Abstract
Recent studies have revealed that the gut microbiota modulates brain development and behavior, but the underlying mechanisms are still poorly understood. Here, we show that bacterial peptidoglycan (PGN) derived from the commensal gut microbiota can be translocated into the brain and sensed by specific pattern-recognition receptors (PRRs) of the innate immune system. Using expression-profiling techniques, we demonstrate that two families of PRRs that specifically detect PGN (that is, PGN-recognition proteins and NOD-like receptors), and the PGN transporter PepT1 are highly expressed in the developing brain during specific windows of postnatal development in both males and females. Moreover, we show that the expression of several PGN-sensing molecules and PepT1 in the developing striatum is sensitive to manipulations of the gut microbiota (that is, germ-free conditions and antibiotic treatment). Finally, we used the PGN-recognition protein 2 (Pglyrp2) knockout mice to examine the potential influence of PGN-sensing molecules on brain development and behavior. We demonstrate that the absence of Pglyrp2 leads to alterations in the expression of the autism risk gene c-Met, and sex-dependent changes in social behavior, similar to mice with manipulated microbiota. These findings suggest that the central activation of PRRs by microbial products could be one of the signaling pathways mediating the communication between the gut microbiota and the developing brain.
Introduction
During birth and rapidly thereafter, the intestinal tract is massively colonized by trillions of microorganisms, collectively referred to as the gut microbiota. This evolution-driven process has been shown to contribute to the developmental programming of epithelial barrier function, gut homeostasis and angiogenesis, as well as the development and function of the gut immune system.1, 2, 3 A growing number of studies have revealed that the gut microbiota has effects on host physiology and development outside the gastrointestinal system, including the early-life programming of brain circuits involved in the control of emotions, motor activity, and cognitive functions.4, 5, 6, 7, 8 We have previously shown that mice raised under germ-free (GF) conditions exhibit more motor activity and less anxiety-like behavior than do mice with normal gut microbiota (specific pathogen-free, SPF).5 However, the cellular and molecular mechanisms mediating interactions between the gut microbiota and the developing brain remain poorly understood. Here, we postulate that microbial products derived from the commensal gut microbiota can activate pattern-recognition receptors (PRRs) of the innate immune system in the developing brain.
PRRs have a key role in the innate immune responses by recognizing conserved microbial molecules derived from a diverse variety of microbial pathogens.9 However, these motifs are not exclusive to pathogens and are abundantly produced by the commensal gut microbiota during the postnatal colonization process. Previous studies have shown that the gut microbiota is a source of peptidoglycan (PGN; a major component of the bacterial cell wall), which is translocated from the intestinal mucosa into circulation under basal conditions in the absence of pathogens.10 This microbial product was found to influence the function of bone-marrow-derived neutrophils, a type of cell residing at a distance from the gut. The PRR nucleotide-binding oligomerization domain-containing protein-1 (Nod1; which recognizes mesoDAP-containing PGN found predominantly in Gram-negative bacteria) was identified as the homeostatic regulator mediating the systemic effects of PGN.10 These novel findings raise the possibility that PGN may also cross the blood–brain barrier and influence the developing brain via activation of PRRs. In the present study, we sought to investigate whether PGN and its sensing molecules can be detected in developing brain (for example, prefrontal cortex, striatum and cerebellum). Subsequently, we determined whether the expression of PGN-sensing molecules and PGN transporter in the developing brain is sensitive to manipulations of the gut microbiota. For this purpose, we focused on the striatum since we have previously shown that this brain region is very sensitive to the absence of gut microbiota.5 Finally, we used the PGN-recognition protein 2 (Pglyrp2) knockout (KO) mice to test the potential influence of PGN-sensing molecules on brain development and behavior.
Materials and methods
Animals
Pregnant C57BL/6N female mice were obtained from Charles River Laboratories (Sulzfeld, Germany) and housed individually in standard plastic cages (Makrolon Type III, Tecniplast, Buguggiate, Italy) under controlled temperature, humidity and light (12:12 h light–dark cycle) conditions. Food and water were available ad libitum. After birth, brain tissues were collected from naive C57BL/6N mice at various postnatal ages (postnatal days 1, 3, 5, 7, 14, 21 and 60) from multiple litters; we defined the day of birth as postnatal day (P) 0.
Perturbations of gut microbiota
C57BL/6 GF mice. 3-day-old C57BL/6 SPF and GF male and female mice from multiple litters were obtained from the Core Facility for Germ Free Research, Karolinska Institutet, Sweden.
Perinatal antibiotic treatment. Pregnant C57BL/6N female mice were exposed to ampicillin (cat no. A8351, 0.6 mg ml−1; Sigma-Aldrich, St Louis, MO, USA; freshly made every day) in their drinking water, beginning 5 days before delivery. The dams continued to receive antibiotics in their drinking water during the first three postnatal days. Thus, perinatal antibiotic treatment refers to antibiotic exposure both in utero and during early postnatal life. In all experiments, multiple litters (n=6 per treatment) were used to avoid any potential litter effects. This low-antibiotic regimen does not produce changes in the body weight of the dams or of their offspring (data not shown). In addition, we have not observed any alterations in maternal behavior.
Transgenic mouse model
Pglyrp2 KO mice. Heterozygous Pglyrp2-breeding pairs were obtained from the Jackson laboratory (Bar Harbor, ME, USA).11 Animals were bred and housed in our animal facility under SPF conditions. Wild-type (WT) and mutant mice were obtained from multiple heterozygous Pglyrp2-breeding pairs (n=6). The Pglyrp2 mice used were backcrossed to C57BL/6 mice for at least 10 generations in the original laboratory where they were generated, as well as backcrossed for at least one generation in the Jackson Laboratory (Stock No. 000664).
All experiments were conducted according to a protocol approved by the Ethics Committee on Animal Research, Stockholm North and in accordance with the European Communities Council Directive of 24 November 1986 (86/609/EEC).
PGN detection assay
To detect and quantify PGN levels in the developing brain, various brain regions (that is, prefrontal cortex, striatum and cerebellum) were rapidly dissected out on an ice cold surface, wet weighed, frozen on dry ice and stored at −80 °C until use. Frozen samples were thawed on ice for 10 min, and then homogenized in 400 μl of sterile, cold phosphate-buffered saline (PBS), pH 7.4 for 20 s. For serum samples, blood samples were collected by cardiac puncture prior to death, and allowed to clot for 3 hours at 2–8 °C, and then centrifuged at 1800 g (or 4000 rpm) at room temperature for 15 minutes. The serum was immediately removed and stored at −80 °C until use. Thereafter, 50 μl of each homogenate or serum sample was assayed using the silkworm larvae plasma (SLP) detection kit (Wako Pure Chemical Industries, Osaka, Japan).12 The SLP method allows the rapid, simultaneous detection of PGN components of both Gram-positive and Gram-negative bacteria. PGN derived from Micrococcus luteus (Str. IF03333; Wako Pure Chemical Industries) was used as reference, and PBS as negative control. Optical density measurements were performed using an xMark Microplate Absorbance Reader (Bio-Rad Laboratories, Sundbyberg, Sweden). All data were then processed using Microplate Manager 6, Version 6.3 software. The microplate reader was set up as follows: Assay absorbance: 650 nm, Assay mode: kinetic/integral, Duration: 30 s interval, 180 reads (total 90 min), Assays temperature: 30 °C, Auto mix: once at low speed. The standard was plotted with a regression of Log–Log horizontal axis-concentration of SLP activation material and vertical axis-onset time, according to the manufacturer's specifications. In all brain tissue samples, the PGN concentrations were normalized for wet weight.
Quantitative real-time polymerase chain reaction
Expression of genes of interest were quantitatively determined by using a SYBR Green Real-Time PCR Detection System (Bio-Rad, Sundbyberg, Sweden) as previously described.13 The housekeeping gene, Hsp90ab1, was used for normalization, as its expression is stable throughout postnatal development in all tissues examined. The Primer-BLAST web-based software was used to design gene-specific primers (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). All primer sequences, annealing temperatures, and gene accession numbers are listed in Supplementary Table 1.
Western immunoblotting
Western immunoblotting was performed as previously described.14 The membranes were immunoblotted using polyclonal antibodies against Pglyrp2 (cat no. sc-50471, 1:5000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and Pglyrp3 (cat no. MBS852923, 1:5000; MyBioSource, San Diego, CA, USA). Rabbit polyclonal antibody against Heat Shock Protein 90 (Hsp90; cat no. 4874, 1:10 000; Cell Signaling Technology, Beverly, MA, USA) served as a loading control.
Immunohistochemistry
Mice were anaesthetized either by hypothermia-induced anesthesia (that is, 3-day-old mice) or with an intraperitoneal injection of sodium pentobarbital (100 mg kg−1 of body weight; Apoteket, Stockholm, Sweden) and perfused via the ascending aorta with a mixture of paraformaldehyde and picric acid (4% paraformaldehyde and 0.2% picric acid, in 0.01 m phosphate-buffered saline (PBS), pH 7.35). The brains were rapidly dissected, immersed in the same fixative for 90 min and then cryoprotected in 10% sucrose, 0.02% bacitracin (Sigma), and 0.01% sodium azide (Sigma), in PBS for 24–36 h. Brain sections were made using a cryostat, collected on SuperFrost Plus slides (VWR, Stockholm, Sweden) and stored at −20 °C until additional processing.
Brain sections from 3-day-old mice were immunostained with a polyclonal rabbit antibody against Pgrp2 (cat no. SC-50471, 1:100; Santa Cruz Biotechnology). Brain sections from 3-week-old mice were stained with a polyclonal rabbit antibody against Nod1 (cat no. bs-7085R, 1:100; Bioss Antibodies, Woburn, MA, USA). The secondary antibody was Donkey Anti-Rabbit Cy2 antiserum. For double labelling, the following antisera were used: mouse monoclonal antibody against NeuN to label mature neurons (cat no. MAB377, 1:250; Merck Millipore, Billerica, MA, USA), mouse monoclonal antibody against Calbindin to label Purkinje cells in the cerebellum (cat no. AB82812, 1:500; Abcam; Cambridge, UK), goat polyclonal antibody against GFAP to label astrocytes (cat no. AB53554, 1:500; Abcam), and goat polyclonal antibody against Iba1 to label microglial cells (cat no. AB5076, 1:800; Abcam), and were detected using either Donkey Anti-Mouse Cy5 antiserum or Donkey Anti-Goat Cy5 antiserum (1:250; Jackson ImmunoResearch Europe, Suffolk, UK).
For immunofluorescence techniques, sections were dried at room temperature for 1 h, rehydrated in 0.01m PBS (pH 7.4) for 5 min, and then incubated for 60 min in 3% bovine serum albumin in PBS. All primary and secondary antibodies were diluted in 1% bovine serum albumin, and 0.2% Triton-X in 0.01m PBS. The sections were then incubated with the primary antibodies for 36 h at 4 °C, and an additional incubation at room temperature for 1 h. After washing five times with PBS for 5 min each, sections were incubated with the secondary antibodies for 2 h in darkness at room temperature. Finally, sections were rinsed five times in PBS and coverslipped in glycerol containing 2.5% diazabicyclooctane (Sigma).
Photomicrographs were taken with an Olympus XM10 (Olympus Sverige, Stockholm, Sweden); digital camera Illustrations were prepared using the Cell Sens Imaging Software ver. 1.12 (Olympus, Tokyo, Japan). Images were only adjusted for brightness and contrast.
Behavioural studies
Testing took place between 0900 and 1600 h under low illumination to reduce stress. On the day of testing, animals were brought in their home cages to the testing room and allowed to rest for at least 1 h before testing. Test chambers were cleaned first with disinfectant and then with 70% ethanol and water after each animal. All animals were naive to the tasks. The experimenter was blind to the genotype during behavioral testing.
Open-field test
Animals were placed individually in the center of an open-field box (48 cm × 48 cm; Acti-Mot detection system; TSE, Bad Homburg, Germany), and their spontaneous motor activity was recorded as described.15 The computer program automatically recorded the following parameters: distance traveled in the center, periphery and total (entire box) and a count of rearing activity (vertical infrared photo beam breaks).
Dark–light box test
In this test, the mouse was placed into the dark compartment and allowed to freely explore the apparatus (48 × 48 cm; with two zones of equal areas) for 5 min. Time spent in the dark and light compartments were measured by using photocells (TSE).
Elevated plus maze test
Mice were individually placed in the center of an elevated plus maze apparatus (Kinder Scientific, Poway, CA, USA), facing an open arm and allowed to explore for 5 min. The time spent in, and entries into, the open and closed arms, and total ambulation were recorded via infrared photobeams and analyzed with Motor Monitor software (Kinder Scientific).
Three-chamber social approach task
Social preference was evaluated in a three-chambered apparatus as previously described.16 The rectangular three-chambered box was made of clear Plexiglas and was divided into three equally sized chambers (43.7 cm long, 20 cm wide and 35 cm high). The walls dividing the chambers contained a small rectangular opening (10 cm wide and 6.5 cm high) to allow the test mouse access to the adjacent chambers. Clear rectangular Plexiglas doors were used to close the openings when required. The test session began with a short habituation (that is, 1 min) in the center chamber without access to the side chambers, followed by a 10 min habituation session to the entire empty box, with access to all three chambers. The test mouse was then briefly confined to the center chamber while the experimenter placed the stimuli. To test for social approach behaviors towards a live unfamiliar mouse (that is, same age, sex and strain), a novel C57BL/6 mouse previously habituated to a grid enclosure (that is, inverted wire pencil cup, Galaxy, Kitchen Plus, http://www.kitchen-plus.com) was placed in one of the side chambers. A control novel object, an identical empty grid enclosure devoid of social odors, was placed in the other side chamber. The location of the novel object and the novel mouse alternated between the left and right side chambers across subjects. After both stimuli were positioned in the side chambers, the two doorways were simultaneously opened and the test mouse was allowed access to all three chambers for 10 min. Time spent in each chamber, and time spent around enclosures (that is, near the novel object or novel mouse), was recorded by a video camera positioned directly over the testing apparatus (EthoVision XT version 7; Noldus, Wageningen, The Netherlands).
Statistical analysis
Several pilot studies were conducted (for example, gene expression, SLP assay and behavioral experiments) to estimate the sample size and aimed at detecting differences between groups at the 0.05 level. Data were expressed as mean±s.e.m. Statistical analysis was performed using STATVIEW version 5.1 software. Data from gene and protein expression studies were analyzed using one-way ANOVA. Post hoc comparisons were made using Fisher's Least Significant Difference test when significant analysis of variance (ANOVA) effects were found. Data from behavioral studies were analyzed using either repeated-measures ANOVA or factorial ANOVA when appropriate. Post hoc comparisons were made using a Bonferroni/Dunn test when significant ANOVA effects were found. The threshold for statistical significance was set as P⩽0.05.
Results
Can bacterial PGN from the commensal gut microbiota cross the blood–brain barrier?
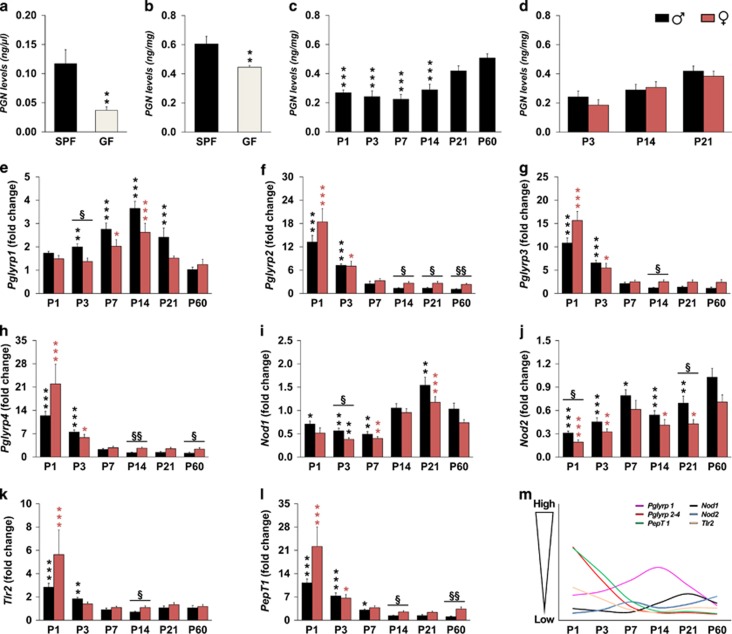
To ascertain whether bacterial PGN can be translocated into the blood under normal conditions, we examined the levels of PGN in serum of SPF and GF juvenile (that is, P 25) male mice, using the SLP assay. We detected the presence of PGN in the serum of SPF juvenile male mice (Figure 1a). As expected, we found that the PGN levels were significantly lower in the serum of GF juvenile male mice compared to SPF male mice (Figure 1a). We next determined whether PGN can cross the blood–brain barrier (BBB), and examined the levels of PGN in the developing brain using the same assay. To validate the SLP assay for the detection of PGN in the brain, we first compared PGN levels in brain tissue of juvenile SPF and GF male mice. However, given that L-DOPA (L-3,4-dihydroxyphenylalanine) is part of the activation mechanisms of SLP assay and GF mice show elevated levels of L-DOPA in the striatum,5 we decided to evaluate the cerebellum of juvenile SPF and GF male mice (that is, P25), in a brain region with a relative low dopamine innervation. The PGN levels were significantly lower in the cerebellum of juvenile GF male mice compared with SPF (Figure 1b), thus suggesting that PGN can cross the BBB under normal conditions. We next investigated the levels of PGN during postnatal brain development in various brain regions (that is, prefrontal cortex, striatum and cerebellum) of C57BL/6 male mice. Interestingly, we observed an age-dependent increase in PGN levels in the cerebellum (Figure 1c), paralleling the postnatal bacterial colonization process.17 A similar pattern was also observed in the prefrontal cortex and striatum (data not shown). To determine potential sex differences in the levels of PGN in the developing brain, we compared the levels of PGN in the cerebellum of C57BL/6 male and female mice at P3, P14 and P21. However, no significant sex-differences were found (Figure 1d).
Figure 1.
Bacterial peptidoglycan from commensal gut microbiota and its sensing molecules are present in the developing brain. Peptidoglycan (PGN) levels were determined in serum (a) and the cerebellum (b) of specific pathogen free (SPF) and germ-free (GF) juvenile (that is, P25) male mice, as determined using the silkworm larvae plasma assay (serum: n=5 for SPF and n=7 for GF, and cerebellum: n=8 per group; **P<0.01 when compared with SPF control group). (c) The bar graphs show PGN levels in the developing cerebellum of C57BL/6 male mice (n=6 per group; ***P<0.001 when compared with the adult group). (d) PGN levels were measured during cerebellar development in both male (black bars) and female (red bars) mice at P3, P14, and P21 (n=5 for females and n=6 for males). Quantitative RT-PCR was used to examine the mRNA expression levels of Pglyrp1 (e), Pglyrp2 (f), Pglyrp3 (g), Pglyrp4 (h), Nod1 (i), Nod2 (j), Tlr2 (k) and PepT1 (l), which were examined in the striatum of male (black bars) and female (red bars) C57BL/6 mice during postnatal development (n=4 for P1 females and n=5 for the other postnatal age groups). Graphical visualization of PGN-sensing molecules and PepT1 expression levels during striatal development (m). The expression level of each gene examined was normalized to Hsp90ab1 levels and expressed relative to adult male levels (that is, P60). All data (a–l) are presented as means (±s.e.m.). (e–l) *P<0.05, **P<0.01, and ***P<0.001 when compared with their respective adult male group. Differences between male and female mice are indicated as follows: §P<0.05, §§P<0.01. The label is as follows: postnatal day (P).
Having established that PGN is translocated into the blood circulation and the developing brain, we used quantitative real time PCR to assess whether PGN-sensing molecules are also expressed during early postnatal brain development. The expression profiles of two families of PRRs that specifically recognize PGN, namely the PGN-recognition proteins (PGRPs)18 Pglyrp1, Pglyrp2, Pglyrp3 and Pglyrp4 and the intracellular NOD-like receptors (NLRs)19 Nod1 and Nod2, were investigated during postnatal mouse brain development. In addition, we investigated the expression profile of the membrane-bound Toll-like receptor 2 (Tlr2), which recognizes PGN and other microbial components.20 We found that all of these PGN-sensing molecules were expressed in the developing brain during specific temporal windows of postnatal development. Interestingly, we also found several significant sex-differences in their expression levels. In the striatum, the gene expression levels of Pglyrp1 increased gradually to peak levels at P14 in both male and female mice, after which they decreased to adult levels (Figure 1e). However, males had significantly higher levels of Pglyrp1 than females at P3 (Figure 1e). In contrast, the gene expression levels of Pglyrp2, Pglyrp3 and Pglyrp4 were significantly higher during the first few days of life in both sexes, thereafter decreasing to adult levels (Figures 1f–h). Interestingly, during the second week and thereafter, females had significantly higher levels of Pglyrp2 (that is, P14, P21 and P60), Pglyrp3 (that is, P14) and Pglyrp4 (that is, P14 and P60) than males (Figures 1f–h). The gene expression levels of Nod1 were significantly lower during the first postnatal week than in adults, after which they increased to peak levels at P21 in both sexes (Figure 1i). Males showed significantly higher levels of Nod1 than females at P3. In contrast, the gene expression levels of Nod2 increased gradually with age in both males and females (Figure 1j). Overall, there was a tendency for males to express higher levels of Nod2 during all postnatal ages, which reached significance at P1 and P21 (Figure 1j). Similar to the gene expression levels of Pglyrp2–4, Tlr2 mRNA levels were significantly higher during the first few days of life (Figure 1k). At P14, females showed significantly higher expression levels of Tlr2 than males.
We also examined the expression profile of peptide transporter 1 (PepT1; also known as soluble carrier family 15 member 1, Slc15a1), which has been previously shown to efficiently translocate PGN fragments (for example, muramyl dipeptide) into colonic cells.21 In the striatum, PepT1 mRNA levels were significantly higher during the first postnatal days of life than in adults in both sexes (that is, P60; Figure 1l). Interestingly, females had significantly higher levels of PepT1 at P14 and P60 (Figure 1l). These results suggest that when PGN levels are low during early postnatal life, PepT1 is abundant in the developing brain, enhancing the capacity to transport PGN fragments into nerve cells.
In the prefrontal cortex and cerebellum, the expression patterns of Pglyrp2, Pglyrp3, Pglyrp4, Tlr2 and PepT1 were very similar to those found in the striatum (Supplementary Figures 1 and 2). Moreover, these genes have significantly higher expression levels in newborn females compared to males. This sex difference is more evident in the prefrontal cortex than in the cerebellum or striatum. In contrast, we found brain regional differences in the expression patterns of Pglyrp1, Nod1 and Nod2 (Figure 1 and Supplementary Figures 1 and 2). Similar to the striatum, in the cerebellum, Pglyrp1, Nod1 and Nod2 have generally higher expression in males than females (Supplementary Figure 2). These results indicate that a highly dynamic and sensitive period exists, during which microbial colonization of the gut can influence brain development in an age-, region- and sex-specific manner.
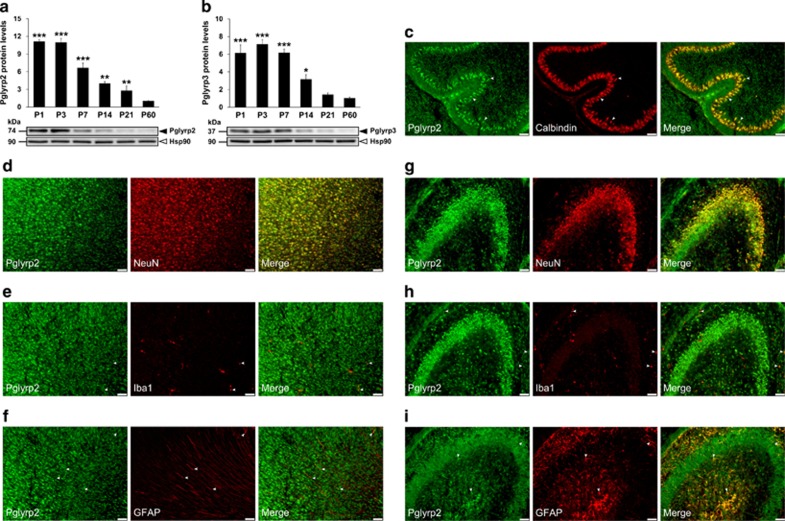
To characterize the protein expression levels of some PGN-sensing molecules (that is, Pglyrp2 and Pglyrp3) in the developing brain, we performed western blot analysis on striatal tissue of male mice at the same time points that were selected for the gene expression analysis (that is, P1, P3, P7, P14, P21 and P60). In the striatum, both Pglyrp2 and Pglyrp3 protein levels were significantly higher during early postnatal life, thereafter decreasing to adult levels (Figures 2a and b, respectively). Overall, the protein profiles of Pglyrp2 and Pglyrp3 were found to be similar to that of their mRNA expression levels.
Figure 2.
Expression and cellular localization of PGRPs in the developing brain. Changes in striatal Pglyrp2 (a) and Pglyrp3 (b) protein levels during postnatal development. Protein levels were normalized to the Hsp90 protein levels and expressed relative to the adult group (that is, P60; n=4 per group). All data (a and b) are presented as means (±s.e.m.). *P<0.05, **P<0.01 and ***P<0.001 when compared with their respective adult groups (that is, P60). Distribution of Pglyrp2 in the cerebellum (c), frontal cortex (d–f) and hippocampus (g–i) at P3. Double immunofluorescence staining shows staining of Pglyrp2 (green) combined with either NeuN (a neuronal marker; red), Iba1 (a microglial marker; red), GFAP (an astrocyte marker; red) or calbindin (a marker for purkinje cells; red). The merged images show the colocalization of Pglyrp2 (green) and NeuN (red) in the frontal cortex (d) and hippocampus (g) and with calbindin (red) in the cerebellum (c). Note that Pglyrp2 is moderately colocalization with astrocytes (f and i), and with microglia to a lesser extend (e and h). Arrows indicate Pglyrp2-positive Purkinje cells (c), Pglyrp2-positive microglia (e and h) and Pglyrp2-positive astrocytes (f and i). The scale bars represent 50 μm. Hsp, heat shock protein; P, postnatal day; PGRP, peptidoglycan-recognition protein.
Next, we examined the cellular expression and localization of Pglyrp2 and Nod1 in various brain regions of male mice, at their high level of expression (that is, P3 and P21, respectively), using immunohistochemical methods. Consistent with our expression data, both Pglyrp2 and Nod1 protein were expressed in multiple brain regions, including the prefrontal cortex, hippocampus and cerebellum (Figure 2 and Supplementary Figure 3, respectively). To determine whether these PGN-sensing molecules are expressed by neurons, astrocytes and/or microglia, we performed double staining with anti-NeuN (neuron-specific marker) antibody, anti-GFAP (a marker for astrocytes) or anti-Iba1 (a marker for microglia) antibody. In all regions examined, we found that both Pglyrp2 and Nod1 were highly expressed in the cell membrane and cytoplasm of neurons (for example, pyramidal neurons in both the cortex and hippocampus). In addition, Pglyrp2 and Nod1 were moderately expressed in astrocytes (Figure 2 and Supplementary Figure 3), and in microglia (albeit at very low levels; Figure 2 and Supplementary Figure 3). In the cerebellum, both Pglyrp2 and Nod1 were colocalized with Calbindin-positive Purkinje cells (Figure 2c and Supplementary Figure 3f, respectively).
Can the expression of PGN-sensing molecules in the neonate brain be altered by manipulations of gut microbiota?
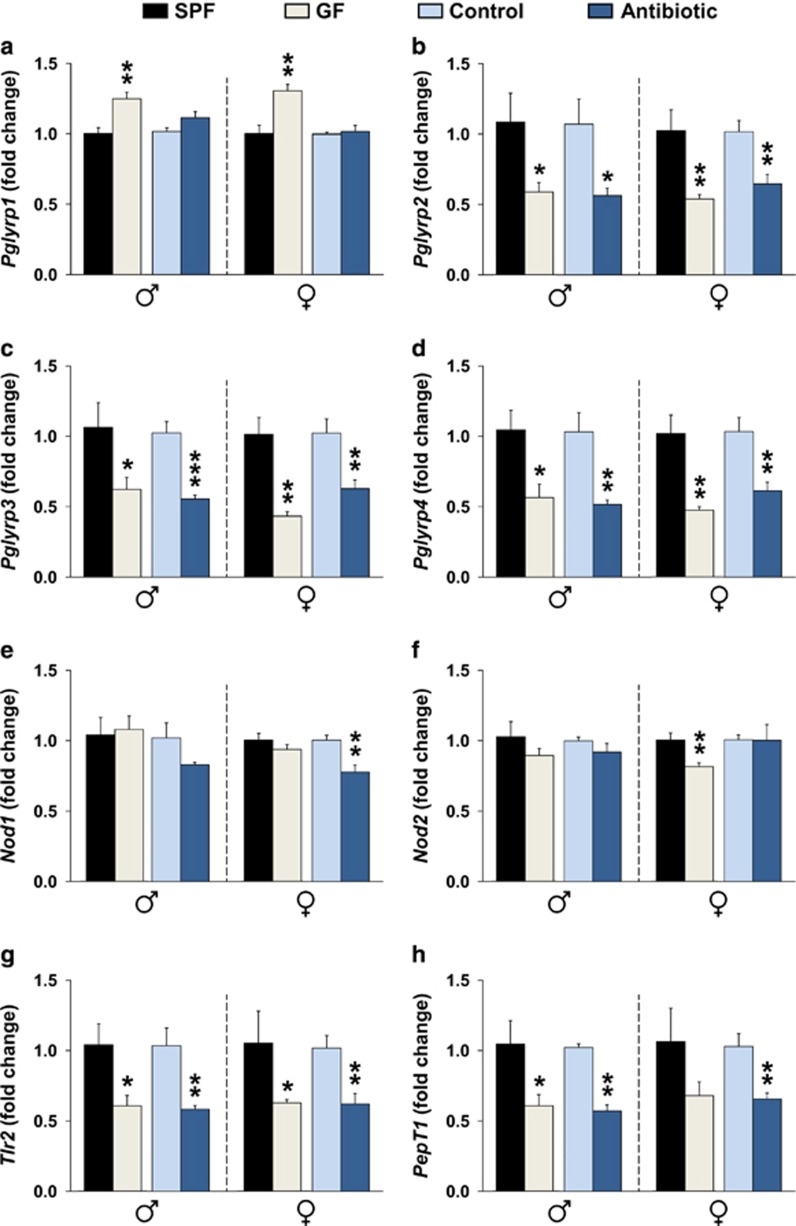
We next evaluated whether the brain-specific expression of PGRPs, NLRs, Tlr2 and PepT1 is sensitive to manipulations of the gut microbiota. For this purpose, we compared the expression of these molecules in two experimental models, namely, C57BL/6 GF mice and maternal antibiotic exposure during the last week of gestation and first 3 days postpartum. The latter model is more clinically relevant as GF conditions are not translatable to humans. Moreover, there is increasing concern that exposure to antibiotics during pregnancy and/or lactation may impact the maternal microbiome and thereby influence the assembly of the infant microbiome (via vertical mother-neonate transfer) and subsequent 'early-life' microbiome–brain interactions.22, 23, 24, 25 Importantly, we found that striatal tissues derived from 3-day-old offspring of antibiotic-treated dams had lower PGN levels than did 3-day-old offspring of control dams (Supplementary Figure 4).
In the striatum of 3-day-old mice, we found that the expression levels of Pglyrp2–4, and Tlr2 were significantly lower in both GF and perinatal antibiotic-treated male and female mice than in their respective control groups (Figures 3b–d, g). Similarly, the expression levels of PepT1 were significantly lower in perinatal antibiotic-treated male and female mice, and in GF male mice compared with their respective control groups (Figure 3h). In contrast, the expression levels of Pglyrp1 were significantly higher in GF male and female mice, but not in the perinatal antibiotic model. The expression levels of Nod1 were significantly lower in perinatal antibiotic-treated female (Figure 3e), whereas the levels of Nod2 were significantly lower in GF female mice (Figure 3f). No other significant changes in NLR expression were found. Taken together, these results indicate that several PGN-sensing molecules in the brain are sensitive to perturbations of the gut microbiota early in life.
Figure 3.
Manipulation of the gut microbiota alters the brain-specific expression of peptidoglycan-sensing molecules early in life. Gene expression levels of Pglyrp1 (a), Pglyrp2 (b), Pglyrp3 (c), Pglyrp4 (d), Nod1 (e), Nod2 (f), Tlr2 (g) and PepT1 (h) in the striatum of three-day-old germ-free (GF) and perinatal antibiotic-treated male and female mice. All data (a–h) are presented as means (±s.e.m.). Significant differences between GF (n=6 for both males and females) and specific pathogen free (SPF; n=5 for both males and females) mice and between perinatal antibiotic-treated (n=5 for males, and n=6 for females) and control (n=5 for males, and n=6 for females) mice are indicated as *P<0.05, **P<0.01 and ***P<0.001.
Does genetic disruption of the PGN recognition protein 2 alter the expression of synapse-related genes and behavior?
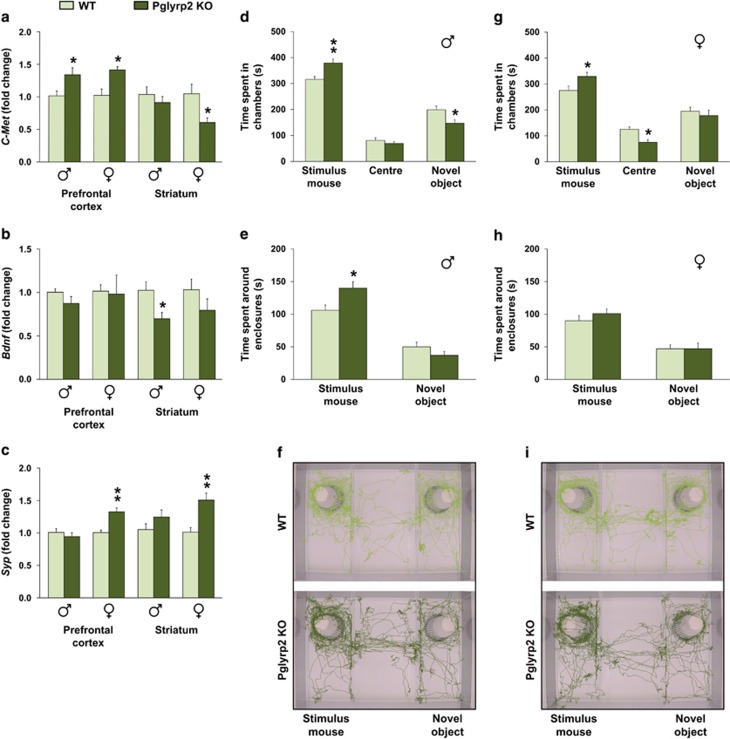
To begin addressing the potential influence of PGN-sensing molecules on brain development and behavior, we decided to focus on Pglyrp2 given that this molecule is an N-acetylmuramyl-l-alanine amidase that hydrolyzes bacterial PGN between the sugar backbone and the peptide chain (which is unique among the mammalian PGRPs26). In addition, Pglyrp2 is highly expressed during the first few days of life and also sensitive to manipulations of the gut microbiota. Using transgenic mice, we first investigated the effect of deleting Pglyrp2 on the expression of the autism risk gene c-Met and brain-derived neurotrophic factor (Bdnf), both implicated in the formation and modulations of brain circuits.27, 28 The expression of synaptophysin (Syp) was also examined, as an indirect marker of synaptogenesis in the developing brain.29 Importantly, both c-Met and Bdnf are highly sensitive to manipulations of the gut microbiota (Supplementary Figure 5).
In the developing prefrontal cortex, the expression levels of c-Met were significantly increased in Pglyrp2 KO male and female mice compared with their respective controls (Figure 4a). In contrast, in the striatum, c-Met levels were significantly reduced in Pglyrp2 KO female mice (Figure 4a), but not in males. On the other hand, the expression levels of Bdnf were significantly reduced only in the striatum of Pglyrp2 KO male mice (Figure 4b). The expression levels of Syp were significantly increased in both the prefrontal cortex and striatum of Pglyrp2 KO female mice compared with their respective controls (Figure 4c). Taken together, these results suggest that Pglyrp2 is involved in regulating the development and formation of brain circuits (for example, striatum) in a sex-dependent manner.
Figure 4.
Genetic disruption of the PGN recognition protein 2 alters the expression of synapse-related genes and social behavior. Gene expression levels of c-Met (a), Bdnf (b) and Syp (c) in the prefrontal cortex and striatum of 3-day-old Pglyrp2 knockout (KO) and wild-type (WT) male and female mice (n=5 for male Pglyrp2 KO mice, n=6 for female Pglyrp2 KO mice and n=6 for WT mice). Bars show time (seconds) spent in the different chambers during the social approach session by Pglyrp2 KO and WT male (d) and female (g) mice (n=8 per group). Bars show time (seconds) spent interacting with the stimulus mouse or in close proximity to the novel object by male (e) and female (h) mice. Representative traces of movement patterns of Pglyrp2 KO (dark green) and WT (light green) male (f) and female (i) mice during the 10-min sociability test. All data (a–e, g and h) are presented as means (±s.e.m.). *P<0.05, **P<0.01 compared with their respective WT mice. PGN, peptidoglycan.
We then subjected juvenile (that is, P22–24) Pglyrp2 KO male and female mice to a battery of tests for exploratory activity, anxiety and social behavior (that is, similar battery of tests that we previously used in GF mice5). In the open field, light–dark box and elevated plus maze tests, no significant changes were observed in Pglyrp2 KO male and female mice compared with their respective WT controls (data not shown). In the three-chamber social approach test, however, juvenile Pglyrp2 KO male mice spent significantly more time in the chamber containing the novel mouse than in the chamber with the novel object, compared with control mice (P<0.01; Figure 4d). Moreover, they also spent significantly more time around the novel mouse (for example, sniffing and/or interacting with the stimulus mouse) than the wild-type males did (P<0.05; Figure 4e), indicating high sociability. Although juvenile Pglyrp2 KO female mice spent significantly more time in the chamber containing the novel mouse than in the chamber with the novel object, compared with control mice (P<0.05; Figure 4g), they spent a similar amount of time as the wild-type females around the novel mouse (Figure 4h). These results indicate that Pglyrp2 modulates the development of social behavior in mice. Given that amygdala is a key brain region involved in processing of social stimuli, we investigated the expression levels of c-Met, Bdnf and Syp in the amygdala of the behaviorally assessed juvenile Pglyrp2 KO and WT mice, as well as in the prefrontal cortex and striatum regions. Interestingly, we found that the expression levels of c-Met were altered in a region- and sex-dependent manner (Supplementary Figure 6). In the amygdala, the expression levels of c-Met were significantly increased in juvenile Pglyrp2 KO female mice compared with their WT controls, but not in juvenile Pglyrp2 KO male mice. In the striatum, we found the exact opposite pattern. No other changes were observed.
To determine whether the levels of PGN levels in the brains of Pglyrp2 KO male and female mice are altered, we used the SLP assay and assed PGN levels in the cerebellum of Pglyrp2 KO and WT male mice. The levels of PGN in the brains of Pglyrp2 KO mice were similar to that of WT mice (Supplementary Figure 7).
Discussion
Our data challenge the traditional view that microbial products translocate into the brain only under conditions of compromised BBB function (for example, bacterial or viral infections), by revealing that PGN from the commensal gut microbiota, PGN-sensing molecules, and PGN transporter are present in the normal developing brain. In addition, we show that perturbation of the gut microbiota (for example, GF condition or antibiotic treatment) alters the expression of several PGN-sensing molecules in the developing brain. Moreover, we demonstrate sex-dependent alterations in social behavior of juvenile mice lacking Pglyrp2. Based on these results, we propose that one potential mechanism mediating the interactions between the commensal gut microbiota and the developing brain, influencing life-long behavior, involves the activation of PRRs (for example, PGRPs and NLRs) of the innate immune system in the brain.
Several decades ago, PGN fragments were found in the cerebral spinal fluid of patients with sleeping disorders.30, 31 It has since been speculated that bacterial products may regulate central nervous system function in mammals, and that dysbiosis of the gut microbiota could contribute to the pathophysiology of psychiatric and neurological disorders. However, the presence of PGN in healthy brain tissues in the absence of infections has been a matter of debate.32, 33 Nevertheless, recent studies strongly indicate that PGN fragments from indigenous bacteria translocate from the intestinal gut mucosa into circulation and have the potential to affect the immune system, and homeostasis at distal sites (that is, bone marrow).10 In the present study, we were able to confirm the presence of PGN in serum of juvenile mice. Moreover, we believe our results are the first to demonstrate that PGN fragments can cross the BBB during postnatal development in healthy animals. It is worthwhile emphasizing here that the age-dependent increase in brain PGN levels, observed during postnatal development, parallels the postnatal bacterial colonization process, rather than the maturation of the BBB. Indeed, brain PGN levels are low during the first week of life at the time when the BBB is still developing. Critically, PGN-sensing molecules (that is, PGRP1-4, Nod1, Nod2 and Tlr2) are highly expressed during specific windows of postnatal brain development, suggesting that PGN fragments (for example, muramyl dipeptides and mesoDAP-containing PGN) may differentially affect brain development depending upon the postnatal age.
Importantly, we found that both Pglyrp2 and Nod1 are highly expressed in neurons in the developing mouse brain, suggesting potential direct effects of PGN on neurons. In addition, these molecules are moderately expressed in astrocytes and also in microglia, but to a much lesser extent. Previous studies have detected Nod2 receptors primarily in microglia and astrocytes in the adult rodent brain, where they play an important role in the inflammatory responses to bacterial pathogens.34 In addition, there is evidence that neonatal microglia exposed to high PGN levels produce conditioned media that promotes excessive numbers of cholinergic neurons and levels of choline acetyltransferase.35 Thus, the relatively low expression of Pglyrp2 and Nod1 in microglia is rather surprising and requires further detailed investigation. Although the Allen Mouse Brain Atlas provides some gene expression data on PGRPs (for example, Pglyrp1 and Pglyrp2), we are still lacking information about the cellular localization of Pglyrp1, Pglyrp3 and Pglyrp4 at the protein level in the developing brain. Unfortunately, we found very few reliable antibodies commercially available to examine PGRPs in brain tissues.
PRRs have long been recognized as important mediators for the initiation of immune responses during infections. However, there is growing appreciation that in addition to their immune function, PRRs are able to modulate neurodevelopmental processes. The pioneering work of Carla Shatz and collaborators clearly demonstrated that genes involved in immunity such as the major histocompatibility complex class I and PirB are also present in neurons, where they have a role in synaptic plasticity, learning, memory and neurological disorders.36, 37, 38 Moreover, several studies have demonstrated that some Toll-like receptors (TLRs) that are strongly expressed during mouse brain development are able to modulate key neurodevelopmental processes. For example, Tlr3 and Tlr8 have been implicated in neurite outgrowth and neuronal death,39, 40 whereas Tlr2 and Tlr4 have been shown to regulate neuronal differentiation and cell proliferation.41, 42 The growing body of evidence regarding the involvement of TLRs in several dynamic processes in the developing brain suggests that TLRs might also impact motor, social and cognitive functions later in life. Indeed, it was recently shown that working memory,43, 44 motor learning and anxiety are broadly influenced by Tlr3, and that Tlr4 has a role in shaping spatial reference memory, fear learning and memory. The present results expand previous findings by showing that PGN-sensing molecules are also highly expressed in the developing mouse brain in an age-, region- and sex-specific manner. Interestingly, some PGRPs (that is, Pglyrp2, Pglyrp3 and Pglyrp4) are expressed at higher levels in females than in males, whereas NLRs and Pglyrp1 are expressed at higher levels in males. These findings may have clinical implications in terms of sex-specific susceptibility observed in many neurodevelopmental disorders and psychiatric disorders.
The exact function of PGRPs in mammals remains poorly understood. The available evidence from non-mammalian animals suggests that some PGRPs may promote development of the immune system and host tissues, as well as protection from pathogen infections.9, 26 To our knowledge, this is the first study to demonstrate that Pglyrp2 has an impact on brain development and social behavior. In the absence of Pglyrp2, mice exhibit higher levels of sociability in the three-chamber social approach test. This phenotype is more pronounced in male than in female Pglyrp2 KO mice. In line with these findings, we recently found that adult GF mice exhibit increased levels of sociability compared to SPF mice13 (but see the study by Desbonnet and colleagues8). Moreover, GF mice show reduced striatal expression levels of Pglyrp2 early in life. Interestingly, we also discovered that the absence of Pglyrp2 leads to alterations in the expression of genes involved in the formation of synapses (for example, Bdnf and c-Met), suggesting that PGN-sensing molecules may have a neurodevelopmental role, as previously shown for some members of the TLR family.45 Given that the striatum has been implicated in integrating social information into coding of social action and reward,46 it is tempting to speculate that the aforementioned changes in synapse-related gene expression in the developing striatum could partly underlie alterations in social behavior later in life. Interestingly, juvenile Pglyrp2 KO male, but not female, mice exhibit alterations in c-Met gene expression in the striatum. Surprisingly, we found very few changes in the amygdala, a key brain region involved in social behavior. However, we only examined a limited number of candidate genes (that is, c-Met, Bdnf and Syp), and further studies using unbiased, whole-transcriptome profiling are needed to identify other target genes regulated by Pglyrp2.
The regulation of c-Met is of particular interest given that this gene has been associated with a subset of individuals with co-occurring autism spectrum disorder and gastrointestinal conditions.47 Previous studies have shown that c-Met is involved in multiple biological process, including cell proliferation, angiogenesis, immune function, intestinal epithelial development48 and brain development.27, 49 The observation that the expression of c-Met was altered in the striatum of neonate GF mice and perinatal antibiotic-treated mice raises the possibility that alterations in the gut microbiota, during key developmental time windows, may increase the risk of neurodevelopmental disorders via regulation of the expression of c-Met and other risk genes.
Pglyrp2 is the only mammalian PGRP with bacterial PGN hydrolyzing activity.26 We therefore expected Pglyrp2 KO mice to have higher PGN levels than WT. However, we observed similar PGN levels in the cerebellum of Pglyrp2 KO and WT mice by means of the SLP assay. Using commercially available PGN ligands, we were able to confirm that the SLP assay detects muramyl peptides (for example, MDP, MurNAc-L-Ala-D-isoGln), but not the Nod1 ligand Tri-DAP (L-Ala-γ-D-Glu-mDAP). Therefore, the SLP assay is not optimal to detect the products of hydrolysis by Pglyrp2 (for example, Nod1-activating peptides).
Although the present study provides, to our knowledge, the first evidence supporting a new connection between microbiome, bacterial PGN-sensing molecules of the innate immune system, brain development and behavior, it also has several limitations that must be acknowledged. First, as mentioned earlier, the SLP assay has important limitations with regards to its use in the brain, especially in high dopamine innervated regions. Therefore, there is a great need in the microbiome field to develop more sophisticated assays, such as liquid chromatography coupled to mass spectrometry, to determine the specific type of PGN fragments that are found in the developing brain. Second, the effects of antibiotics are complex (for example, can affect production of short-chain fatty acids, and mitochondrial function)50 and we cannot exclude the possibility that other indirect effects on the brain may affect the expression of PRRs in the developing brain. Third, future research is needed to more fully characterize the influence that the natural ligands of PGN-sensing molecules have on brain development and behavior, as well as their downstream gene targets.
It is becoming increasingly clear that a more complete understanding of the mechanisms mediating the early life microbiome gut–brain communication is required to develop more promising microbiota-modulating-based therapeutic interventions for neurodevelopmental and psychiatric disorders. Several mechanisms have been proposed to mediate the communication between the commensal gut microbiota and the developing brain, and these include (i) signals via neuronal circuits (for example, bidirectional vagus nerve-to-brain communication), (ii) activation of immune responses within the gut or elsewhere that consequently influence the brain and (iii) the production of microbiota-derived metabolites that directly influence the brain, including bacterial fermentation products, such as butyrate and propionate.51, 52, 53 The present study suggests a new signaling pathway in which neuronal PRRs are activated by microbial products (for example, PGN fragments). This is supported by the following observations: (1) PGN from the non-pathogenic gut microbiota can cross the BBB, and parallel to the postnatal microbial colonization process, PGN levels increase with age; (2) PGN-sensing molecules are highly expressed in neurons during early postnatal development, and their expression is sensitive to manipulations of the gut microbiota (that is, GF conditions and perinatal antibiotic treatment); (3) interruption of one of the PRR pathways (for example, by using genetic manipulation of the Pglyrp2) induces behavioral changes and alterations in the expression of the autism risk gene c-Met that are similar to those observed in GF mice and perinatal antibiotic-exposed mice. Given the fact that PRRs seem to have a dual role in immunity and in neural development, it is tempting to speculate that disruption of the gut microbiota may render the developing brain not only more susceptible to neurodevelopmental disorders, but also increase the risk for immune disturbances. In conclusion, our findings suggest that a role of non-pathogenic bacterial-PGN should now also be considered in the context of normal central nervous system development and function.
Acknowledgments
This work was supported by the Olle Engkvist Byggmästare Foundation, Swedish Research Council (grant no. K2015-62X-22745-01-4), Strategic Research Program in Neuroscience at Karolinska Institutet, Swedish Brain Foundation and Knut and Alice Wallenberg Foundation. KU acknowledges support from the Swedish Research Council (#621-2012-3564). TA is the recipient of a PhD training grant from the Karolinska Institutet.
Author contributions
TA, HF and RDH designed the study. TA performed the gene, protein expression profiling, the behavioral experiments and SLP assay. YQ performed and analyzed the immunohistochemistry experiments and contributed to the gene profiling experiments. SG and KU contributed to the initial studies relating to the SLP assay. TF contributed to pilot studies relating to the antibiotic treatment. TW contributed to the immunohistochemistry experiments. TA, HF and RDH wrote the paper with input from all authors.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol 2004; 12: 129–134. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science 2001; 292: 1115–1118. [DOI] [PubMed] [Google Scholar]
- Cash HL, Hooper LV. Commensal bacteria shape intestinal immune system development. ASM News 2005; 71: 6. [Google Scholar]
- Sommer F, Backhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 2013; 11: 227–238. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 2011; 108: 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil 2011; 23: e119. [DOI] [PubMed] [Google Scholar]
- Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry 2012; 18: 666–673. [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry 2014; 19: 146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 2013; 14: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010; 16: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wang Z, Locksley RM. Innate immune responses in peptidoglycan recognition protein L-deficient mice. Mol Cell Biol 2004; 24: 7949–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Asahi N, Suzuoki F, Ashida M, Matsuura S. Detection of peptidoglycan and beta-glucan with silkworm larvae plasma test. FEMS Immunol Med Microbiol 1996; 15: 129–134. [DOI] [PubMed] [Google Scholar]
- Arentsen T, Raith H, Qian Y, Forssberg H, Diaz Heijtz R. Host microbiota modulates development of social preference in mice. Microb Ecol Health Dis 2015; 26: 29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Forssberg H, Diaz Heijtz R. Motor skill learning is associated with phase-dependent modifications in the striatal cAMP/PKA/DARPP-32 signaling pathway in rodents. PLoS One 2015; 10: e0140974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Chen M, Forssberg H, Diaz Heijtz R. Genetic variation in dopamine-related gene expression influences motor skill learning in mice. Genes Brain Behav 2013; 12: 604–614. [DOI] [PubMed] [Google Scholar]
- Yang M, Silverman JL, Crawley JN. Three-chambered social approach task for mice. Curr Protoc Neurosci 2011. Chapter 8, Unit 8.26. [DOI] [PMC free article] [PubMed]
- Hasegawa M, Osaka T, Tawaratsumida K, Yamazaki T, Tada H, Chen GY et al. Transitions in oral and intestinal microflora composition and innate immune receptor-dependent stimulation during mouse development. Infect Immun 2010; 78: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. Review: Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate immun 2010; 16: 168–174. [DOI] [PubMed] [Google Scholar]
- Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol 2014; 14: 9–23. [DOI] [PubMed] [Google Scholar]
- Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol 2007; 5: 264–277. [DOI] [PubMed] [Google Scholar]
- Ingersoll SA, Ayyadurai S, Charania MA, Laroui H, Yan Y, Merlin D. The role and pathophysiological relevance of membrane transporter PepT1 in intestinal inflammation and inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol 2012; 302: G484–G492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M. Antibiotic overuse: stop the killing of beneficial bacteria. Nature 2011; 476: 393–394. [DOI] [PubMed] [Google Scholar]
- Ledger WJ, Blaser MJ. Are we using too many antibiotics during pregnancy? BJOG 2013; 120: 1450–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 2010; 51: 77–84. [DOI] [PubMed] [Google Scholar]
- Stokholm J, Schjorring S, Eskildsen CE, Pedersen L, Bischoff AL, Folsgaard N et al. Antibiotic use during pregnancy alters the commensal vaginal microbiota. Clin Microbiol Infect 2014; 20: 629–635. [DOI] [PubMed] [Google Scholar]
- Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol 2011; 11: 837–851. [DOI] [PubMed] [Google Scholar]
- Peng Y, Huentelman M, Smith C, Qiu S. MET receptor tyrosine kinase as an autism genetic risk factor. Int Re Neurobiol 2013; 113: 135–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci 2013; 14: 7–23. [DOI] [PubMed] [Google Scholar]
- Becher A, Drenckhahn A, Pahner I, Margittai M, Jahn R, Ahnert-Hilger G. The synaptophysin-synaptobrevin complex: a hallmark of synaptic vesicle maturation. J Neurosci 1999; 19: 1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Karnovsky ML, Krueger JM, Pappenheimer JR, Biemann K. Peptidoglycans as promoters of slow-wave sleep. I. Structure of the sleep-promoting factor isolated from human urine. J Biol Chem 1984; 259: 12652–12658. [PubMed] [Google Scholar]
- Krueger JM, Karnovsky ML, Martin SA, Pappenheimer JR, Walter J, Biemann K. Peptidoglycans as promoters of slow-wave sleep. II. Somnogenic and pyrogenic activities of some naturally occurring muramyl peptides; correlations with mass spectrometric structure determination. J Biol Chem 1984; 259: 12659–12662. [PubMed] [Google Scholar]
- Sen Z, Karnovsky ML. Qualitative detection of muramic acid in normal mammalian tissues. Infect Immun 1984; 43: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R, Chevalier G, Eberl G, Gomperts Boneca I. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell Microbiol 2014; 16: 1014–1023. [DOI] [PubMed] [Google Scholar]
- Liu X, Chauhan VS, Marriott I. NOD2 contributes to the inflammatory responses of primary murine microglia and astrocytes to Staphylococcus aureus. Neurosci Lett 2010; 474: 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Acevedo G, Muralidharan B, Padala N, To J, Jonakait GM. Toll-like receptor ligands and CD154 stimulate microglia to produce a factor(s) that promotes excess cholinergic differentiation in the developing rat basal forebrain: implications for neurodevelopmental disorders. Pediatr Res 2007; 61: 15–20. [DOI] [PubMed] [Google Scholar]
- Lee H, Brott BK, Kirkby LA, Adelson JD, Cheng S, Feller MB et al. Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature 2014; 509: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djurisic M, Vidal GS, Mann M, Aharon A, Kim T, Ferrao Santos A et al. PirB regulates a structural substrate for cortical plasticity. Proc Natl Acad Sci USA 2013; 110: 20771–20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger LM, Shatz CJ. Immune signalling in neural development, synaptic plasticity and disease. Nat Rev Neurosci 2004; 5: 521–531. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Okun E, Tang SC, Griffioen K, Cheng A, Mughal MR et al. Toll-like receptor 3 is a negative regulator of embryonic neural progenitor cell proliferation. J Neurosci 2008; 28: 13978–13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Li J, Chiu I, Wang Y, Sloane JA, Lu J et al. Toll-like receptor 8 functions as a negative regulator of neurite outgrowth and inducer of neuronal apoptosis. J Cell Biol 2006; 175: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun E, Griffioen KJ, Son TG, Lee JH, Roberts NJ, Mughal MR et al. TLR2 activation inhibits embryonic neural progenitor cell proliferation. J Neurochem 2010; 114: 462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter R, Ronen A, Rolls A, London A, Bakalash S, Young MJ et al. Toll-like receptor 4 restricts retinal progenitor cell proliferation. J Cell Biol 2008; 183: 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun E, Barak B, Saada-Madar R, Rothman SM, Griffioen KJ, Roberts N et al. Evidence for a developmental role for TLR4 in learning and memory. PLoS One 2012; 7: e47522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun E, Griffioen K, Barak B, Roberts NJ, Castro K, Pita MA et al. Toll-like receptor 3 inhibits memory retention and constrains adult hippocampal neurogenesis. Proc Natl Acad Sci USA 2010; 107: 15625–15630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun E, Griffioen KJ, Mattson MP. Toll-like receptor signaling in neural plasticity and disease. Trends Neurosci 2011; 34: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez-Mendoza R, Schultz W. The role of the striatum in social behavior. Front Neurosci 2013; 7: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM et al. Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions. Pediatrics 2009; 123: 1018–1024. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol 2003; 4: 915–925. [DOI] [PubMed] [Google Scholar]
- Qiu S, Lu Z, Levitt P. MET receptor tyrosine kinase controls dendritic complexity, spine morphogenesis, and glutamatergic synapse maturation in the hippocampus. J Neurosci 2014; 34: 16166–16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFabe DF. Enteric short-chain fatty acids: microbial messengers of metabolism, mitochondria, and mind: implications in autism spectrum disorders. Microb Ecol Health Dis 2015; 26: 28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 2012; 10: 735–742. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 2012; 13: 701–712. [DOI] [PubMed] [Google Scholar]
- Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest 2015; 125: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.