Abstract
Diffuse large B-cell lymphoma (DLBCL) is an aggressive cancer that affects ~22,000 people in the United States yearly. Understanding the complex cellular interactions of the tumor microenvironment is critical to the success and development of DLBCL treatment strategies. In vitro platforms that successfully model the complex tumor microenvironment without introducing the variability of in vivo systems are vital for understanding these interactions. To date, no such in vitro model exists that can accurately recapitulate the interactions that occur between immune cells, cancer cells, and endothelial cells in the tumor microenvironment of DLBCL. To that end, we developed a lymphoma-on-chip model consisting of a hydrogel based tumor model traversed by a vascularized, perfusable, round microchannel that successfully recapitulates key complexities and interactions of the in vivo tumor microenvironment in vitro. We have shown that the perfusion capabilities of this technique allow us to study targeted treatment strategies, as well as to model the diffusion of infused reagents spatiotemporally. Furthermore, this model employs a novel fabrication technique that utilizes common laboratory materials, and allows for the microfabrication of multiplex microvascular environments without the need for advanced microfabrication facilities. Through our facile microfabrication process, we are able to achieve micro vessels within a tumor model that are highly reliable and precise over the length of the vessel. Overall, we have developed a tool that enables researchers from many diverse disciplines to study previously inaccessible aspects of the DLBCL tumor microenvironment, with profound implications for drug delivery and design.
Introduction
Non-Hodgkin lymphoma (NHL) is a group of lymphocyte-derived cancers, of which the majority are B-cell in origin. Diffuse large B-cell lymphoma (DLBCL) is one of the most common B-cell lymphomas, accounting for approximately 30% of all NHL cases1,2, or approximately 22,000 people per year. DLBCL is an aggressive cancer that has been cured with a combination of chemotherapy and immunotherapy3,4, but remains fatal in ~40% of patients due to therapy resistance.5 Immunotherapy with anti-CD20 monoclonal antibody in combination with chemotherapy, has significantly improved the overall survival in patients with DLBCL.6 However, this disease results in death for a significant proportion of patients and durable remission rates for advanced stages of DLBCL remain at best50–60%. In patients refractory to conventional therapies, long-term remissions have been observed with various immunotherapy approaches, such as allogenic stem cell transplantation7–10 or CAR T cell therapies,11–13 suggesting that better understanding of the tumor-immune microenvironment is necessary, and that the targeting of critical immune components could be an effective therapeutic strategy for these patients. Therefore, novel methods, that are both economical and broadly applicable, are needed to study the role of immune cells in DLBCL.
In vivo cancer models are hindered by the inability to isolate specific biological systems or cellular interactions within the body.14,15 It is therefore necessary to develop in vitro models that recapitulate in vivo systems while mitigating the biological variability (e.g. 3 dimensionality, tumor-endothelial interactions, etc.) inherent to living systems. While numerous in vitro “tumor-on-a-chip” devices have been reported for studying cancer and for detecting chemotherapy sensitivity,16–18 controlled on-chip systems have not been created to study immune system-tumor interactions, especially the complex cross-talk between endothelial cells, various immune cells of the tumor niche, and neoplastic cells, nor have they allowed for the quantitative analysis of immunotherapeutic agents. By minimizing such critical immune-vascular interactions, previously reported tumor-on-chip models are not able to effectively recapitulate the biological system they aim to represent.19 Typical tumor-on-chip systems developed to date aim to study the complex interactions between tumor cells and the surrounding endothelium by placing a semipermeable membrane or hydrogel between the two cellular components20,21, allowing cellular signals and infused reagents to diffuse through the barrier before reaching the artificial tumor niche. However, this barrier does not exist in vivo: endothelial cell lined blood vessels directly traverse tumors, supplying them with nutrients and facilitating endothelial cell – tumor cell interactions. Further, while several groups have reported systems that allow for co-culture of an endothelialized lumen with other cell types, these systems have yet to be adapted for the study of DLBCL.22,23 An in vitro system that features a blood vessel flowing through a tumor would be a powerful tool for understanding the complex crosstalk between endothelial cells and various immune cells of the DLBCL tumor niche, especially as it applies to cancer metastasis.24,25 To that end, we have developed a novel, hydrogel based device with integrated, perfusable, endothelialized microvasculature, and modeled tumor-immune crosstalk in DLBCL (Fig. 1). This microvasculature possesses a rounded lumen, which has been proven to be more physiologically relevant than traditional in vitro models.26 This technology places the various cells of the DLBCL tumor niche directly in contact with the endothelium of a perfusable microvessel, thereby more accurately recapitulating the DLBCL tumor microenvironment.
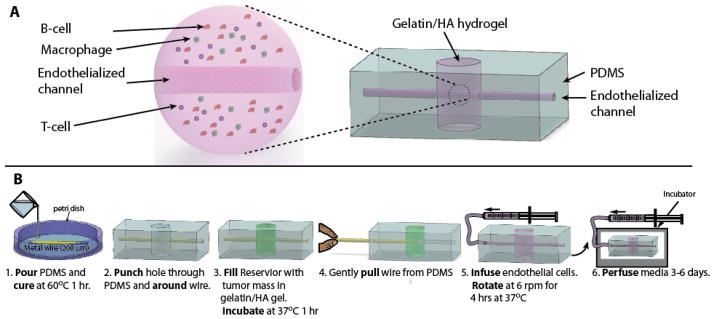
Fig. 1. The DLBCL-on-a-chip microvasculature model is fabricated using common laboratory items.
A) An overall schematic highlighting the embedded DLBCL hydrogel (Left) within the PDMS macrostructure (Right) B) Schematic of the fabrication process. PDMS was molded around a stainless steel wire, and a hole was punched out. This hole was filled in with the DLBCL hydrogel and allowed to polymerize. The hydrogel was then sealed with a plasma bonded glass coverslip. The stainless steel wire was removed and the resulting microchannel was cultured with endothelial cells.
Furthermore, this lymphoma-on-chip model was developed using a unique fabrication technique27 that obviates the need for photolithography-based microfluidic technologies typical of tumor-on-chip models, which require advanced engineering training and access to state-of-the-art microfabrication facilities.28 This novel fabrication method facilitates economical and widely accessible use of the model. Our lymphoma-on-chip system comprises multiple key technical innovations that allow for the investigation of spatiotemporal aspects of DLBCL pathophysiology and drug delivery response and that represent advantages over previously published work: namely, the inclusion of a fully perfusable endothelialized microvessel within a hydrogel-based model of lymphoma, as well as the ability to create multiple, distinct, microenvironments connected to one another in the same multiplex system. This represents a highly under-studied area in tumor research and especially in NHL. Additionally, the ability to create and continuously monitor spatially distinct microniches of different cellular populations (neoplastic B cells, stromal cells, T cells, endothelial cells) that are in close proximity to one another will provide a new level of understanding as to how these cells interact with each other and with the vascular space over time. Furthermore, the development of multiplex microvascular environments with spatial distinct microniches will facilitate the study of global as well as local interactions within the tumor microenvironment.
Experimental
Fabrication of DLBCL-on-chip model and multiplex system
A ~10 cm long strand of 200 μm diameter stainless steel wire was placed atop a ~1 mm thick layer of cured polydimethylsiloxane (PDMS) (Dow Corning, Midland, MI) in a 150 mm petri dish and covered with an uncured layer of PDMS of ~1 mm thickness (Fig. 1B). This first cured layer of PDMS sets the imaging depth of the device. The second layer of PDMS is then cured. The PDMS macrostructure encasing the stainless steel wire was extracted from the petri dish and cut into rectangular prism shapes at a distance of 1 cm from each end of the wire. Cuts were made to the depth of the stainless steel wire on each side of the macrostructure, allowing the wire to protrude 1 cm from each end of the PDMS. A hydrogel reservoir was created by punching out a hole in the PDMS around the stainless steel wire using a 4 mm diameter biopsy punch. The wire was left undisturbed by punching on both sides of the wire and pulling out the plug. Any deformations that the hole punching process imparted on the stainless steel wire were removed by pulling the bent section of the stainless steel wire, via the protruding end section, approximately 5 mm into the PDMS, leaving the section that was previously encased in PDMS (and therefore not subject to deformations during the reservoir punching process) exposed in the hydrogel reservoir. A 12 mm diameter circular glass coverslip was plasma bonded to the bottom surface of the device, leaving an open reservoir to be filled with the hydrogel. Prior to injection of the hydrogel, the top surface of the device, as well as a second 12 mm diameter circular coverslip, was plasma treated. Hydrogel containing tumor cells was added to the hydrogel reservoir to simulate a 3D tumor microenvironment. The hydrogel solution was added until the meniscus disappeared and the fluid was as close to planar with the surface of the PDMS as possible. This minimized the introduction of air into the device and precluded the overfilling of the well that could inhibit the glass – PDMS bond. Hydrogel addition is done quickly (~2 min) to avoid disturbing the plasma treated surface of the devices. The second 12 mm diameter circular coverslip was bonded over the top of the hydrogel reservoir, sealing off the reservoir. After waiting approximately 30 min for the hydrogel to cure, the stainless steel wire was removed, leaving a microchannel in the tumor microenvironment as well as in the PDMS. Tubing was connected to the device through the channel left by removing the stainless steel wire and was used to infuse media was into the lymphoma-on-chip device to begin delivering nutrients to the tumor cells. In order to fabricate the multiplex system, an additional, identical, hole punch step was added. The holes were punched such that the resulting wells were spaced 1 cm apart. The second well was filled with the same hydrogel formulation, containing explanted healthy lymph node cells rather than DLBCL tumor cells.
Hydrogel composition and loading – HyStem-C hydrogel (ESI BIO, Alameda, CA) was used for the lymphoma-on-chip device. Briefly, Glycosil (thiol-modified hyaluronan) and Gelin-S were dissolved in degassed water and mixed at 1:1 (v/v) ratio. Tumor and immune cells (A20, CD3+, and CD11b+ cells) were added to Glycosil+ Gelan-S mixture just before loading. PEG di-acrylate (PEG-DA) solution (9% wt/vol in degassed water) was added to Glycosil+Gelan-S+cell mixture at 1:4 (v/v) and quickly loaded into the device.
Endothelialization of DLBCL-on-chip model and multiplex system
Completed devices were coated with 5% fibronectin (Sigma-Aldrich, St. Louis, MO) and 1 mg/mL thiolated arginylglycylaspartic acid (RGD-SH) (Genemed Synthesis, San Antonio, TX) in mouse endothelial cell media (Cell Biologics, Chicago, IL) to promote endothelial attachment to the hydrogel and PDMS, and incubated at 37°C for 1 hour. Mouse lung microvascular endothelial cells (MLMVECs) (Cell Biologics, Chicago, IL) were injected into the device at a seeding density of approximately 5 million cells/mL. Endothelialized devices were incubated at 37°C for 2 hours while rotating the device about the horizontal axis at 8 rpm using a Hulamixer (Life Technologies, Carlsbad, CA) in order to ensure uniform seeding within the microchannel. After rotation, MLMVEC media was injected into the device at 2 uL/min using a syringe pump (Harvard Apparatus, Holliston, MA). The endothelial cells were allowed to grow to confluence (2–6 days) before conducting any experiments. The healthy lymph node model was positioned downstream of the media flow from the tumor model in the multiplex system.
Validation of the DLBCL-on-chip model
VE-cadherin expression - endothelialized devices were cultured under flow for approximately 72 hours, exposing the endothelium to wall shear stresses of ~1 dyn/cm2. Cells were washed with PBS, fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), blocked with 1% bovine serum albumin, and stained with a rabbit polyclonal antibody specific to VE-cadherin (Santa Cruz Biotechnology, Santa Cruz, CA). A secondary goat anti-mouse antibody tagged with Alexa Fluor 488 (Life Technologies, Carlsbad, CA) was added. VE-cadherin expression was visualized via confocal microscopy.
Diffusion and endothelial channel permeability–Endothelialized devices were cultured under flow to confluence. A fluorescently tagged secondary antibody (Alexa Fluor 488 goat anti-rabbit antibody, Invitrogen, Carlsbad, CA) was perfused through the device and allowed to diffuse through the endothelial monolayer into the in vitro tumor microenvironment. Diffusion of the secondary antibody (i.e. permeability of the hydrogel and endothelial monolayer) was measured via fluorescence intensity at specific distances away from the endothelialized channel. Fluorescence intensity was visualized via epifluorescence microscopy.
Macrophage depletion - Anti-mouse colony stimulating factor-1 receptor (CSF-1R) antibody (Biolegend, San Diego, CA) was infused into the device for 48 hours. After treatment with anti-CSF-1R, CD11b antibody pre-labeled cells were stained with cytox orange and assessed for cell death.
Tumor generation - A20 (TIB208, ATCC), a mouse B-cell lymphoma cell line equivalent to human DLBCL was used for tumor generation in Balb/C mice. Briefly, 1 × 107 A20 B lymphoma cells were injected subcutaneously in Balb/C mice and tumor growth was closely monitored by measuring length (longest axis) and width (shortest axis) of the tumor. The mice were sacrificed when the tumor size reached 1.5 cm in any direction. All animal experiments were approved by Institutional Animal Care and Use Committee (IACUC) of Georgia Institute of Technology.
Cell separation-Before loading cells in lymphoma-on-chip device, single cell suspension of tumor cells was prepared from explanted A20 tumor. Also, CD3+ T cells and CD11b+ cells were isolated from naïve Balb/C mice splenocytes. Briefly, whole tumor or spleen were minced into small pieces and digested in collagenase D (Roche Diagnostics, Indianapolis, IN) for 1 hour. The digested tissue was then passed through a cell strainer and the red blood cells (RBCs) were lysed using RBC lysis buffer. Finally, individual cell types such as CD3+ T cells and CD11b+ cells were isolated using Miltenyi magnetic cell separation kits (Miltenyi Biotec, San Diego, CA).
Lymph node extraction and processing - Balb/C mice were sacrificed and both inguinal lymph nodes were carefully extracted. A single cell suspension was achieved by pressing the lymph nodes against a 40um cell strainer and filtering the obtained cell solution once again.
Hydrogel digestion and cell recovery – Hydrogel was first carefully collected from the device by removing its top and bottom coverslips, and then incubated in collagenase D (2 mg/mL in Opti-MEM media) and Hyaluronidase (0.5 mg/mL in Opti-MEM media) solution for 2.5 hours for digestion. Cells were finally collected by centrifugation at 500 g for 5 minutes and used for flow cytometry.
Flow Cytometry-Cells were stained with various fluorescent labeled anti-mouse antibodies (eBioscience, San Diego, CA) and analyzed in a BD Accuri flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
Viability Assay - Live cells were determined via calcein (Invitrogen, Carlsbad, CA) staining. Total cell number was determined via epifluorescence microscopy.
Imaging
Confocal microscopy – Fluorescent images were collected on a LSM 700 confocal microscope (Zeiss, Jena, Germany), Fig. 3: Endothelial cells were stained with CellMask Orange (plasma membrane) (Life Technologies, Carlsbad, CA) and Hoechst (nucleus) (Life Technologies, Carlsbad, CA). B-cells, T-cells, and macrophages were pre-labeled with CellTrace dyes of different wavelengths (Thermo Fisher Scientific, Waltham, MA). Fig. 5: Endothelial cells were stained with CellMask Orange (plasma membrane) (Life Technologies, Carlsbad, CA). Explanted tumor and healthy lymph node lymphocytes were stained with Hoechst (nucleus) (Life Technologies, Carlsbad, CA).
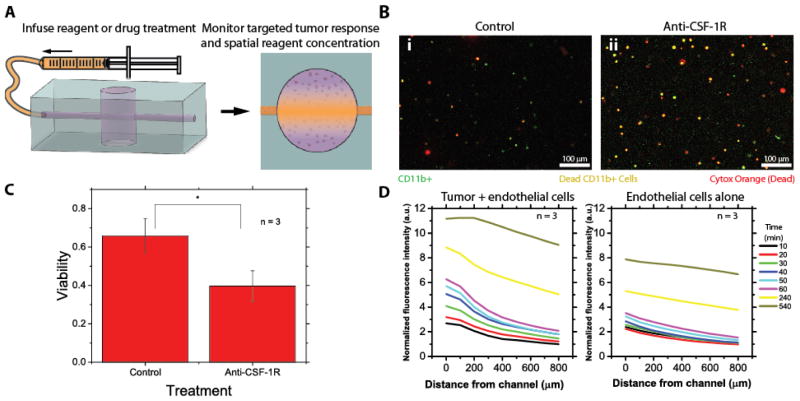
Fig. 3. Treatment with an anti-CSF-1R antibody preferentially causes macrophage death in the DLBCL –on-chip model and diffusion and endothelial permeability can be spatiotemporally modeled.
A) Drug treatments can be infused into the DLBCL-on-chip model and reagent diffusion can be analyzed in conjunction with tumor cell response. B-i) Untreated control devices experience little cell death amongst any DLBCL cell type, B-ii) while anti-CSF-1R antibody treated devices experience preferential cell death of macrophages (CD11b+). Green cells correspond to macrophages (alive or dead), red cells correspond to dead cells of any type and orange cells correspond to dead macrophages. C) Treatment with an anti-CSF-1R antibody causes a nearly 50% reduction in macrophage viability. Statistical significance was determined via t-test (P<0.05). D) Diffusion curves with respect to time and distance of a fluorescently tagged antibody within the lymphoma-on-chip model with and without tumor cells after perfusion for different lengths of time show saturation of the channel within the time scale of the experiment and follow Fick’s law of diffusion. Average normalized fluorescence intensity of 3 experiments is ultimately lower in the case when tumor cells are absent, indicating an increase in endothelial permeability in the presence of tumor cells.
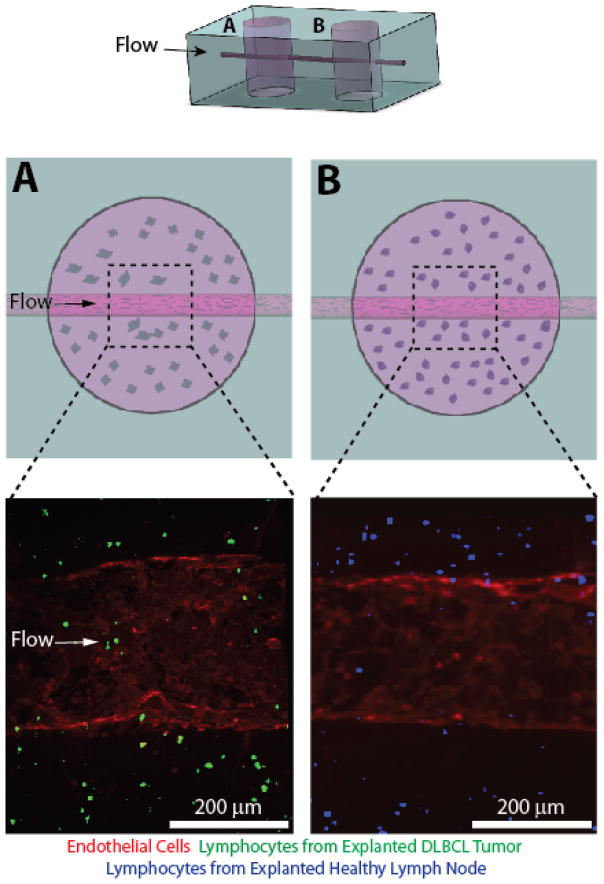
Fig. 5. A multiplex DLBCL-on-a-chip model was developed with a downstream healthy lymph node microvascular model connected via an endothelialized microchannel.
The addition of an additional fabrication step allows the DLBCL tumor hydrogel model (A) to be placed upstream of a healthy lymph node model comprising lymphocytes extracted from an explanted healthy lymph node (B). The top cartoon represents the overall PDMS macrostructure of the multiplex system. The middle row depicts cartoons of the DLBCL tumor hydrogel (A) and the healthy lymph node hydrogel (B). The bottom row depicts confocal micrographs of the distinct microvascular regions illustrated in the row above.
Epiflourescence microscopy – Epifluorescence images were taken on a TE2000-u microscope (Nikon, Tokyo, Japan): CellMask orange and Syto-13 (nucleus) (Life Technologies, Carlsbad, CA). Images were taken using a 10× objective (NA: .3, Nikon, Tokyo, Japan).
Spinning disk confocal microscopy - Fluorescent images were collected on a PerkinElmer UltraVIEW VoX spinning disk confocal microscope at the Petit Institute’s Optical Microscopy Core. Image processing, stitching, and three-dimensional reconstruction were done with Volocity (PerkinElmer Inc., USA).
Statistics
We conducted t-tests using MATLAB assuming unequal variance reported statistical significance at P < 0.05.
Results
This lymphoma-on-chip model recapitulates the 3D lymphoma tumor microenvironment
Here we report the first in vitro system that captures the complex interactions between immune cells and endothelial cells in the lymphoma microenvironment. This lymphoma-on-chip model consists of a tumor-cell-containing hyaluronic acid-based hydrogel embedded within a PDMS structure. A perfusable, fully endothelialized, microchannel traverses the hydrogel, recapitulating a blood vessel within a tumor (Fig. 1A). This technique is highly reproducible considering the lack of advanced microfabrication technology necessary. Although the removal of stainless steel wire from the hydrogel leads to some inconsistency in the shape of the channel, we report an average channel diameter of 328 ± 51 μm. Fluid flow within the deformable hydrogel leads to channel dilation, resulting in a channel larger than the stainless steel wire used to create it. Furthermore, the diameter varies by only ~8 μm over the length of the channel.
The inner surface of the fully perfusable hydrogel-embedded microchannel is coated with an endothelial cell monolayer, bringing immune cells in lymphoma and endothelial cells in close proximity
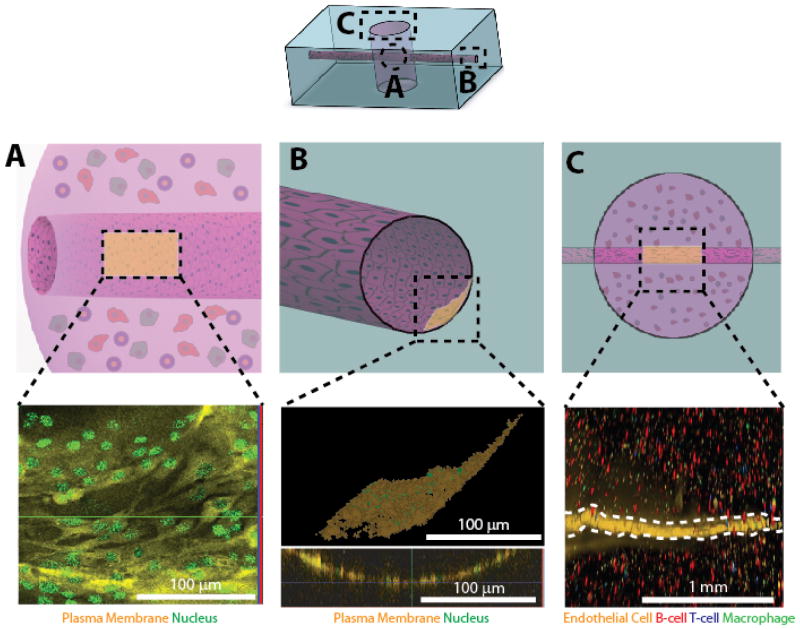
The proximity of the endothelial channel to the “tumor” allows this system to truly recapitulate the DLBCL tumor microenvironment. We were able to generate a perfusable vessel lined with a cylindrical endothelial monolayer that passes through both PDMS (Fig. 2A top) and a hyaluronic acid-based hydrogel embedded with tumor and immune cells (Fig. 2B top). The presence of PDMS is necessary to provide structural support for the DLBCL tumor, as well as to provide inlet and outlet ports to the microchannel, allowing us to deliver reagents directly to the tumor hydrogel. We show a confluent endothelial monolayer (Fig. 2A bottom) in close proximity to tumor cells within the DLBCL hydrogel (Fig. 2B bottom). Additionally, we report the presence of viable MLMVECs, T cells, B cells, and macrophages all operating within this in vitro tumor microenvironment (Fig. 2C).
Fig. 2. The DLBCL-on-a-chip microvasculature mode can be cultured with a confluent endothelial cell monolayer.
MLMVECs were confluent cultured in the microchannels of the system both in the (A) DLBCL hydrogel as well as the (B) PDMS macrostructure. Scale bar is 100 μm. C) The endothelial monolayer (white dotted line) lies in close proximity to T cells, B Cells, and macrophages within the DLBCL hydrogel, recapitulating the DLBCL tumor microenvironment. The top row depicts an overall schematic of the system, the middle row depicts specific regions of interest within the system, and the bottom row shows confocal micrographs of those regions. The endothelial monolayers shown in the confocal micrographs (lower panels) correspond to the shaded region in the PDMS macrostructure schematics (upper panels). Scale bar is 1 mm.
Blocking CSF-1 with an anti-CSF-1R antibody perfused through the endothelialized microchannel causes targeted macrophage killing in the lymphoma-on-chip model
To demonstrate that this system is capable of recapitulating targeted drug delivery to the tumor microenvironment, we tested the effect of anti-CSF-1R antibody for its ability to specifically kill macrophages embedded within the DLBCL hydrogel in our model. CSF-1 binds to CSF-1R and is required for the growth and survival of CD11b+ macrophages.29 An antibody blocking CSF-1R was perfused into the system via the endothelial microchannel and was shown to preferentially cause statistically significant macrophage cell death (Fig. 3A–C) relative to the control, in which the viability of CD11b+ cells was similar to the overall tumor cell viability (~70%) (Fig. S1C–D). Additionally, by infusing a fluorescent antibody and measuring its diffusion into the DLBCL hydrogel over time, we can model diffusion of reagents delivered via the endothelial channel according to Fick’s law of diffusion31 (Fig. 3D). Over time, we are able to show that the area surrounding the microchannel, and eventually the entire lymphoma-on-chip model, becomes saturated with the reagent. The ability to spatially and temporally model diffusion of a perfused reagent through this system has profound implications to study drug delivery and design. Furthermore, we report that the diffusion of a fluorescent antibody was increased in devices where tumor cells were included in the hydrogel as compared to devices in which tumor cells were not included (Fig. 3D, Fig. S2). These results reveal a trend that the presence of tumor cells increased endothelial microchannel permeability in the DLBCL-on-a-chip model. This effect of tumor cells on endothelial permeability has been reported in vivo in DLBCL.30 These findings were supported by minimal expression of VE-cadherin by endothelial cells in the system (Fig. S1B), and indicate the ability of the system to recapitulate cell-cell interactions with in the tumor microenvironment.
Cellular components of the lymphoma-on-chip model can be extracted from the hydrogel for post-treatment analysis
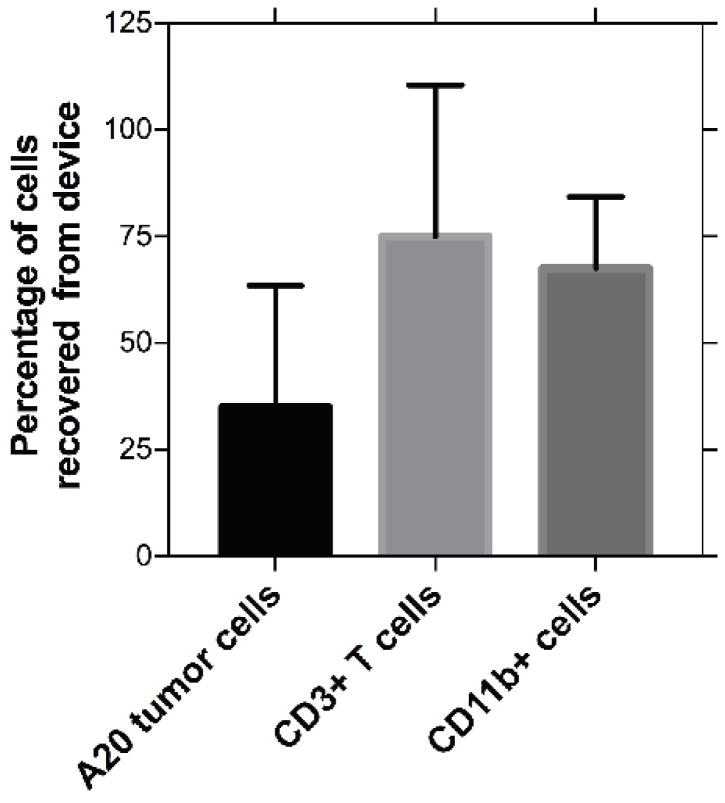
The unique fabrication method of the in vitro lymphoma-on-chip model allows for post-experiment collection and analysis of various cell types within the model. By carefully removing the coverslips used to seal the tumor-containing hydrogel, we were able to extract the tumor mass from the device. The hydrogel was digested and cellular components were analyzed via flow cytometry. We were able to show via flow cytometry analysis that a significant percentage of each cellular component can be recovered after experimentation (Fig. 4). Additionally, we were able to isolate and differentiate between the different cell types using immunostaining. These results suggest that we can quantitatively assess the presence of various surface proteins in response to a particular treatment. This highlights the versatility of this experimental platform, as cellular analysis is possible in addition to various real-time imaging techniques.
Fig. 4. Recovery of cellular components of tumor microenvironment for post treatment analysis is possible from the DLBCL-on-chip model.
Cellular components of the DLBCL tumor microenvironment were harvested after culture of endothelial cells for 48 hours and analyzed for cell type via flow cytometry. Percentages refer to the percentage of each specific cell type recovered from the total initial number of that cell type within the device. These results indicate that we were able to specifically extract and analyze various cellular components post-treatment.
Simple alterations to the fabrication protocol allow for the development of distinct microenvironments connected via an endothelial microchannel
The simple fabrication technique presented herein allows for dynamic alterations to the system. We report a slight modification of the previously described device design to include an additional hydrogel well which contains explanted cells from a healthy lymph node. This vascularized “healthy lymph node” was placed downstream of the tumor cell containing well and the wells are connected by an endothelialized microchannel. This confluent endothelial channel traversed both the DLBCL tumor microenvironment (Fig. 5A, Supplemental Movie 1) as well as the healthy lymph node microenvironment (Fig. 5B, Supplemental Movie 2) channels, with a PDMS portion separating the two distinct vascularized microenvironments. Fabrication of this multiplex system serves to further highlight the adaptability of this system to potentially investigate the downstream effects of DLBCL.
Discussion
Our DLBCL-on-a-chip model provides unique insight into the complex cell – cell interactions of the DLBCL tumor microenvironment by spatially recapitulating a blood vessel within a tumor mass. Our results show that this fabrication technique can be accurately and reliably reproduced to facilitate mass-experimentation without requiring the more traditional, photolithography-based techniques used to fabricate microchannels in hydrogels. By simply using PDMS, a high gauge stainless steel wire, hydrogel components, and standard laboratory equipment, this technique offers an approach accessible to tumor immunologists who may not have access to advanced microfabrication facilities. We show that this model is capable of recapitulating the tumor microenvironment, with viable tumor cells cultured around a cylindrical, endothelialized, microchannel. We show that this model can potentially be used to spatially deliver reagents to a tumor mass, as demonstrated by preferential and specific killing of macrophages in the tumor model utilizing perfusion of anti-CSF-1R antibody. Furthermore, the increase in endothelial permeability in the presence of tumor cells highlights the similarities between this system and the in vivo DLBCL tumor microenvironment. Additionally, we characterize the spatial diffusion of reagent throughout the tumor microenvironment. We show that cells can be safely extracted and isolated from the device after experimentation, allowing for single cell analysis of cells post-experiment. Finally, we demonstrate a multiplex system that allows for the potential study of local and global cellular interactions, as well as the impact that drug treatments may have on the various, distinct, microenvironments within DLBCL. This unique, versatile, fabrication method may also be adapted to study other malignancies. This technology is not without limitations, however. The primary weakness of this technique stems from the friction forces of the stainless steel wire exerted on the hydrogel upon removal of the wire from the device. These forces deform the hydrogel when creating the void that eventually becomes the endothelialized microchannel, leading to channel sizes and shapes that vary slightly between different devices. However, these slight variations do detract from the devices ability to recapitulate the DLBCL tumor microenvironment.
These results indicate that the versatile technique we have presented accurately recapitulates the complex tumor microenvironment, allowing us to easily visualize the interplay between lymphoma, immune, and endothelial cells in vitro in a way never before possible with conventional microfluidics. The key innovation of this technology is the ability to study the spatial interactions of immune cells within the microenvironment provided by the endothelialized microchannel traversing a 3D tumor hydrogel. This model allows us to study the spatial interactions between different cellular components of a tumor. Our results indicate that delivery of a therapeutic agent can be modeled spatiotemporally in our model, potentially leading to novel advancements in drug delivery and design. Overall, this in vitro model of the tumor microenvironment represents an easily accessible tool that enables researchers to spatially investigate the complex cellular and molecular interactions that occur within the tumor microenvironment, particularly in the regime of tumor microvasculature.
Conclusion
In this study, we have described a novel lymphoma-on-chip platform for the study of the complex cellular interaction within the tumor microenvironment in DLBCL. This system utilizes a novel, easily accessible fabrication technique to recapitulate the lymphoma tumor microenvironment surrounding tumor microvasculature. We show that this system has expansive experimental capabilities and can be used to model the delivery and effect of targeted drug treatments within the tumor vasculature. We believe that this technology fulfills a thus unmet need for an economical, accessible, platform to study the complex cellular interactions in the tumor microenvironment, and that scientists lacking specialized engineering training (e.g. experimental biologists, hematologists, immunologists, etc) will be able to utilize this tool to make fundamental advances in the fields of drug delivery and design as it relates to DLBCL and possibly other malignancies.
Supplementary Material
Acknowledgments
Financial support for this work was provided by a National Science Foundation CAREER Award 1150235 (to W.A.L.); an American Heart Association Innovative Research Grant (to W.A.L.); National Institutes of Health Grants 5U01-HL117721 (to W.A.L.), U54HL112309 (to W.A.L.), R01HL121264 (to W.A.L), a National Science Foundation Graduate Research Fellowship DGE-1650044 (to R.G.M, J.C.M, and A.S). We wish to acknowledge the core facilities at the Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology, as well as the Institute for Electronics and Nanotechnology at the Georgia Institute of Technology for the use of their shared equipment, services and expertise. The authors would like to acknowledge J. Ciciliano for careful proofreading of the final manuscript, as well as thoughtful discussions.
References
- 1.Armitage JO. Blood. 1997;89:3909–3918. [Google Scholar]
- 2.National Cancer Institute. [Accessed September 2016]; http://seer.cancer.gov/statfacts/html/nhl.html.
- 3.Feugier P, Hoof A, Sebban C, Solal-Celigny P, Bouabdallah R, Fermé C, Christian B, Lepage E, Tilly H, Morschhauser F, Gaulard P, Salles G, Bosly A, Gisselbrecht C, Reyes F, Coiffier B. J Clin Oncol. 2005;23:4117–4126. doi: 10.1200/JCO.2005.09.131. [DOI] [PubMed] [Google Scholar]
- 4.Roschewski M, Dunleavy K, Wilson W. Leukemia Lymphoma. 2014;55:2428–2437. doi: 10.3109/10428194.2014.883075. [DOI] [PubMed] [Google Scholar]
- 5.Shankland K, Armitage J, Hancock B. Lancet Lond Engl. 2012;380:848–857. doi: 10.1016/S0140-6736(12)60605-9. [DOI] [PubMed] [Google Scholar]
- 6.Cheson BD, Leonard JP. New England Journal of Medicine. 2008;359:613–626. doi: 10.1056/NEJMra0708875. [DOI] [PubMed] [Google Scholar]
- 7.Khouri I, Saliba R, Erwin W, Samuels B, Korbling M, Medeiros L, Valverde R, Alousi A, Anderlini P, Bashir Q, Ciurea S, Gulbis A, Lima M, Hosing C, Kebriaei P, Popat U, Fowler N, Neelapu S, Samaniego F, Champlin R, Macapinlac H. Blood. 2012;119:6373–6378. doi: 10.1182/blood-2012-03-417808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosing C, Saliba R, McLaughlin P, Andersson B, Rodriguez M, Fayad L, Cabanillas F, Champlin R, Khouri I. Ann Oncol. 2003;14:737–744. doi: 10.1093/annonc/mdg200. [DOI] [PubMed] [Google Scholar]
- 9.Michallet M, Sobh M, Milligan D, Morisset S, Niederwieser D, Koza V, Ruutu T, Russell N, Verdonck L, Dhedin N, Vitek A, Boogaerts M, Vindelov L, Finke J, Dubois V, Biezen A, Brand R, Witte T, Dreger P C. EBMT. Leukemia. 2010;24:1725–1731. doi: 10.1038/leu.2010.165. [DOI] [PubMed] [Google Scholar]
- 10.Peniket A, Elvira M, Taghipour G, Cordonnier C, Gluckman E, Witte T, Santini G, Blaise D, Greinix H, Ferrant A, Cornelissen J, Schmitz N, Goldstone A, Registry E. Bone Marrow Transpl. 2003;31:667–678. doi: 10.1038/sj.bmt.1703891. [DOI] [PubMed] [Google Scholar]
- 11.Porter D, Levine B, Kalos M, Bagg A, June C. New Engl J Medicine. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochenderfer J, Wilson W, Janik J, Dudley M, Stetler-Stevenson M, Feldman S, Maric I, Raffeld M, Nathan D, Lanier B, Morgan R, Rosenberg S. Blood. 2010;116:4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kochenderfer JN, Dudley ME, Feldman SA. Blood. 2012;119:2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardenas M, Yu W, Beguelin W, Teater M, Geng H, Goldstein R, Oswald E, Hatzi K, Yang SN, Cohen J, Shaknovich R, Vanommeslaeghe K, Cheng H, Liang D, Cho H, Abbott J, Tam W, Du W, Leonard J, Elemento O, Cerchietti L, Cierpicki T, Xue F, MacKerell A, Melnick A. J Clin Invest. 2016;126:3351–3362. doi: 10.1172/JCI85795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qing K, Jin Z, Fu W, Wang W, Liu Z, Li X, Xu Z, Li J. J Hematology Oncol. 2016;9:72. doi: 10.1186/s13045-016-0303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polini A, Prodanov L, Bhise NS. Expert Opinion on Drug Discovery. 2014;9:335–352. doi: 10.1517/17460441.2014.886562. [DOI] [PubMed] [Google Scholar]
- 17.Chang T, Mikheev A, Huynh W, Monnat R, Rostomily R, Folch A. Lab on a Chip. 2014;14:4540–4551. doi: 10.1039/c4lc00642a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaira V, Fedele G, Pyne S, Fasoli E, Zadra G, Bailey D, Snyder E, Faversani A, Coggi G, Flavin R, Bosari S, Loda M. P Natl Acad Sci Usa. 2010;107:8352–8356. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh D, Hamilton G, Ingber D. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du Y, Ghodousi M, Qi H, Haas N, Xiao W, Khademhosseini A. Biotechnol Bioeng. 2011;108:1693–1703. doi: 10.1002/bit.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zervantonakis I, Kothapalli C, Chung S, Sudo R, Kamm R. Biomicrofluidics. 2011;5:13406. doi: 10.1063/1.3553237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herland A, Meer A, FitzGerald E, Park T-E, Sleeboom J, Ingber D. Plos One. 2016;11:e0150360. doi: 10.1371/journal.pone.0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchanan C, Voigt E, Szot C, Freeman J, Vlachos P, Rylander M. Tissue Eng Part C Methods. 2013;20:64–75. doi: 10.1089/ten.tec.2012.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orr FW, Wang HH, Lafrenie RM. The Journal of Pathology. 2000;190:310– 329. doi: 10.1002/(SICI)1096-9896(200002)190:3<310::AID-PATH525>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 25.Hamada J, Cavanaugh P, Lotan O, Nicolson G. Brit J Cancer. 1992;66:349–54. doi: 10.1038/bjc.1992.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Forouzan O, Burns J, Shevkoplyas S. Lab on a Chip. 2011;11:3231–3240. doi: 10.1039/c1lc20293f. [DOI] [PubMed] [Google Scholar]
- 27.Mannino R, Myers D, Ahn B, Wang Y, Rollins M, Gole H, Lin A, Guldberg R, Giddens D, Timmins L, Lam W. Sci Reports. 2015;5:12401. doi: 10.1038/srep12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin D, Xia Y, Whitesides GM. Nat Protoc. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 29.Pixley F, Stanley E. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Aoki Y, Tosato G. Leukemia Lymphoma. 2009;41:229–237. doi: 10.3109/10428190109057978. [DOI] [PubMed] [Google Scholar]
- 31.Lin CC, Metters AT. Advanced drug delivery reviews. 2006;58:1379–1408. doi: 10.1016/j.addr.2006.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.