaction rescued neurogenesis. This work reveals that ECM stiffness dictates NSC lineage commitment by signaling via a YAP and β-catenin interaction during a defined temporal window.
Introduction
Stem cell self-renewal and differentiation are tightly controlled by the cellular microenvironment or “niche,” which presents a spectrum of soluble and immobilized biochemical signals and biophysical cues4. Among this latter class of inputs, mechanical cues encoded in the extracellular matrix (ECM) are increasingly recognized as important regulators of lineage commitment in many types of stem cells1. For example, we previously showed that ECM stiffness variations in the range of 1 – 10 kPa strongly bias the neuronal vs. astrocytic differentiation of adult neural stem cells (NSCs)3,5. These findings are important both in designing material scaffolds for biomedical applications and for our understanding of NSC biology in vivo, where hippocampal NSCs are exposed to anatomic stiffness gradients as they migrate into the dentate gyrus and undergo neuronal differentiation and maturation4,6.
Despite several recent advances, much remains unknown about the molecular mechanisms that connect mechanical inputs to stem cell differentiation. As a prominent example, cells typically sense ECM stiffness cues over seconds to minutes7, yet it is unclear over what time scales stem cells integrate these cues and execute lineage commitment decisions that only fully declare themselves with changes in marker expression and cellular function days to weeks later. For instance, we recently showed that NSCs adapt their intrinsic mechanical properties to those of the ECM within 12 hours of seeding and that transient pharmacological inhibition of mechanosensing pathways can abrogate downstream marker expression and differentiation that occurs 4–6 days after adhesion5. Similarly, traction forces generated by human mesenchymal stem cells (MSCs) within 12 hours of adhesion can predict lineage specification measured 14 days later87. Consistent with such observations, studies with photoactive materials that can be either irreversibly softened9 or stiffened10 have revealed that the influence of ECM stiffness on MSC differentiation depends strongly on the initial duration of exposure to the stiffness cue. These approaches have introduced the concept that stem cells may possess “mechanical memory,” such that initial exposure to instructive ECM stiffnesses for critical lengths of time shifts differentiation programs even when the ECM is subsequently softened8.
Collectively, these observations raise the possibility that stem cells may be maximally primed within some temporal window to receive, remember, and act upon ECM stiffness inputs, and that a “point of no return” may exist beyond which stem cells no longer respond to these signals. However, a key limitation to definitively identifying the beginning and end points of these windows, and the molecular mechanisms that act therein, is the lack of a single ECM platform that can be reversibly stiffened or softened on cue. The absence of such materials represents a broad and important unmet need in the fields of stem cell engineering and mechanobiology.
Results
As an initial, motivating study, we conducted serial adhesion experiments in which NSCs were first cultured on polyacrylamide (PA) ECMs of a given stiffness under media conditions that give rise to a mixture of neurons and glia (i.e. mixed differentiation – 1 μM retinoic acid and 1% fetal bovine serum11), and then dissociated and re-seeded on an ECM of a different stiffness. If NSCs were re-seeded onto a stiff ECM (72kPa, 10% acrylamide & 0.3% bis) more than 3 days after initially being seeded on a soft ECM (0.7kPa, 4% acrylamide & 0.05% bis), the downstream lineage distribution matched that of cells cultured on soft substrates for the entire time course (Supplementary Figure S1). Conversely, cells cultured on stiff ECMs and then re-seeded onto soft ECMs exhibited lineage distributions characteristic of a soft ECM phenotype (i.e., high neurogenesis) only if the replating occurred earlier than 2 days in culture. These results began to bracket a critical temporal window of mechanosensitivity, where mechanical instruction of lineage commitment occurs no later than 2–3 days after initial adhesion. However, cell detachment induces the risk of disrupting cellular mechanosensing mechanisms that are linked to cytoskeletal organization as well as selecting for particular cellular subpopulations12,13. We therefore sought to create a material system in which ECM stiffness could be dynamically and reversibly modulated without removing cells from the surface.
A mechanically-tunable ECM system capable of delivering temporally-defined stiffness cues to stem cells must meet several critical requirements: minimal swelling and contraction of the gel upon stiffening or softening to avoid cellular deformation, reversible stiffness modulation over a relevant stiffness range that encompasses stem cell mechanosensitivity to analyze the effects of temporal stiffening and softening on lineage specification, and functionalization with ECM adhesion proteins or peptides. Prior materials systems have met some but not all of these requirements. Several systems exhibit extensive swelling or shrinking upon stiffness-switching14–17, and photo-cleavable18,19 or photo-crosslinkable10 gels are an advance, but their irreversible nature limits the scope of biological questions that can be asked.
We therefore engineered a PA ECM system based on prior PA hydrogels in which stiffness can be reversibly manipulated with oligonucleotide-based crosslinks15–17 (Figure 1a). Two distinct, acrydite-functionalized DNA-oligonucleotides (“sidearms”) are co-polymerized into a PA hydrogel, and subsequent addition of a soluble linker DNA strand (L) containing sequences complementary to the sidearms hybridizes with two sidearms and thereby adds additional crosslinks to stiffen the gel. Furthermore, the L strand also contains a “toehold” region that binds to neither of the sidearms, and thus subsequent addition of a “release strand” (R) that is fully complementary to L (including the toehold) can competitively hybridize with L to remove crosslinks and soften the gel (Figure 1a). A further, critical innovation in this system is that DNA crosslinks are formed via incubation of the two acrydite-functionalized DNA oligonucleotides with the linker strand to allow hybridization before their co-polymerization into hydrogel. This enables substantially higher control over DNA crosslink concentrations than prior work and a much greater range of possible stiffness values.15,16
Figure 1. A DNA-Bis crosslinked PA gel enabled dynamic and reversible modulation of stiffness.
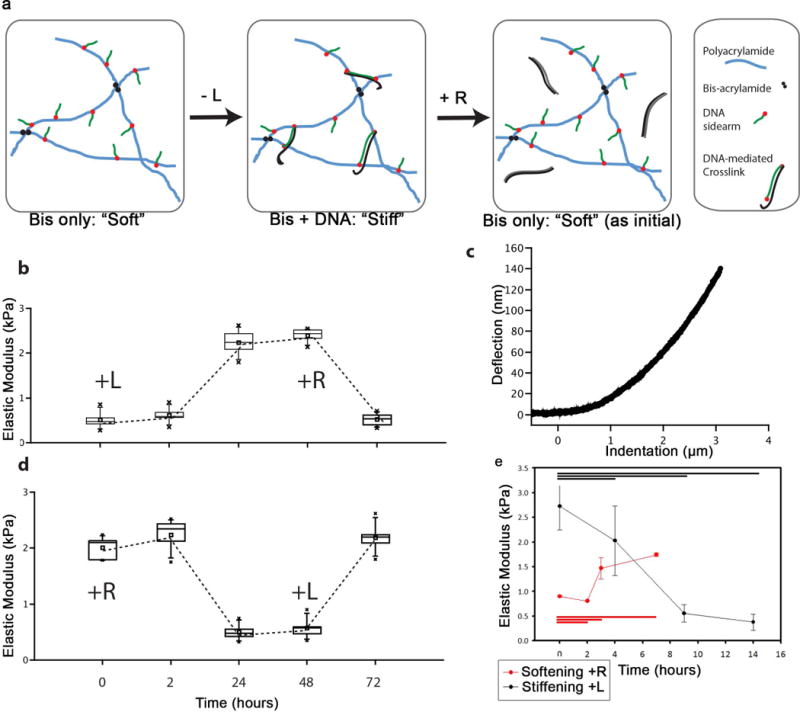
A: Molecular schematic of DNA-Bis-PA gels. PA-copolymerized acrydite-DNA sidearms hybridize to a shared, linker DNA-strand (L) to stiffen the gel. A release strand (R) can subsequently soften the gel by hybridizing to and removing L. B–E: AFM measurements of DNA-Bis-PA gel stiffening (B). After overnight incubation in R, L strand was added (time 0) to DNA-Bis-PA gel. AFM indentation measurements were conducted at various times, with a representative curve illustrated in (C). At 48 hours, R strand was added to induce softening. We observed softening to approximately the same stiffness value as at t=0. Measurement of the analogous, reverse case of softening revealed similar behavior (D). The stiffening of the gel was followed in higher temporal resolution for the first 14 hours after addition of L strand, and a plateau was reached by 9 hours (E). B, D, – box and whisker plots in this and subsequent figures represent a seven-number summary: square – mean, bar – median, box edges – 1 standard deviation, whiskers – 95th and 5th percentile, x – min and max, n = 3. E – 1-way ANOVA, n = 3, error bars represent 1 standard deviation.
The elastic behavior of a DNA-bis crosslinked gel was characterized by atomic force microscopy (AFM) and parallel plate rheology under various L and R strand conditions (Figure 1b, d, Supplementary Table 1). Addition of L to fully uncrosslinked gels led to an onset of stiffening within 2 hours (Figure 1e), and the switch was complete within 9 hours, a timescale compatible with cell differentiation and other processes. Likewise, addition of the release strand R to initially stiff gels led to softening, which is a slightly slower process but still finishes by 9 hours. Finally, reversal of these mechanical switching steps enabled reversion to the original material properties within the same time frame (Figure 1b, d). Another feature that enables investigation of many stiffness regimes was the additional use of “background” covalent bis-acrylamide crosslinks (i.e. DNA-bis-PA), which allows the material’s dynamic range to span many starting stiffness setpoints and prevents material liquefaction upon reversal of DNA crosslinks. For DNA-PA gels crosslinked solely by oligonucleotide hybridization, shear rheometry while heating above the melting temperature of DNA crosslinks (Supplementary Figure S2a, b, c) led to liquefaction above 70°C (Supplementary Figure S2a), indicated by the crossover of the storage modulus and the loss modulus. In contrast, gels with the background DNA-bis-PA crosslinking softened to a minimal level, set by the level of bis-PA links, but did not liquefy upon heating (Supplementary Figure S2b). The material is analogously stable to addition of excess release strand.
DNA-bis-PA gels tuned for reversible switching over the relevant NSC mechanosensitive range3,5 of 0.3–3 kPa (DNA-bis-PA) were synthesized and conjugated with laminin to facilitate cell adhesion. Laminin conjugation for all gels was conducted with reaction of the surface with 50 μg/mL sulfo-SANPAH under UV for 8 minutes, and incubation overnight at 4°C with 20 μg/mL full length laminin, as previously published5. We first validated this material system by reproducing our earlier discovery that soft ECMs bias NSCs towards neurogenic differentiation: ~60–70% β-III tubulin+ on soft compared to ~40% β-III tubulin+ on stiff gels (0 days of gel softening/stiffening controls in Figure 2a, representative images in Figure 2c).3,5 We next conducted “step-stiffness” studies in which NSCs were first cultured on stiff (3 kPa) DNA-bis-PA hydrogels under mixed differentiation conditions, softened between 1–5 days later by addition of the R strand, and analyzed for lineage distributions on day 6 (Figure 2a). The degree of neurogenesis depended strongly on the time at which the ECM was softened, with switching at earlier time points favoring neurogenesis and later time points not permitting it. Astrocytic differentiation within this relatively soft stiffness range was comparatively stiffness-insensitive, consistent with our prior results3,5. Finally, a “point of no return” was observed at approximately 3 days, since materials softened after this time supported similar, lower levels of neurogenesis (<30%) as materials that were maintained at stiff values throughout.
Figure 2. Changing stiffness of differentiating NSCs biased lineage commitment during the first two days after seeding.
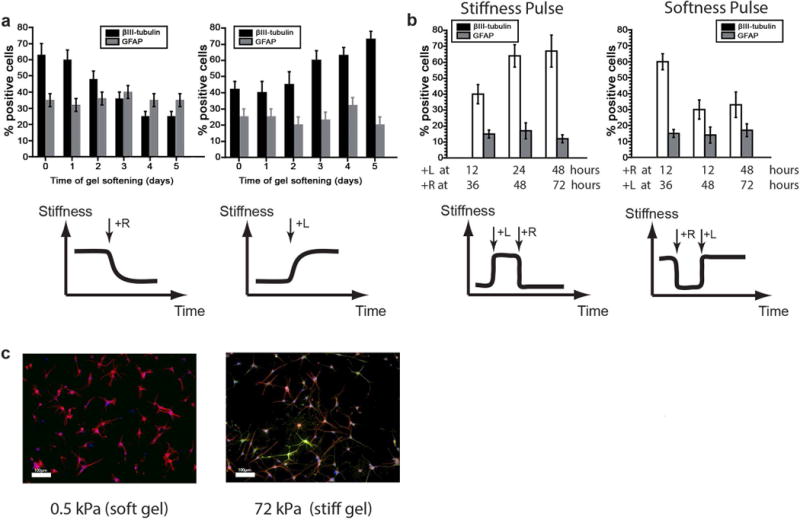
A: NSC differentiation under step-change stiffness conditions. NSCs were seeded on an initially stiff DNA-Bis-PA gel, which was then softened after different intervals of time. Softening early increased the percentage of βIII-tubulin positive cells. However, there was no significant difference in lineage commitment if the substrate was softened later than 3 days after induction of differentiation. Error bars show 1 standard deviation, n=3. B: NSC differentiation under stiffness “pulse” conditions. DNA-Bis-PA gels were seeded with NSCs, stiffened at 12–48 hours, then stiffened at 36–72 hours. Similarly, NSCs were seeded on stiff gels, which were softened and then later stiffened. at specified times from day 1 to day 5 after onset of differentiation. Mechanically instructed lineage commitment was plastic before 48 hours. Error bars show 1 standard deviation, n=3. C: Representative images for neurogenesis on 300 Pa (soft) and 3 kPa (stiff) gels (DAPI: Blue, βIII-tubulin: Red, GFAP: Green), scale bar: 100µm, 10× magnification.
In such experiments involving a single, unidirectional change in stiffness, it is unclear whether the mechanical instruction of cell fate depends on the total time a differentiating cell is exposed to a given stiffness, the time point at which the stiffness is changed, or both. To distinguish among these possibilities, we took advantage of the reversibility of this material and performed stiffness and softness “pulse” experiments in which we were able to either transiently soften stiff gels by sequential addition of R and later L, or stiffen soft gels by addition of L and later R, at a range of times from 1 to 5 days after onset of differentiation (Figure 2b). We found more β-III tubulin+ cells on the substrates that were soft within a 12–36 hour window vs. later time windows, and less neuronal differentiation occurred when cells were exposed to stiff ECMs within this window. These results thus revealed that cells are most mechanosensitive and plastic during their first 12 to 36 hours of differentiation and become insensitive to stiffness changes (in either direction) once this mechanosensitive time window closes, narrowing the window significantly from our previous estimate of ~3 days.
The existence of a critical time window for mechanosensitive NSC differentiation raises the intriguing question of what molecular mechanisms are activated during this window and how they coordinate with signals traditionally understood to control NSC neurogenesis. The transcriptional co-activator Yes-associated protein (YAP) has previously been implicated as a regulator of a range of mechanosensitive behaviors, including mesenchymal stem cell differentiation,13 where it functions as a molecular rheostat8. We first investigated whether total intracellular YAP levels depended on substrate stiffness and found levels substantially higher on stiff (75 kPa) vs. soft (700 Pa) gels, in particular during the first 48 hours of differentiation (Figure 3a, b).
Figure 3. YAP dynamically varied with stiffness, and was necessary and sufficient for directing stiffness-mediated neurogenesis.
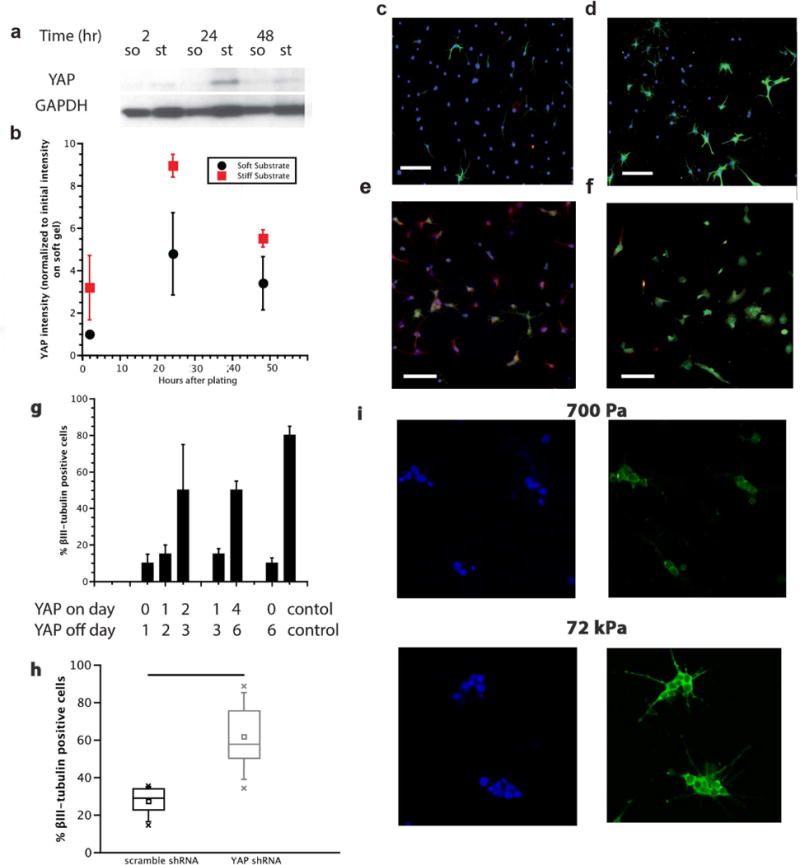
A, B: Western blot of total YAP following onset of differentiation. Total YAP levels showed a significant difference (B, normalized to initial YAP intensity on soft gel) on 700 Pa (soft) versus 75 kPa (stiff) gels. On stiff gels, relative levels of YAP were higher between 24 to 48 hours, a period that corresponded with the mechanosensitive window described in Figure 2, error bars represent 1 standard deviation, n=3. Data is normalized to soft YAP intensity at 2 hours after plating. C,D: shRNA knockdown of YAP. After 6 days of differentiation on laminin-coated tissue culture plastic, shRNA YAP knockdown (D) showed a higher ratio of βIII-tubulin positive cells than the control (C), quantified in (H) as a seven-number summary with n=3, p<0.01 1-way ANOVA. DAPI: blue, βIII-tubulin: green. Scale bar: 100µm, 10× magnification. E,F: YAP-GFP overexpressing NSCs on 700 Pa (E, soft) and 75 kPa (F, stiff) gels. DAPI: blue, βIII-tubulin: red, YAP-GFP: green. Scale bar: 100µm, 10× magnification. G: Quantification of βIII-tubulin positive cells after pulsed YAP overexpression. Tetracycline was added to and withdrawn from soft gel cultures at the indicated times. Cells that were exposed to increased YAP-GFP levels early in the differentiation process (day 0 and 1, day 1 and 2) were much less likely to commit to the neuronal lineage than cells that are exposed to YAP-GFP at later stages (day 2 and 3, or day 4, 5, 6), n=3, error bars represent 1 standard deviation, p<0.01 1-way ANOVA. I: NSCs on both soft (top) and stiff (bottom) substrates showed no difference in the nuclear vs cytoplasmic localization of YAP. DAPI: blue, left. YAP: green, right, mean ratio ~.3 N/C, image magnification 60×.
Given that matrix stiffness strongly regulates YAP expression, we next asked whether YAP may play a causal role in stiffness-mediated NSC lineage commimtent. To do this, we manipulated YAP levels by generating and expressing a YAP-GFP fusion construct (Supplementary Figure S3). First, under the same soft gel (0.7 kPa) differentiation conditions that had earlier supported robust neurogenesis in control NSCs (Figure 2c), constitutive overexpression of YAP-GFP reduced neurogenesis (Figure 3e, g). In contrast, when we suppressed YAP by engineering a lentiviral vector carrying a previously validated anti-YAP shRNA20, neurogenesis was strongly promoted, even on very stiff (72 kPa) substrates (Figure 3c, d, f, g, h, Supplementary Figure S3). Thus, YAP is both necessary and sufficient to suppress neurogenesis, and manipulation of YAP expression can override stiffness regulation of lineage commitment.
Intriguingly, the mechanosensitive time window of 12–36 hours identified using ECM stiffness pulses (Figure 2b) correlates closely with the observed YAP expression dynamics on stiff surfaces (Figure 3a, b). To determine whether dynamic modulation of ectopic YAP expression (to emulate the YAP profile observed on stiff surfaces) could phenocopy the impact of stiffness on differentiation, we used a tetracycline-regulated gene expression system21 that we introduced into a retroviral vector to mirror our stiffening/softening experiments by introducing pulses of YAP expression at critical time points throughout the lineage commitment process. We found that YAP-GFP overexpression exerted a particularly strong impact on lineage commitment early in differentiation (Figure 3e, g). Specifically, YAP-GFP expression during day 1, day 2, or both days reduced neurogenesis to similarly low levels as if YAP-GFP were expressed during the whole 6-day differentiation window. These experiments implicate the same 12–36 hour window identified by both endogenous YAP expression in cells on a stiff substrate (Figure 3b) and stiffness pulses introduced using the reversibly switchable gels (Figure 2b).
With the knowledge that YAP is necessary and sufficient for mechanosensitive suppression of neurogenesis (Figure 3), we next asked what mechanism drives this behavior. An obvious possibility based on previous studies with mesenchymal stem cells13 would be that stiffness drives the nuclear translocation of YAP, which in turn may contribute to lineage commitment through YAP/TEAD co-regulation of gene expression. Intriguingly, however, when we examined YAP localization via immunofluorescence, we did not observe differences in nucleo-cytoplasmic distribution as a function of matrix stiffness (Figure 3i). Since YAP is regulated not only by expression levels and nuclear shuttling13 but also by cytosolic interactions22, we hypothesized that YAP could conceivably be functioning by interfacing with the transcriptional activity of other important signaling pathways. We thus investigated whether YAP could interact with effectors of neurogenesis. β-catenin is a critical component of the Wnt signaling pathway that plays critical roles in organismal development, stem cell differentiation, and cancer23–26, and we recently found that β-catenin is activated downstream of ephrin signaling to induce NSC differentiation25. Upon upstream signal activation, this protein translocates to the nucleus, where it acts as a transcriptional co-activator of target genes, including the proneuronal transcription factor NeuroD1 in NSCs27. Interestingly, β-catenin has also been reported to bind to YAP within the Hippo signaling pathway, either directly or through co-association with the β-catenin destruction complex22,28. Thus, we hypothesized that YAP may sequester and thereby reduce β-catenin activity.
To investigate this possibility, we first asked if YAP expression had an effect on β-catenin transcriptional activity. Expression of a 6xTCF luciferase reporter29 showed that overexpression of YAP yielded a sharp reduction in β-catenin transcriptional activity (Figure 4a.) Addition of the GSK-3β inhibitor CHIR99021, which inhibits degradation of β-catenin and thereby potentiates its downstream signaling, significantly enhanced neuronal differentiation, and even rescued this differentiation in YAP-GFP overexpressing NSCs (Figure 4b, c).
Figure 4. YAP interacted with β-catenin to bias stiffness-mediated NSC lineage commitment.
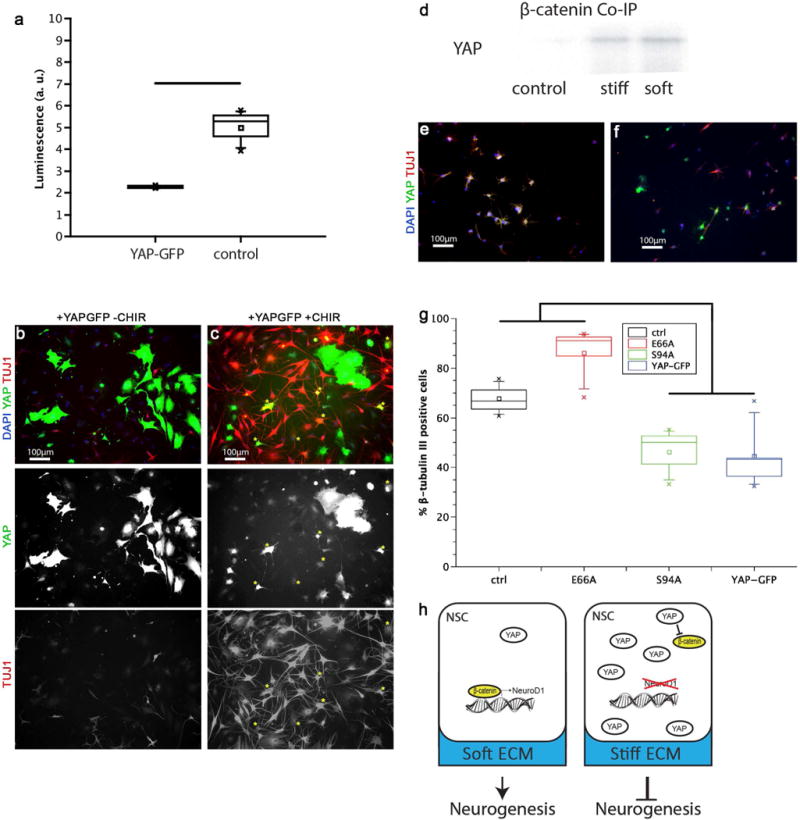
A: YAP-GFP decreased β-catenin transcriptional activity. The activity of a β-catenin responsive promoter-reporter was significantly reduced in YAP-GFP cells compared to control cells (n = 3, p<0.01 by t-test). B, C: In control cells (B), YAP-GFP cells did not stain positive for βIII-tubulin, but GSK3β inhibition restored neural differentiation in some YAP-GFP cells (C). Yellow stars indicate cells that stain positive for both YAP-GFP and βIII-tubulin. D: YAP and β-catenin interact in differentiating NSCs. Co-immunoprecipitation with a β-catenin antibody and probing with a YAP antibody showed that on both 300 Pa (soft) and 72 kPa (stiff) samples, YAP co-precipitated with β-catenin (control: no β-catenin antibody on stiff surface). E,F: Binding mutants of YAP-GFP showed differentiation rescue of neurogenesis on soft (200 Pa) substrates. A YAP mutant unable to bind β-catenin (E66A) (E) showed the same level of neuronal differentiation as naïve, non-YAP expressing cells, compared to suppressed neurogenesis by YAP-GFP (compare with B). Numerous cells stained positive for both βIII-tubulin and E66A YAP-GFP; in contrast, in the control YAP-GFP case there were no cells that stained positive for both βIII-tubulin and GFP. The S94A YAP mutant that lacked TEAD binding (F) suppressed neurogenesis to the same extent as wild type YAP-GFP (compare B). G: Quantification of levels of neurogenesis in the naïve, E66A, S94A, and YAP-GFP cases. Naïve and E66A were significantly different from S94A and YAP-GFP cases on soft substrates (200 Pa) (n=3 gels, p < 0.005 by 1-way ANOVA). H: Schematic of the proposed effect of substrate stiffness on NSC lineage commitment. On soft substrates, β-catenin drives transcription of neurogenesis effectors, such as NeuroD1. On stiff substrates, YAP levels are sufficiently high to sequester and inhibit available β-catenin, thereby preventing β-catenin dependent transcription.
The above results are consistent with a model in which YAP antagonizes β-catenin to suppress neural lineage commitment. To investigate whether these two molecules biochemically associate within NSCs, either by direct binding22 or mutual association for example to Axin and the β-catenin destruction complex22, we performed co-immunoprecipitation of β-catenin and YAP during the lineage commitment process (Figure 4d). Pulldown of β-catenin and probing for YAP showed association between these two molecules on both very soft (200 Pa) and stiff (72 kPa) substrates. To investigate the functional consequences of YAP association with β-catenin, we performed loss-of-function studies in which we overexpressed a YAP mutant20,22 that lacks the ability to associate with β-catenin (YAP E66A). In parallel, a YAP mutant unable to bind TEAD proteins (YAP-S94A)22 was investigated to assess the direct transcriptional role of YAP. Interestingly, the YAP-E66A mutant, whose interaction with β-catenin is ablated, exhibited similar, high neurogenesis on soft substrates as control cells not expressing YAP (Figure 4e, g). In contrast, YAP-S94A overexpressing cells exhibited similar, low levels of neuronal differentiation as NSCs overexpressing wild type YAP (Figure 4f, g). This result indicates that YAP’s β-catenin binding activity plays a much stronger functional role in NSC mechanoregulation than YAP’s transcriptional co-regulatory activity. This is fundamentally distinct both from previous observations in MSCs and the nuclear and TEAD-mediated mechanism of action classically portrayed in the Hippo pathway. Therefore, we propose a model wherein matrix stiffness controls neurogenesis by regulating a balance between pro-neurogenic β-catenin activity and anti-neurogenic YAP activity, an antagonism that depends primarily on a direct binding interaction between β-catenin and YAP (Figure 4h).
Discussion
In this study, we have identified a 12–36 hour mechanosensitive time window during which NSC lineage commitment is maximally receptive to ECM stiffness inputs. In particular, by leveraging a material platform in which stiffness can be dynamically modulated, we found that exposure to a transient stiffness pulse from 12–36 hours was sufficient to suppress neurogenesis on soft matrices, and conversely a “softness pulse” during this same time window rescued neurogenesis on stiff matrices. Furthermore, we identified signaling events that mediate the impact of these stiffness cues on stem cell differentiation. Endogenous expression of the transcriptional co-activator YAP peaked during the 12–36 hour time window, and ectopic, pulsed overexpression of YAP during this window was sufficient to override soft matrix cues and thereby block neuronal differentiation. However, counter to the proposed model of YAP regulation in stiffness-dependent differentiation of mesenchymal stem cells, stiffness did not lead to nuclear vs cytoplasmic partitioning of YAP. Furthermore, YAP co-precipitated with the pro-neurogenic effector β-catenin, and disruption of this interaction reversed YAP-mediated suppression of neurogenesis. Analogously, pharmacological activation of β-catenin restored NSC neurogenic lineage commitment even with YAP overexpression. Our results therefore support a model in which matrix stiffness acts during a critical time window to regulate a balance between Wnt/β-catenin vs. YAP signaling, whereby YAP antagonizes the neurogenic effects of β-catenin through a binding interaction. This model is compatible with direct binding and sequestration of β-catenin20, or suppression of β-catenin through association of both of these proteins with Axin within the β-catenin destruction complex28,30. Further resolution of the precise mechanism of this interaction, and of the temporal dynamics of the balance between YAP and β-catenin during the critical time window, represent intriguing future possibilities for investigation.
A critical innovation and enabling technology in this study was the utilization of bis-acrylamide/oligonucleotide PA hydrogels in which stiffness could be dynamically and reversibly modulated. The underlying covalent crosslinks provide a basal structural stability (also reducing shrinking/swelling), on top of which the fully reversible DNA hybridization crosslinks can dynamically and reversibly modulate stiffness at a timescale that enables investigation of stem cell lineage commitment (Figure 1e). Hydrogel materials in which stiffness can either be irreversibly increased10 or decreased9 using photoirradiation have been previously employed to investigate stem cell lineage commitment; however, a two-way mechanical switch in which stiffness may repeatedly increased and/or decreased was a key advance that enabled us to define a critical window during which lineage commitment is maximally sensitive to mechanical inputs. Our work also suggests that the specific temporal window of exposure to a defined stiffness cue beginning and ending at specific times, rather than the duration of exposure, governs lineage decisions. Stiffness step changes that occurred before the window opened (<24 hours after seeding) did not bias differentiation strongly, but pulsed stiffnesses of the same duration (24hr) inside of the window strongly influenced fate distributions. Isolation of this defined temporal window enabled us to connect these changes with time-dependent signaling events and thus offer a mechanistic model for mechanosensitive differentiation. Importantly, these materials are uniquely well suited to interrogate a low-stiffness regime (0.1 – 10 kPa), which corresponds well to the elasticity of hippocampal tissue31 and may be of value for investigating lineage commitment in other soft tissues. With additional tuning to reduce DNA exclusion and to promote DNA (e.g. shortened oligos or smaller gels) diffusion into the gel, stiffer regimes could even further broaden the range of stem cell and mechanobiology investigations enabled by this system.
Materials & Methods
Cell Culture
Adult rat neural stem cells were cultured as described previously3,11. Briefly, cells were cultured in DMEM-F12 (Invitrogen) + N2 Supplement (Life Technologies) and 20ng/mL FGF-2 on laminin-coated polystyrene plates. For differentiation experiments, cells were cultured in mixed differentiation media (DMEM-F12 + N2, 1 μg/ml retinoic acid, 1% fetal bovine serum) if not otherwise noted.
Polyacrylamide Hydrogels
Non-switchable PA hydrogels were produced as described previously5. Briefly, acrylamide and bisacrylamide precursor solutions were degassed by nitrogen bubbling, and polymerized onto PlusOne Bind-Silane treated glass coverslips with 0.1% ammonium persulfate and tetramethylethylenediamine. DNA gels were produced using sidearm and linker sequences as previously described17,31,32. The polymerization method was slightly optimized compared to the previously published method: Both sidearm sequences were added into the polymerization mix, and the solution was allowed to polymerize under degassing sonication for 15 minutes, before being mixed with linker DNA strand and pipetted onto Bind-Silane activated glass coverslips.
DNA/bis-crosslinked hydrogels were produced by adding additional acrylamide, bis-acrylamide, and APS/TEMED to the freshly combined DNA-acrydite-acrylamide polymer (final composition: 3.13% acrylamide & .008% bis reacted with 1mM hybridized SA1+SA2+L). For all gels, laminin functionalization was achieved by incubating gels in 50μg/ml sulfo-SANPAH and irradiating with a UV flood lamp for 8 minutes. The gels were then washed once in PBS, and incubated with 20μg/ml full length mouse laminin (EHS sarcoma derived) for 24 hours at 4°C5.
Rheological Characterization
Rheological characterization of all hydrogels was performed using an Anton Paar Physica MCR 301 rheometer with an 8 mm parallel plate geometry. The solvent trap around the sample location was filled with water to prevent sample dehydration. The temperature of the sample was controlled by a Peltier element (Anton Paar).
Atomic Force Microscopy
Atomic force measurements were performed on a Veeco (Bruker) Catalyst Bioscope instrument. All measurements of gel properties were done at room temperature in cell culture medium (DMEM-F12). Briefly, the deflection sensitivity of each MLCT-BIO cantilever used was measured against a glass cover slide, and the spring constant was determined by thermal tune. Force indentation curves were fitted against a Sneddon indentation model (cone indenting infinite half-space) to obtain elastic moduli33.
Immunoblotting
Electrophoresis was performed using NuPAGE 4–12% Bis-Tris Gels, and transfer to PVDF membranes for Western Blotting was done in a tank blotting cell for 3 hours on ice. Primary antibody dilutions were 1:1000. Horseradish-peroxidase conjugated secondary antibodies (Sigma-Aldrich) were used at 1:100000 dilutions. Quantification was performed by imaging SuperSignal West Dura ECL substrate (Pierce) in a Kodak Bio-Rad imager. Co-immunoprecipitation assays were prepared using a commercial kit (Pierce Classic Immunoprecipitation Kit) according to manufacturer’s instructions. Briefly, cell lysates were pre-cleared against an agarose control resin, then incubated with 1% anti-β-catenin antibody (BD Biosciences 610154) overnight. Antibody complexes were captured using a protein A/G resin and eluted in NuPage LDS sample buffer (Life Technologies).
Immmunofluorescence
Cells were fixed in 4% paraformaldehyde in PBS for 10 minutes. After washing thoroughly with PBS, cells were permeabilized and blocked with 0.3 % Triton-X and 5% goat serum (room temperature, 1h). Hereafter, samples were incubated with primary antibodies for 24 hours at 4 °C. After several washes, samples were incubated with secondary antibodies for 2 hours at room temperature. After two final washes, DAPI was added as a nuclear marker. Primary antibodies used were βIII-tubulin (Covance MMS-435P), GFAP (Abcam 7260), YAP (Cell Signaling 4912), β-catenin (BD 610154). Secondary antibodies from Life Technologies were obtained for the appropriate species conjugated with Alexa dyes.
Viral Vectors
The rat YAP cDNA sequence was synthesized by IDT and inserted into a pCLPIT retroviral vector5. β-catenin reporter constructs (7xTGP) were used as described previously26. shRNA against YAP constructs (pLKO.1 vector system) were also obtained from Addgene. In all three cases, cell lines were established by selection with 1 ug/ml puromycin for 48 hrs and 0.6 ug/ml puromycin thereafter.
YAP E66A and YAP S94A were generated via Quickchange PCR. YAP knockdown by shRNA was confirmed by Western blot (Figure S3), and an 85% reduction of expression levels could be achieved in the high multiplicity of infection case.
Luciferase Assay
Cells were transfected with a 6xTCF luciferase reporter construct28. 105 cells per condition were lysed in 20 μl of lysis buffer (Promega), mixed with 100 μl of Luciferase Assay substrate (Promega), and immediately measured in a luminometer (Promega TD 20/20).
Supplementary Material
Figure S1. Replating studies.
Replating cells from soft to stiff and vice versa shows that they were susceptible to the stiffness of the environment predominantly in early timepoints. On the horizontal axis, the day of replating is shown, and on the vertical axis the percentage of marker-positive cells is shown. The earlier the cells are replated (day 1–2), the closer their population of neurons matches the day 6 control (always at final stiffness). Error bars represent 1 standard deviation, n=3.
Figure S2. Temperature-dependent rheology of DNA-crosslinked PA gels.
To characterize elastic behavior of a DNA-only crosslinked gel (DNA-PA), shear rheometry was performed over a temperature range spanning the predicted denaturation (melting) temperature of these DNA crosslinks (Figure S1 A,B,C). DNA-PA gels crosslinked purely by DNA oligomers turned completely fluid at temperatures over 70 °C (Figure S1A), indicated by the crossover of the storage modulus and the loss modulus. In contrast, DNA-Bis-PA gels significantly softened but did not liquefy upon heating (Figure S1B). Polyacrylamide-Bis gels (Figure S1C) showed no temperature-dependent change in stiffness.
Figure S3. Western blot and quantification of shRNA knockdown of YAP. The high MOI shYAP was used for all experiments. The lanes of the gel correspond to the graph.
Acknowledgments
The authors gratefully acknowledge the California Institute for Quantitative Biosciences and the California Institute for Regenerative Medicine for their support through the CIRM/QB3 Shared Stem Cell Facility. This work was supported by a grant from the NIH (1R01NS074831 to D.V.S. and S.K.) as well as fellowships from the Human Frontier Science Program (S.R.) and the NSF (K.G.)
Footnotes
Author Contributions
Sebastian Rammensee: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Michael Kang: Collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Katerina Georgiou: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript.
Sanjay Kumar: Conception and design, Financial support, administrative support, data analysis and interpretation, manuscript writing, final approval of manuscript.
David Schaffer: Conception and design, Financial support, administrative support, data analysis and interpretation, manuscript writing, final approval of manuscript.
References
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–81. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saha K, et al. Substrate Modulus Directs Neural Stem Cell Behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim DA, Huang YC, Alvarez-Buylla A. The adult neural stem cell niche: lessons for future neural cell replacement strategies. Neurosurg Clin N Am. 2007;18:81–92, ix. doi: 10.1016/j.nec.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Keung AJ, de Juan-Pardo EM, Schaffer DV, Kumar S. Rho GTPases mediate the mechanosensitive lineage commitment of neural stem cells. Stem Cells. 2011;29:1886–97. doi: 10.1002/stem.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elkin BS, Azeloglu EU, Costa KD, Morrison B. Mechanical heterogeneity of the rat hippocampus measured by atomic force microscope indentation. J Neurotrauma. 2007;24:812–22. doi: 10.1089/neu.2006.0169. [DOI] [PubMed] [Google Scholar]
- 7.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy Proteins on Stretchy Substrates: The Important Elements of Integrin-Mediated Rigidity Sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–6. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014 doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 11.Peltier J, Agrawal S, Robertson MJ, Schaffer DV. In vitro culture and analysis of adult hippocampal neural progenitors. Methods Mol Biol. 2010;621:65–87. doi: 10.1007/978-1-60761-063-2_5. [DOI] [PubMed] [Google Scholar]
- 12.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–59. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 13.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 14.Previtera ML, et al. Fibroblast morphology on dynamic softening of hydrogels. Ann Biomed Eng. 2012;40:1061–72. doi: 10.1007/s10439-011-0483-2. [DOI] [PubMed] [Google Scholar]
- 15.Lin DC, Yurke B, Langrana NA. Inducing reversible stiffness changes in DNA-crosslinked gels. J Mater Res. 2005;20:1456–1464. [Google Scholar]
- 16.Jiang FX, Yurke B, Firestein BL, Langrana NA. Neurite outgrowth on a DNA crosslinked hydrogel with tunable stiffnesses. Ann Biomed Eng. 2008;36:1565–1579. doi: 10.1007/s10439-008-9530-z. [DOI] [PubMed] [Google Scholar]
- 17.Lin D, Yurke B, Langrana NA. Mechanical Properties of a Reversible, DNA-Crosslinked Polyacrylamide Hydrogel. J Biomech Eng. 2004;126:104. doi: 10.1115/1.1645529. [DOI] [PubMed] [Google Scholar]
- 18.Kloxin AM, Tibbitt MW, Anseth KS. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nat Protoc. 2010;5:1867–87. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science (80-) 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKay JL, Keung AJ, Kumar S. A Genetic Strategy for the Dynamic and Graded Control of Cell Mechanics, Motility, and Matrix Remodeling. Biophys J. 2012;102:434–442. doi: 10.1016/j.bpj.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 2012;31:1109–22. doi: 10.1038/emboj.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 24.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–79. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 25.Ashton RS, et al. Astrocytes regulate adult hippocampal neurogenesis through ephrin-B signaling. Nat Neurosci. 2012;15:1399–406. doi: 10.1038/nn.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–52. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 27.Kuwabara T, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12:1097–105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai J, Maitra A, Anders Ra, Taketo MM, Pan D. β-Catenin destruction complex-independent regulation of Hippo – YAP signaling by APC in intestinal tumorigenesis. Genes Dev. 2015:1–14. doi: 10.1101/gad.264515.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azzolin L, et al. YAP/TAZ Incorporation in the β-Catenin Destruction Complex Orchestrates the Wnt Response. Cell. 2014;158:157–70. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Chippada U, Yurke B, Langrana NA. Simultaneous determination of Young’s modulus, shear modulus, and Poisson’s ratio of soft hydrogels. J Mater Res. 2010;25:545–555. [Google Scholar]
- 32.Liedl T, Dietz H, Yurke B, Simmel F. Controlled trapping and release of quantum dots in a DNA-switchable hydrogel. Small. 2007;3:1688–93. doi: 10.1002/smll.200700366. [DOI] [PubMed] [Google Scholar]
- 33.Butt HJ, Cappella B, Kappl M. Force measurements with the atomic force microscope: Technique, interpretation and applications. Surf Sci Rep. 2005;59:1–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Replating studies.
Replating cells from soft to stiff and vice versa shows that they were susceptible to the stiffness of the environment predominantly in early timepoints. On the horizontal axis, the day of replating is shown, and on the vertical axis the percentage of marker-positive cells is shown. The earlier the cells are replated (day 1–2), the closer their population of neurons matches the day 6 control (always at final stiffness). Error bars represent 1 standard deviation, n=3.
Figure S2. Temperature-dependent rheology of DNA-crosslinked PA gels.
To characterize elastic behavior of a DNA-only crosslinked gel (DNA-PA), shear rheometry was performed over a temperature range spanning the predicted denaturation (melting) temperature of these DNA crosslinks (Figure S1 A,B,C). DNA-PA gels crosslinked purely by DNA oligomers turned completely fluid at temperatures over 70 °C (Figure S1A), indicated by the crossover of the storage modulus and the loss modulus. In contrast, DNA-Bis-PA gels significantly softened but did not liquefy upon heating (Figure S1B). Polyacrylamide-Bis gels (Figure S1C) showed no temperature-dependent change in stiffness.
Figure S3. Western blot and quantification of shRNA knockdown of YAP. The high MOI shYAP was used for all experiments. The lanes of the gel correspond to the graph.


