Abstract
Cellular membranes display a diversity of functions that are conferred by the unique composition and organization of their proteins and lipids. One important aspect of lipid organization is the asymmetric distribution of phospholipids across the plasma membrane. The unequal distribution of key phospholipids between the cytofacial and exofacial leaflets of the bilayer creates physical surface tension that can be used to bend the membrane; and like Ca2+, a chemical gradient that can be used to transduce biochemical signals. Phospholipid flippases in the type IV P-type ATPase (P4-ATPase) family are the principle transporters used to set and repair this phospholipid gradient, and the asymmetric organization of these membranes are encoded by the substrate specificity of these enzymes. Thus, understanding the mechanisms of P4-ATPase substrate specificity will help reveal their role in membrane organization and cell biology. Further, decoding the structural determinants of substrate specificity provides investigators the opportunity to mutationally tune this specificity to explore the role of particular phospholipid substrates in P4-ATPase cellular functions. The present work reviews the role of P4-ATPases in membrane biology, presents our current understanding of P4-ATPase substrate specificity, and discusses how these fundamental aspects of P4-ATPase enzymology may be used to enhance our knowledge of cellular membrane biology.
Keywords: P4-ATPase, phospholipid flippase, membrane biology, membrane asymmetry, protein engineering, phospholipid transport
Introduction
Cellular membranes are the key medium that eukaryotic cells use to communicate with their external environment and segregate internal processes. The lipid components of the membrane are largely amphipathic glycerophospholipids (GPLs), sphingolipids (SLs), and sterols, which self-associate into bilayers in an aqueous environment. These membrane bilayers facilitate a variety of essential cellular functions such as encapsulating organelles for partitioned function, forming discrete vesicles for trafficking between organelles, and establishing the chemical environment that encompasses ion channels, transporters, and transmembrane receptors alike. Cellular membranes achieve this diversity of functions through differences in chemical composition and thus physical properties (van Meer et al., 2008). For example, sterols are enriched in the plasma membrane (PM) to rigidify and improve the seal of this important protective barrier. The endoplasmic reticulum (ER) is a more malleable structure rich in cis-unsaturated GPLs that produce a fluid environment for protein insertion and membrane synthesis (Holthuis and Menon, 2014). Further, as membrane flows through the Golgi toward the cell surface, it is substantially remodeled through SL synthesis, sterol loading, and the generation of phospholipid (PL) asymmetry (Graham and Burd, 2011). These examples and more underline the clear influence of membrane biology over cellular function, aspects that we are just beginning to fully appreciate.
Cellular membranes are capable of influencing biology through their organization as well as their composition. Studies over the last 40 years have established that different lipid species are asymmetrically distributed across cellular membranes, highlighting the importance of the trans-bilayer chemical composition of the PM and trans-Golgi network (TGN) (Devaux, 1991, Panatala et al., 2015). For example, the asymmetric distribution of phosphatidylserine (PS) has been demonstrated to be a critical signaling molecule in numerous cellular processes. PS is an anionic GPL that is typically maintained on the cytofacial aspect of the trans-Golgi, endosomes, and PM (Fairn et al., 2011). The dynamic trans-bilayer regulation of PS distribution across membranes influences the formation of transport vesicles (Chen et al., 1999, Gall et al., 2002), blood clotting mechanisms (Bevers et al., 1982, van Geffen et al., 2016), recognition of apoptotic cells (Fadok et al., 1992, Fadok et al., 1993), cell-cell fusion (van den Eijnde et al., 2001), cytoskeletal scaffolding (Comfurius et al., 1989, Manno et al., 2002), signal transduction (Yeung et al., 2006, Kay and Grinstein, 2013), phagocytosis (Fadok et al., 1998, Hoffmann et al., 2001), and viral-host entry (Chen et al., 2015, Richard et al., 2015). The cell achieves this dynamic regulation of PS asymmetry through a series of lipid transporters.
Three key classes of transporters have been demonstrated to influence the asymmetric distribution of lipids within the PM, i) flippases, ii) floppases, and iii) scramblases. PL flippases are type IV P-type ATPases (P4-ATPases), a family of enzymes that translocate PL substrates from the exofacial or lumenal aspect to the cytofacial leaflet of the membrane (Coleman et al., 2009, Zhou and Graham, 2009). Floppases are members of the ABC transporter family that have been demonstrated to translocate lipid species from the cytofacial to the exofacial aspect of the PM (Hamon et al., 2000, Pohl et al., 2005, Coleman et al., 2013). Scramblases, including Xkr8 and TMEM16F, have been demonstrated to non-discriminately dissipate the asymmetric phospholipid (PL) gradient between membrane leaflets (Bevers and Williamson, 2010, Suzuki et al., 2013, Fujii et al., 2015). Interestingly, a recent study has even highlighted the possibility that membrane asymmetry may also be more broadly influenced through a scramblase-like activity elicited by G-protein coupled receptors (Goren et al., 2014). Together, these three types of transporters set, repair, and dissipate the trans-bilayer distribution of lipids across the plasma membrane.
The asymmetric distribution of lipids across cellular membranes forms a chemical gradient that the cell uses for signaling. The establishment and dissipation of this chemical gradient is a dynamic process, as illustrated by the translocation of PS during apoptosis. Under healthy cell conditions a PS gradient is generated by flippases that maintain the lipid on the cytofacial leaflet of the PM. Yet during apoptotic cascades, caspase signaling events cooperate to inhibit the activity of the flippases while activating PM scramblases, thereby dissipating the trans-bilayer distribution and extruding PS to the outer leaflet (Segawa et al., 2014, Segawa and Nagata, 2015, Nagata et al., 2016). Once on the outer leaflet of the PM, PS is recognized by a diverse collection of receptors on macrophages that induce phagocytosis of the apoptotic cell (Poon et al., 2014, Amara and Mercer, 2015). These signaling events serve as examples of how membrane architecture can encode important chemical gradients, and are not unlike how the cell uses ion gradients to transduce signals.
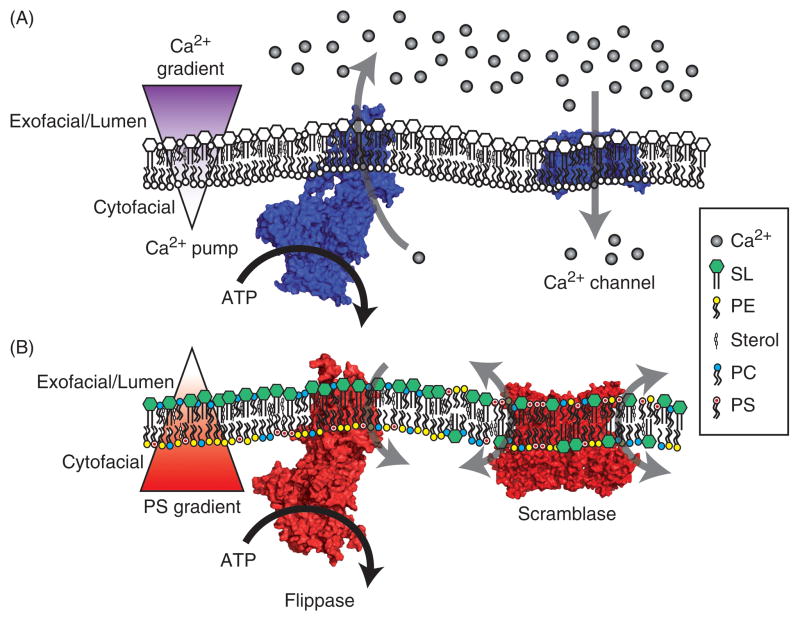
Eukaryotic cells regulate trans-bilayer PL distribution in a way that is similar to how Ca2+ gradients are established and used to transduce signals. In each case there are substrate pumps actively working against substrate gradients to achieve polarized distributions, with Ca2+ cleared from the cytosol and PS pumped to the inner leaflet of the PM [Figure 1]. Both mechanisms use P-type ATPases as the key enzymes that help set the PL and Ca2+ gradients of a cell. Type-IIA/B P-type ATPases (P2A/B-ATPases such as SERCA1 or PMCA1) actively transport Ca2+ from the cytosol [Figure 1A], while the aforementioned P4-ATPases translocate PLs [Figure 1B] (Palmgren and Nissen, 2011). Once the gradients are established, Ca2+ channels and scramblases are the mediators that transduce this chemical potential into a signaling event, opening ion selective channels [Figure 1A] (Simms and Zamponi, 2014) or facilitating the non-discriminate equilibration of PLs across the bilayer [Figure 1B] (Montigny et al., 2016). The affectors and effectors of Ca2+ depolarization are diverse and well-described; however, far less is known regarding the initiation and consequences of PL depolarization.
Figure 1. A comparison of Ca2+ and phospholipid gradients.
A) Cellular Ca2+ is tightly regulated. At rest, Ca2+ is actively transported into the lumen of the ER and across the PM to the extracellular fluid by Ca2+ pumps, creating a polarized gradient of Ca2+ ions across the membrane. When activated, Ca2+ channels open to harness this chemical potential and transduce cellular signals by rapidly increasing the cytosolic Ca2+ concentration. B) Similar to the establishment of the Ca2+ gradient, lipid flippases (P4-ATPases) will selectively transport glycerophospholipid species from the lumenal or exofacial aspect to the cytofacial leaflet, creating an asymmetric distribution of phospholipids across the membrane. In this example, PS is retained on the cytofacial leaflet of the membrane. Once this PS gradient is set, scramblase activation will non-selectively translocate lipid species between the leaflets of the bilayer to transduce cellular signals. In legend: calcium (Ca2+), sphingolipid (SL), phosphatidylethanolamine (PE), cholesterol in mammals or ergosterol in yeast (sterol), phosphatidylcholine (PC), phosphatidylserine (PS). Ca2+ pump: sarco/endoplasmic reticulum Ca2+ ATPase (PDB: 3W5D). Scramblase: TMEM16F (PDB: 4WIT). Ca2+ channel: voltage-gated Ca2+ channel (PDB: 4MS2). Flippase: Dnf1 homology model (Roland & Graham, 2016). Color figures can be found online.
Calcium gradients and their signaling potential have been examined extensively, in part because a host of technologies are available for the measurement and modulation of Ca2+. Calcium chelators, small molecules that bind and sequester Ca2+ from its target molecules, have been leveraged for nearly half a century to blunt the effects of Ca2+ when dissecting cellular pathways (Bers et al., 1994). Further, various pharmacological strategies have been developed for the manipulation of Ca2+ channels, transporters, and signaling pathways (Bers et al., 1994). Comparatively, the field of membrane biology has been limited by the molecules that we can uniquely perturb and the tools for quantifying lipid species in subcellular compartments of living cells. These studies have often relied on metabolic strategies, such as knockdown or inhibition of biosynthetic enzymes, to study the influence of individual lipid molecules on a pathway of interest. Yet with more than 1,000 different lipid species in a eukaryotic cell sharing metabolic pathways and shunts, these approaches can lead to uncertainty surrounding the affector lipid species (Han, 2016).
P4-ATPases provide an opportunity to specifically modulate the architecture of a cellular membrane, rather than the overall composition. P4-ATPases are localized to the Golgi, endosomes, and plasma membrane, giving them broad access to several critical cellular membranes (Lopez-Marques et al., 2014). As will be detailed later, this essential enzyme family translocates specific substrates to remodel the membrane bilayer. In addition, P4-ATPases can be mutationally tuned to alter their PL substrate specificity (Baldridge and Graham, 2012, Baldridge and Graham, 2013, Baldridge et al., 2013, Roland and Graham, 2016). These designer P4-ATPases have already proven useful for determining the influence of flipping specific substrates on vesicle trafficking and lateral membrane organization (Xu et al., 2013, Hankins et al., 2015b). This review will discuss the enzymology of P4-ATPases, their substrate specificities, and how forward genetic approaches have helped generate an increasing number of designer flippases with unique substrate specificities. We discuss an ongoing debate over the physical nature of P4-ATPase substrate transport, and review new literature that provides insight regarding the primary structural mechanisms of substrate discrimination. Finally, we will highlight recent applications of designer P4-ATPases and project possible strategies to address existing challenges within the field of cellular membrane biology.
P4-ATPase domain organization and catalytic cycle
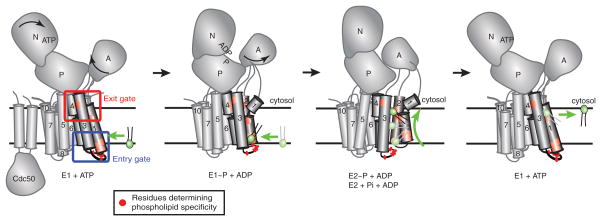
Most P4-ATPases are heterodimers consisting of a catalytic α subunit and a noncatalytic β subunit in the Cdc50 protein family. The α subunits have the same general architecture of all P-type ATPases with cytosolic domains mediating nucleotide binding (N), phosphorylation (P) and dephosphorylation (A). The membrane domain of the P4-ATPase α subunits consists of 10 transmembrane (TM) segments and the β subunits provide two more TM segments along with a glycosylated ectodomain [Figure 2]. The β subunit is required for transport of the P4-ATPase from the ER to the Golgi and also influences the catalytic cycle of the pump (Saito et al., 2004, Lenoir et al., 2009, Coleman and Molday, 2011).
Figure 2. P4-ATPase domain organization and proposed 2-gate mechanism of phospholipid translocation.
Most P4-ATPases are composed of a catalytic α subunit and a noncatalytic β subunit (Cdc50). Movements of the nucleotide binding (N), phosphorylation (P), actuator (A) and membrane domain helices during catalysis are modeled after SERCA1 structures trapped in the indicated conformational states. Clusters of residues that determine substrate specificity in TM1-4 labeled as the entry and exit gates. A substrate phospholipid is shown in green using the “Credit Card” model of transport to the cytosolic leaflet. Color figures can be found online.
ATP hydrolysis by P-type ATPases occurs in two steps: after binding of ATP to the N domain, the γ phosphate group is first transferred to an aspartate in the P domain, and the aspartyl-phosphate bond is then hydrolyzed by the A domain. These chemical events drive conformational changes in the pumps classically described by the Post-Albers cycle (see Figure 2 and (Montigny et al., 2016) for a more complete review of the Post-Albers cycle). For the P4-ATPases, no PL substrate interaction appears to be required for the initial ATP binding and phosphorylation events (E1 → E1~P) (Xie et al., 1989, Coleman et al., 2012), in contrast to the Na+/K+ ATPase (a P2C-ATPase) for which Na+ greatly stimulates phosphorylation (Palmgren and Nissen, 2011). The P4-ATPases can spontaneously transition from E1~P to the lower energy E2~P conformation, but then require PL substrate binding for efficient dephosphorylation (E2~P → E2), which is analogous to the influence of K+ on the Na+/K+ ATPase (Coleman et al., 2012). PL substrate is likely flipped across the membrane to the cytosolic side during the E1~P → E2~P → E2 transitions, and may be released into the cytosolic leaflet from the pump as it resets back to the E1 state [Figure. 2].
Cation pumps bind substrate in the center of the membrane domain using charged and polar residues in TM segments 4, 5, 6, 8 and 9 (Toyoshima, 2009, Palmgren and Nissen, 2011). For SERCA1, two Ca2+ ions enter from the cytosol via an open half-channel to their binding site in the E1 conformation, but they are completely occluded within the membrane domain in the E1~P conformation as the cytosolic access channel closes. Transition to the E2~P conformation opens a half-channel to the ectoplasmic side of the membrane while disrupting the high affinity Ca2+ binding site, allowing egress of Ca2+ from the pump as it is exchanged for protons. The protons are then pumped to the cytosol in the E2~P → E1 steps in the cycle (Toyoshima, 2009).
As crystal structures of SERCA1 with two bound Ca2+ ions appeared in the literature (Zhang et al., 1998, Toyoshima et al., 2000, Toyoshima and Nomura, 2002, Toyoshima and Mizutani, 2004, Toyoshima et al., 2013), it became apparent that the tightly confined and deeply buried canonical substrate binding sites in the membrane domain created a problem for the P4-ATPases and their very large substrates. Two Ca2+ ions occupy a volume of 12.4 Å3. By comparison, a PL substrate occupies an immense ~1300 Å3, and even the PL headgroup alone (204 Å3 for phosphoryl choline) is 16 times larger than the Ca2+ ions (Wiener and White, 1992). Moreover, P4-ATPases lack most of the charged and polar residues in the canonical site used to bind cations in all characterized P2-ATPases, and have hydrophobic residues in their place. Thus, it is difficult to imagine that a PL could bind to a structurally similar canonical binding site in the P4-ATPases and enter into an occluded state inaccessible from either surface of the bilayer. While large amphipathic molecules can be observed intercalating between TM segments in P2-ATPase crystal structures, these are typically inhibitors and are not transport substrates (Montigny et al., 2016). The P4-ATPase “giant substrate problem” initially cast substantial skepticism on whether these enzymes could catalyze PL transport. However, the reconstitution of PL flippase activity with purified P4-ATPases from yeast (Drs2) and mammals (ATP8A1 and ATP8A2) capped a large body of genetic evidence to indicate that P4-ATPases are necessary in vivo and sufficient in vitro for PL flippase activity (Kato et al., 2002, Pomorski et al., 2003, Natarajan et al., 2004, Saito et al., 2004, Elvington et al., 2005, Coleman et al., 2009, Zhou and Graham, 2009, Lee et al., 2015). Now that it is widely accepted that most if not all P4-ATPases are PL flippases, major research directions have turned to deciphering how these pumps evolved the ability flip PL substrates across the membrane and how substrate specificity it achieved.
Eukaryotic organisms typically express multiple P4-ATPases that differ in their subcellular localization and substrate specificity. For simplicity, we will focus this review on a few of the 14 mammalian P4-ATPases (ATP8A1, ATP8A2, ATP8B1-B4, ATP9A, ATP9B, ATP10B-10D, and ATP11A-11C) and the 5 budding yeast P4-ATPases (Drs2, Dnf1-3, and Neo1). With regard to substrate specificity, the P4-ATPases can be roughly divided into three categories: enzymes that preferentially flip PS and to a lesser extent PE (ATP8A1, ATP8A2, ATP11A, ATP11C, Drs2) (Natarajan et al., 2004, Paterson et al., 2006, Coleman et al., 2009, Yabas et al., 2011, Takatsu et al., 2014), enzymes that preferentially flip PC and PE (ATP8B1, ATP8B2, ATP10A, Dnf1,2, and 3) (Pomorski et al., 2003, Alder-Baerens et al., 2006, Takatsu et al., 2014, Naito et al., 2015), and enzymes whose substrate preference is unknown (ATP9A, ATP9B and Neo1). Although PL translocase activity has not been directly observed for the orthologous ATP9/Neo1 group, it was recently shown that genetic manipulations of Neo1 were capable of altering plasma membrane PE and PS asymmetry, and these observations are consistent with a putative Golgi-resident flippase activity (Takar et al., 2016). In addition, loss of an ATP9 ortholog in C. elegans (TAT-5) also causes a loss of membrane PE asymmetry, suggesting this group may be PE/PS flippases (Wehman et al., 2011). The biological role for PC translocation is still unclear for the second group of P4-ATPases and it remains to be tested if PC is restricted to the cytosolic leaflet of cells expressing these particular enzymes. In yeast, however, Dnf1 and Dnf2 appear to preferentially translocate lyso-PC and lyso-PE, phospholipids lacking one of the acyl chains, and perhaps play a role in repairing damaged membrane or scavenging these lipids from the environment as a nutrient source (Riekhof and Voelker, 2006, Riekhof et al., 2007). The PS/PE flippases clearly have a role in establishing the asymmetric distribution of PS and PE to the inner leaflet of the plasma membrane, an organization that appears to be widely conserved across eukaryotic species.
The approaches for characterizing P4-ATPase substrate specificity and the mechanism of translocation will be described below, but the prevailing view is that the P4-ATPases have solved the “giant substrate problem” by using a noncanonical transport pathway relative to the well-characterized P2-ATPases. One version of this “Credit Card” model for substrate transport is shown in Figure 2 and postulates that the PL substrate slides along the protein-lipid interface such that only the headgroup and glycerol backbone, analogous to the magnetic strip of a credit card, is “read” by the P4-ATPase. The fatty acyl tails simply reorient within the hydrophobic core the membrane and the entire molecule would not have to be occluded in the center of the membrane domain. Directionality of the transport process may be driven by sequential binding interactions of the substrate at an exofacial entry gate, followed by transfer to a second cytofacial exit gate, before release to the cytosolic leaflet. Rather than using the canonical substrate binding site (formed by residues in TM segments 4, 5, 6, 8 and 9) the P4-ATPases primarily use residues in TM segments 1 – 4 to select substrate, which define the noncanonical route.
Strategies for assessing P4-ATPase activity
There are inherent challenges in the examination of enzymes that catalyze the translocation of their environment. Floppases, scramblases, and flippases are integral membrane proteins that transport an amphipathic PL substrate approximately 30–50 Å from one side of a cellular membrane to the other while reorienting the molecule 180°. Despite these obstacles, research over the last several decades has yielded a wealth of information on the substrate specificity of these enzymes. Researchers in the P4-ATPase field have used three primary strategies to examine the enzymology of PL flippases: i) transport of chemically labeled PL substrates across biological or artificial membranes, ii) changes in membrane curvature associated with transport of unmodified PL across the erythrocyte membrane, and iii) substrate-stimulated ATPase activity of purified P4-ATPases in detergent micelles. Each method has inherent advantages and disadvantages, and all have contributed to our current understanding of P4-ATPase substrate specificity.
The most widely used method for examining P4-ATPase activity is the use of chemically labeled substrates. Labeled PLs were first used to describe P4-ATPase substrate specificity by the patriarch of P4-ATPase research, Dr. Philippe Devaux. These initial studies described a Mg2+-ATP dependent PL transporter in human red blood cells that preferred the amino-glycerophospholipid substrates, PE and PS (Seigneuret and Devaux, 1984, Zachowski et al., 1985, Zachowski et al., 1986). Recent work in human erythrocytes suggests this aminophospholipid translocase activity is primarily catalyzed by ATP11C (Arashiki et al., 2016). Devaux and colleagues made these initial observations using spin-labeled PLs, a strategy that measures the accumulation/protection of these labeled lipids from extracellular reducing agents as a consequence of their transport to the cytosolic leaflet. Subsequent studies alternatively used fluorescent tags to a similar effect, with the spin and fluorescent labels appended to a short 6-carbon acyl chain on the sn2 position of the PL. The key advantage to a labeled PL approach is that these substrates can be combined with genetic strategies and used in nearly any cellular system. Further, substrate transport can be easily assessed using spectroscopy or flow cytometry methods to obtain robust, quantitative analysis of transport activity and substrate specificity. The two major disadvantages of this approach are the inherent assessment of non-native lipid substrates, and that a limited set of labeled PLs are commercially available.
Measurements of P4-ATPase activity using membrane curvature rely on the physical principles of the membrane as described by the bilayer couple hypothesis (Sheetz and Singer, 1974). This strategy assesses changes in cellular shape by preferentially loading one side of the membrane bilayer with a desired PL substrate. Initial administration of many PLs will overload the outer leaflet of the membrane causing the cell or vesicle to evaginate. Yet upon active transport, these cells or proteoliposomes will transfer this excess lipid to the inner leaflet, causing the membrane to invaginate. Devaux’s seminal 1984 manuscript tied the transport of labeled PLs to dynamic changes in membrane shape, thus correlating transport of specific GPLs to cell membrane morphological changes (Seigneuret and Devaux, 1984). Drs. Wray Huestis and David Daleke have also used this method in a series of elegant studies examining P4-ATPase substrate specificity (Daleke and Huestis, 1985, Daleke and Huestis, 1989). The major advantage of this approach is the capacity to use unlabeled, native PLs, while the major disadvantage has been the qualitative nature of the curvature scoring.
Substrate-stimulated ATPase measurements are a direct assessment of P4-ATPase catalytic activity. This strategy was first used to describe P4-ATPase activity by Xie and colleagues (Xie et al., 1989, Ding et al., 2000) and later adapted for the examination of the P4-ATPase catalytic cycle (Coleman et al., 2012, Vestergaard et al., 2014). These assays require the purification of the P4-ATPase enzyme complex in detergent for substrate-stimulated ATPase measurements. The key advantages of this approach are the capacity to use unmodified PL substrates to measure the apparent Km of a P4-ATPase, the ability to complete ATP catalysis through the advancement of the P4-ATPase cycle as a function of PL substrate concentration. Yet the two important disadvantages of this in vitro assay are the requirements to purify and measure the membrane protein complex in detergent conditions, and the examination of an indirect property of substrate transport. Substrate-stimulated ATPase measurements are an approximation of PL transport activity via their connection to the P4-ATPase catalytic cycle; however, apparent Km data must be carefully interpreted when examining molecular determinants of catalysis given that the Michaelis constant is an aggregate representation of a complex enzymatic process.
An elaborate story of substrate recognition, discrimination, and transport has emerged from the collective use of these P4-ATPase assays. Challenges continue to exist within the field such as determinations of actual PL substrate binding affinities and the molecular mechanisms of substrate translocation, but these goals are increasingly within reach with the refinement of biochemical and genetic strategies. Several important questions in the field will be addressed with a crystal structure of the P4-ATPase enzyme complex, but will still require validation using the strategies outlined above.
Specificity of P4-ATPases
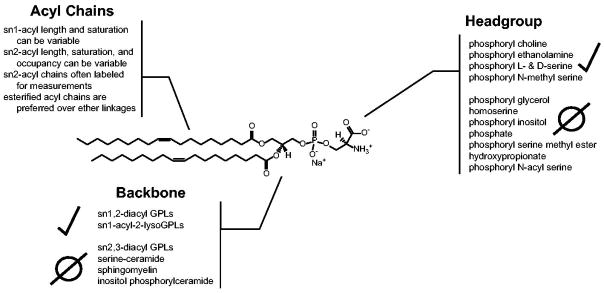
P4-ATPases have been shown to select their PL substrates at three key points: i) the headgroup, ii) the glycerol backbone, and iii) the acyl chain linkages [Figure 3]. Different combinations of headgroup, backbone, and acyl chain selectivity are used to achieve extraordinary substrate specificity. A complete collection of P4-ATPase enzymes and their known substrates was recently reviewed (Lopez-Marques et al., 2015, Andersen et al., 2016). Here we will broadly discuss what PLs have been confirmed as P4-ATPase substrates, and importantly what chemical changes to the PL molecules have been shown to perturb transport.
Figure 3. The anatomy of a P4-ATPase phospholipid substrate.
A dioleoyl phosphatidylserine (DOPS) is used to depict the three key molecular positions that P4-ATPases use to discriminate their substrate: i) the headgroup, ii) the glycerol backbone, and iii) the acyl chain linkages. Presented at each PL molecular position are different modifications that have been tested as P4-ATPase substrates from a variety of enzymes, with positive and negative observations indicated. Selected citations – headgroup: (Seigneuret and Devaux, 1984, Zachowski et al., 1985, Zachowski et al., 1986, Drummond and Daleke, 1995, Paterson et al., 2006, Smriti et al., 2007); backbone: (Morrot et al., 1989, Pomorski et al., 2003, Smriti et al., 2007); acyl chains: (Morrot et al., 1989, Fellmann et al., 1993, Fellmann et al., 2000, Paterson et al., 2006).
Comparisons of the random flip-flop rates of diacylglycerol and phosphatidic acid (PA) establish that the phosphoryl headgroup is the largest impediment in trans-bilayer PL transport (Nieva et al., 1995, Contreras et al., 2010). Thus, the most extensively studied aspect of P4-ATPase substrate specificity has been the coordination of the PL headgroup. Initial observations of headgroup specificity for PL transport in erythrocytes demonstrated a strong preference for PS with a modest activity toward PE, leading to the name “aminophospholipid translocase” (likely ATP11C) (Seigneuret and Devaux, 1984). A later, more extensive evaluation of erythrocyte P4-ATPase substrate selectivity implied dual roles for chemistry and physical size in the selection of PL substrates (Smriti et al., 2007). Both D- and L-serine headgroups were recognized, while methylation of the amine and carboxyl functional groups proved permissive and prohibitive, respectively. The influence of O-linked methyl groups on PS transport could be interpreted as potentially masking an important chemical signature of the molecule; however, a variety of chemical modifications that increase the size of the headgroup perturb transport (Drummond and Daleke, 1995). These physical parameters of headgroup recognition have since been revealed as limiting yet necessary for substrate translocation; no P4-ATPases have been shown to recognize and transport PA substrates (Pomorski et al., 2003, Paterson et al., 2006, Coleman et al., 2009). Finally, examinations of other P4-ATPases have established that choline headgroups can be coordinated, but that the sugar alcohols glycerol and inositol are unable to be transported (Pomorski et al., 2003, Paterson et al., 2006, Coleman et al., 2009). Collectively, these observations indicate that P4-ATPases use complex physical and chemical recognition capacities to coordinate and translocate the PL headgroup.
The selection of the substrate backbone may be perhaps the most rigid of all aspects of P4-ATPase enzymology. Cellular membranes are composed of two major PL species, SLs and GPLs. Early analyses of membrane asymmetry demonstrated that the lumenal/exofacial leaflet is enriched in SLs, while the cytofacial leaflet is populated by aminoGPLs [Figure 1] (Devaux, 1991). To date, only GPLs have been identified as P4-ATPase substrates, with the headgroup in the sn3 position and the acyl chains in the sn1 and sn2 positions [Figure 3] (Smriti et al., 2007). In their work, Smriti and colleagues propose that the discrimination of PL backbone chirality indicates that the backbone and headgroup may form two contacts for paired selection, a hypothesis that recently received additional support (Roland and Graham, 2016). The precise discrimination of substrate backbone is also clearly illustrated when comparing PC and SM selection. The molecular structure of PC and SM are similar, both PLs bear two acyl chains and a phosphoryl choline headgroup, but importantly differ at the PL backbone. The enzymes Dnf1 and Dnf2, ATP8B1 and ATP8B2, and ATP10A have been shown to robustly transport PC, but none have been demonstrated to select SM (Pomorski et al., 2003, Takatsu et al., 2014, Naito et al., 2015). This strict selection is predicted to ensure the integrity of the exofacial leaflet of the PM by preserving SL content, while selectively removing specific GPLs.
P4-ATPases exhibit the least selective pressure on substrate acyl chain composition. Cellular membranes have been shown to display asymmetric differences in membrane fluidity, owing largely to an asymmetric distribution of PL acyl chain composition (Morrot et al., 1986, Cribier et al., 1990, Cribier et al., 1993). These differences are proposed to protect the cell through a tough, exofacial lipid shell of saturated chains, while still providing a malleable, fluid environment of unsaturated chains for the lateral mobility of signaling lipids and the peripheral association of cytosolic proteins (Phillips et al., 2009, Larijani and Poccia, 2012, Czogalla et al., 2014). Studies in erythrocytes using unlabeled substrates demonstrated that acyl chains are discriminated less for their length and saturation, and more for their presence or absence [Figure 3]. The simple addition of an acetyl group in the sn2 position was sufficient to restore recognition and translocation of PLs by P4-ATPases (likely ATP11C) incapable of transporting lysoPLs with a hydroxyl group at sn2 (Morrot et al., 1989, Paterson et al., 2006). In contrast, the budding yeast P4-ATPases Dnf1 and Dnf2 appear to preferentially flip lysoPLs although they will recognize PLs with short acyl chains in the sn2 position (Riekhof and Voelker, 2006). Additionally, the ester linkages connecting the fatty acids to the glycerol backbone also appear to be directly recognized within the enzyme. Ether lipids resembling plasmalogens are transported at a substantially slower rate compared to the typical ester-linked GPLs (Fellmann et al., 1993, Fellmann et al., 2000). These two observations have important implications on membrane biology and the molecular mechanism of P4-ATPase substrate translocation. The non-discriminatory transport of PLs bearing various acyl chains appears to act in a facilitatory rather than active role in asymmetric membrane fluidity (Seigneuret et al., 1984). This asymmetric fluidity of the membrane may be due to the prevalence of cis- vs. trans-unsaturated GPLs and SLs, the selection of GPLs for P4-ATPase transport, and accessibility of inner leaflet PLs to remodeling trans-acylases in the ER (Holthuis and Menon, 2014).
P4-ATPases impose a substantial level of selectivity on their PL substrates from headgroup to acyl chain. The selection of headgroup, backbone, chain occupancy, and ester-linkages – but not acyl chain length or saturation – suggests the P4-ATPase enzyme/substrate complex may extend up to the ester linkages without coordination of the protruding acyl chains themselves. This head-to-tail selection is in agreement with the credit-card hypothesis of substrate transport. Further, this selection implies that some aspect of the enzyme itself is detecting and facilitating, or “gating,” transport at each aspect of substrate chemistry. In the next section, we will review the current literature surrounding what is known about the selection of the PL substrate and how these observations may fit into proposed mechanisms of P4-ATPase translocation.
Approaches to identify molecular basis of substrate discrimination
The divergence of the P4-ATPases from P-type ATPase ion transporters is a curious example of enzyme evolution that parallels the advent of the eukaryotic cell (Axelsen and Palmgren, 1998). It is likely that P4-ATPases were first generated through a gene duplication event with a more ancient ion transporting P-type ATPase family member, yet it is unclear how the P4-ATPases developed the physical capacity to translocate such a large, amphipathic substrate (giant substrate problem). Without a crystal structure, research groups have produced and tested several P4-ATPase homology models using previously crystallized P-type ATPases as templates (Stone et al., 2012, Vestergaard et al., 2014, Roland and Graham, 2016). These studies combined computational modeling with site-directed mutagenesis or forward genetic approaches to each describe important molecular determinants of P4-ATPase substrate transport.
The utility of reverse genetic approaches for determining critical facets of enzymology is manifest, particularly when using disease-associated mutations to guide investigations. Pathology derived from missense mutations assures the investigator that a critical facet of enzymology is being altered. Reverse genetics have been used by the Williamson and Andersen/Molday groups to investigate substitutions leading to hepatic cholestasis and neurologic dysfunction derived from various mutations in the human ATP8B1 and ATP8A2 genes, respectively (Stone et al., 2012, Vestergaard et al., 2014). These investigations have revealed that substitutions at the lumenal aspect of TM4 (N359A – ATP8A2) and lumenal loop of TM5-6 (E981K – ATP8B1, E1261K – Dnf2) were able to decrease the apparent Km for the P4-ATPase substrate. Conversely, a substitution near the cytosolic side of TM3 (I334F – ATP8B1, N601F – Dnf2) enhanced the apparent Km for the PL substrate. Finally, the Andersen group provided compelling evidence for the involvement of TM4 in substrate passage by demonstrating that the apparent Km could be increased or decreased by changing the chemistry of the I364 residue of ATP8A2 (Vestergaard et al., 2014).
Reverse genetic approaches have also taken direction from conserved residues within the P-type ATPase family, as well as chimera-based strategies (Baldridge and Graham, 2012, Coleman et al., 2012). Research into the enzymology of P2-ATPases has highlighted the importance of numerous residues forming an internal cavity within the TM region of the ATPase. Modifications of one of these conserved P-type ATPase residues, a K873 on TM5 of ATP8A2, were shown to substantially decrease the apparent Km of the enzyme (Coleman et al., 2012). It is currently unclear whether this TM5 residue is directly coordinating substrate during transport or whether it may be eliciting a loss-of-function through allosteric interactions within the enzyme (Andersen et al., 2016).
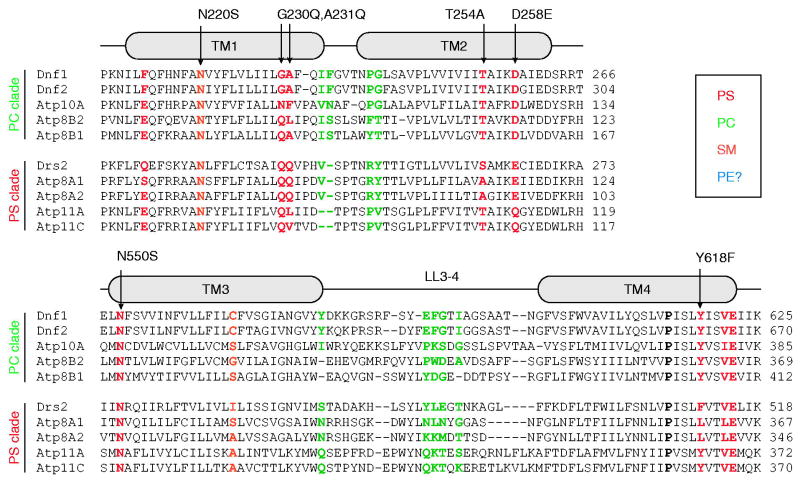
The Graham research group defined residues critical for substrate selectivity by swapping TM domain components between two yeast P4-ATPases, Dnf1 and Drs2, which differ in substrate preference (Baldridge and Graham, 2012). Dnf1 prefers PC and PE substrates while Drs2 preferentially transports PS. These studies have stressed a gain-of-function mutational strategy, identifying Drs2 residues that when transplanted into Dnf1, endowed the chimera with the ability to transport PS. Reciprocal mutations, for which Dnf1 residues were transplanted into Drs2, fortuitously generated separation-of-function mutations whereby Drs2 lost the ability to transport PS while retaining the ability to transport PE [Figure 4]. The substantial advantage of this change-of-function approach is the inherent requirement that the mutations do not disrupt the folding of the enzyme, or its ability to undergo the dramatic conformational changes intrinsic to the catalytic cycle. These examinations revealed the importance of a cytofacial TM4 residue (Y618 – Dnf1, F511 – Drs2), and an exofacial pair of residues in TM1 (Dnf1 – G230,A231; Drs2 – Q237,Q238) that were capable of conferring and ablating PS recognition when exchanged in these two P4-ATPases, respectively [Figure 4] (Baldridge and Graham, 2012, Baldridge and Graham, 2013). These chimera studies were the first examples of primary structural determinants of PL headgroup specificity for a P4-ATPase.
Figure 4. Working toward a P4-ATPase barcode.
A ClustalO alignment of yeast and human P4-ATPases: Uniprot accession numbers S.cerevisiae Dnf1 P32660, S.cerevisiae Dnf2 Q12675, S.cerevisiae Drs2 P39524, H.sapiens ATP8A1 Q9Y2Q0, H.sapiens ATP8A2 Q9NTI2, H.sapiens ATP8B1 O43520, H.sapiens ATP8B2 P98198, H.sapiens ATP10A O60312, H.sapiens ATP11A P98196, H.sapiens APT11C Q8NB49. Shown are alignment fragments highlighting TMs 1,2,3 and 4. The alignments are divided into two clades: the PC transporters and PS transporters. Positions that have been demonstrated to change substrate selectivity are color-coded, with specific gain-of-function mutations indicated with arrows. For example, G230,A231 in Dnf1 prevent PS transport in WT Dnf1, but mutations to the Drs2 residues QQ allows Dnf1 to transport PS. Conversely, the Drs2 QQ → GA mutation disrupts PS transport, but this mutant retains the ability to flip PE. The conserved P-type ATPase TM4 proline is indicated in Bold as a point of reference. Color figures can be found online.
Forward genetic strategies for examining P4-ATPase enzymology have employed directed evolution to isolate gain-of-function substitutions capable of changing P4-ATPase substrate specificity. These studies were performed in yeast using a library of mutagenized Dnf1 enzyme templates, collectively encompassing ~6000 mutations targeted to ~240 codons in TM1 - 6 (1 – 3 mutations per allele). Screens of the library designed to detect Dnf1 mutants that abrogated PC recognition or gained the ability to flip PS identified additional residues at the cytofacial ends of the TM1, 2, and 3 (Baldridge and Graham, 2013, Baldridge et al., 2013). In total, the substrate-determining residues form two clusters at the cytofacial and lumenal aspects of TM1, 2, 3, and 4, and are described as “gates” for substrate entry and exit, respectively [Figures 2,5]. Interestingly, the PS-permissive Dnf1 mutants fell into two groups, those that retained the preference of Dnf1 for lysoPL and flipped lysoPS but not diacyl-PS, and those that were able to flip diacyl-PS (Baldridge et al., 2013). Only the latter group of Dnf1 mutants was capable of replacing the cellular functions of Drs2, emphasizing the importance of substrate discrimination in P4-ATPase biology (Baldridge et al., 2013, Xu et al., 2013, Hankins et al., 2015b).
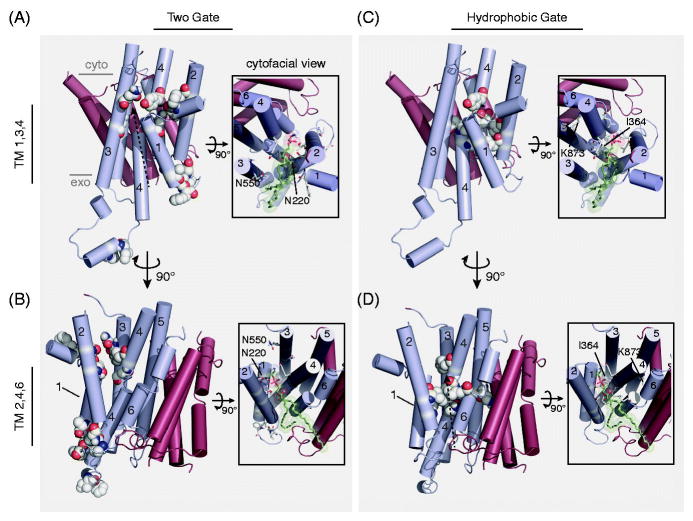
Figure 5. A comparison of putative P4-ATPase substrate pathways.
A,B) The “Two-gate” hypothesis of substrate passage proposes the recognition of PL headgroup at an exofacial/lumenal entry gate (G230, A231, I234, F235, P240, G241, F587, I580) and transport to the cytofacial exit gate (F213, N220, T254, D258, N550, N556, Y618) (numbers from Dnf1). C,D) The “Hydrophobic gate” hypothesis of substrate passage contends that a cluster of hydrophobic residues forming an internal channel to alternate a water-solvated pathway for substrate passage (F88, L112, I115, I362, I364, L367, V908), influenced by the participation of K873 (numbers from ATP8A2). These two hypotheses are not mutually exclusive, and are illustrated by identical dashed-arrows proposing substrate trajectory through the enzyme models. The principle dispute between the two hypotheses is the orientation of the PL substrate during translocation: are the acyl chains directed toward a cleft formed by TM1,3,4 (A,C); or TM2,4,6 (B,D)? Each pathway is depicted using a homology model derived from SERCA (PDB: 3W5D) with a PE molecule docked at the cytofacial aspect in the TM1,3,4 (A,C) and TM2,4,6 (B,D) orientation (Roland and Graham, 2016). Only the TM domain is depicted, with alpha helices shown in cylinders. TM segments 1–6 are colored cyan, while TMs 7–10 are red. Major images: lateral view of the TM domain, substrate-selective residues are colored white/by elements and shown in spheres. Image insets: cytofacial view of the TM domain, substrate-selective residues are colored white/by elements and shown in sticks, docked PE model is colored green/by element and shown in sticks with transparent spheres to illustrate space accommodations. Select residues are numbered and indicated (Dnf1 – Two-gate model; ATP8A2 – hydrophobic gate model). Color figures can be found online.
All of the Dnf1 mutants analyzed in the chimera and initial directed evolution studies retained their strong preference for GPL over SM. To better understand how this enzyme distinguishes PC from SM, two lipids with the same headgroup but different backbones, a directed evolution strategy was used to screen for SM permissive mutants. This study identified an Asn in TM1 that is a major “gatekeeper” of GPL specificity. Specific mutations at this position (Dnf1 – N220S or N220T, but not N220C) significantly enhanced recognition of SM [Figure 4], and combining SM-permissive mutations (Dnf1 –N220S, L1202P) yielded a flippase that preferred SM over PC (Roland and Graham, 2016). N220 is in the same cytofacial cluster of residues (exit gate) now shown to discriminate all three aspects of the PL: headgroup, backbone, and acyl chain occupancy [Figure 3]. While Dnf1 mutations in TM1 (F213S), TM2 (D258E), and TM4 (Y618F) all enhance PS headgroup recognition in Dnf1, other mutations in this cluster alter glycerol to sphingosine preference (TM1 – N220S), or change the selection of acyl chain occupancy (TM2 – T254A and TM3 – N550S). It is currently unclear why PL substrates appear to be most heavily discriminated at the cytofacial aspect of the enzyme, though this may be related to the lack of PL binding requirements for the E1 → E1~P → E2~P transitions.
Model for the structural basis of substrate recognition and translocation
Substrate transport through a P-type ATPase involves four critical enzymatic steps: i) substrate selection, ii) directional passage, iii), catalysis, and iv) release. P4-ATPases tightly regulate headgroup and backbone specificity, with little selective pressure exerted on acyl chain composition. The simplest interpretation of this selectivity suggests that P4-ATPases physically coordinate the headgroup and backbone, while merely positioning the acyl chains in a permissive orientation to facilitate transport. This interpretation is consistent with a “Credit Card” model of substrate transport, wherein the PL substrate slides through the enzyme using a directional transport pathway formed by the enzyme. This model of substrate transport is dramatically different from those established for P1, P2, and P3-ATPases, which use an internal channel that is selectively and directionally coordinated to translocate ion substrates through an occluded state from one side of the membrane to the other. Different experimental approaches have resulted in two competing hypotheses surrounding the substrate pathway of the PL through the P4-ATPase [Figure 5] (Hankins et al., 2015a, Montigny et al., 2016). These proposals have been guided by the experiments described in the previous section of this review. Here we will show the respective models and indicate the proposed substrate pathways.
Substrate selection is a key step in P4-ATPase transport and the “Two-gate” model shown in Figure 5A and B highlights entry and exit gate residues that confer substrate specificity. The key strength of these experiments has been the capacity to alter selection of each of the three major components of the PL: i) headgroup, ii) backbone, and iii) acyl chain. The “Two-gate” model proposes that the PL headgroup binds to the entry (exofacial) gate and slides through a groove formed by TM1, TM2, and TM4 with the hydrophobic acyl chains protruding into the lipid bilayer [Figure 5A]. This model is supported by data that address two of the four key aspects of substrate transport: selection and release.
Directional movement of the substrate is another key aspect of transport, and the “Hydrophobic Gate” model shown in Figure 5B and D highlights the residues proposed to control this process. Mutations in one of these hydrophobic residues (I364, ATP8A2) were capable of changing the apparent Km of the enzyme (Vestergaard et al., 2014). This residue is of particular interest because an I364M mutation causes neurological disease in humans (Onat et al., 2013), and the analogous residue in cation pumps (Glu) is part of the canonical binding site. These targeted analyses were used to propose a “Hydrophobic Gate” model of substrate transport, hypothesizing that the vertical pumping of TM4 controls an internal hydrophobic gate that alternates water penetration from opposite sides of the membrane to achieve directional transport. The “Hydrophobic Gate” model was built on molecular dynamics simulations of a homology model and suggests a “Credit Card” model of transport along a groove formed by TM2, 4 and 6 [Figure 5D]. Thus, the two models are quite similar although they propose transport along opposite sides of the TM domain [Figures 5A and D, arrows].
The “Two-gate” and “Hydrophobic Gate” models address different aspects of P4-ATPase transport with independent experimental systems and approaches, each with its own strengths and weaknesses. First, the “Two-gate” mechanism of substrate selection put forth by the Graham laboratory is the only model to describe how these enzymes select and distinguish their substrates. However, this model highlights exofacial and cytofacial clusters that likely bind substrate, but lacks experimental support for a connecting substrate pathway, a catalytic mechanism, and a mechanism of directionality [Figure 5A,B]. Conversely, the “Hydrophobic Gate” model provides an attractive mechanism of directional transport, and the proposed PL exit pathway partially overlaps with the canonical route traveled by Ca2+ between the cytosol and its binding site [Figure 5C,D]. In addition, a few SERCA1 crystal structures have an annular PL with the headgroup bound to the region analogous to the P4-ATPase exit gate, with fatty acyl chains that appear to protrude into the TM2, TM4, TM6 cleft, suggesting this represents a conserved route for PL exit. However, the “Hydrophobic Gate” model does not provide a mechanism for substrate discrimination. In addition, if one were to draw a straight line connecting the “entry” and “exit” gates on any P4-ATPase homology model, this line would pass directly through the “hydrophobic gate” [Figures 5A and D, arrows]. Thus, the importance of I364 for catalysis does not distinguish the two pathways. Further, there is very little experimental evidence for a direct role of TM6 in PL transport. In contrast, mutations in TM3 residues alter both substrate specificity and apparent Km for the PL substrate (Stone et al., 2012, Baldridge and Graham, 2013, Baldridge et al., 2013).
The primary disagreement between the two “Gate” hypotheses is the orientation of the acyl chains in the hydrophobic bilayer as the hydrophilic aspects of the PL slide through the TM domain [Figure 5, insets]. The “Two-gate” hypothesis proposes that the acyl chains protrude between TM1 and TM3; an assertion based on unbiased gain-of-function substitutions on TM1, TM2, and TM3 that change PL selection at the substrate backbone and acyl chains [Figure 5A, inset]. Of particular relevance are a pair of Asn residues in the Dnf1 “exit gate,” N220 of TM1 and N550 of TM3, which have a substantial influence on distinguishing GPL from SL and lyso-PL from diacyl-PL substrates, respectively [Figure 5A, inset]. We speculate that these Asn residues may form an Asn-clamp to select the substrate backbone and ester linkages, while the TM2 and TM4 “exit gate” residues coordinate the headgroup, thereby orienting the acyl chains toward the TM1,3,4 cleft [Figure 5A, inset] (Roland and Graham, 2016). The opposite orientation of the PL, protruding through TM2,4,6, would move this substrate backbone far from this Asn-clamp [Figure 5B, inset] This leads us to favor a model where the hydrophilic “entry” and “exit” gates guide the selection and transport of the PL substrate along the TM1,3,4 groove, while the movement of the “hydrophobic gate” confers directional transport.
Concluding remarks and future directions
The orchestration of biological membrane composition is a key arbiter of cell biology, and the P4-ATPases are an important conductor within this system. We believe that continued evaluation of P4-ATPase enzymology will reveal additional primary structural determinants of substrate selectivity, a “substrate barcode” [Figure 4]. For example, conserved T and D residues in TM2 play a critical role in preventing PS transport in the yeast PC flippases (Dnf1 and Dnf2), and subtle mutations (e.g. Dnf1 – D258E) are sufficient to allow PS transport. It remains to be determined if these residues exert the same influence on substrate specificity in the human PC flippases. Further, the broad distribution of P4-ATPase expression in animal tissues and subcellular compartments position this enzyme as an attractive candidate for technology development. Indeed, designer substrate-specific P4-ATPases have already been used to reveal novel aspects of vesicle trafficking and membrane organization in yeast. Examinations of molecular determinants of PS specificity produced diacyl PS-specific Dnf1 enzymes that were capable of functionally replacing Drs2 within the secretory pathway, revealing the importance of PS translocation in the budding of vesicles from the TGN (Xu et al., 2013). A similar strategy has also revealed that diacyl PS translocation is required for cargo sorting into exocytic vesicles from the TGN (Hankins et al., 2015b). The key advantage of these manipulations is their circumvention of classical lipid metabolic manipulations that can complicate experimental interpretations with shunted metabolites. Customization of P4-ATPase substrate specificity has currently been limited the yeast system and the molecular dissection of P4-ATPase effectors, but we believe that continued development of these technologies through regulatory and substrate-specific manipulations will yield tools that will have a significant impact on how we examine cellular membranes.
Although P4-ATPases have been shown to transport a variety of different PL substrates, the tight regulation of headgroup and backbone specificity has important implications on cell biology. Cellular consequences of PS translocation have been determined, yet little consideration has been given to the importance of the P4-ATPase’s ability to exclude certain lipids from transport to the cytosolic leaflet. A recent study has revealed a molecular mechanism of PL backbone discrimination, culminating in the development of a SM-specific P4-ATPase (Roland and Graham, 2016). This new designer P4-ATPase now provides an opportunity to examine the trans-bilayer influence of cytofacial SL content within a cellular system, as well as the directed displacement of SLs from exofacial lateral membrane domains. What would happen if SM were translocated to the inner leaflet of the cell? Would the cell generate a bolus of ceramide, thereby inducing PM endocytosis? Would the cell use this metabolite to generate sphingosine-1-phosphate, an important signaling molecule? Would the SM simply accumulate on this leaflet and alter the biophysics of the membrane? How would the altered distribution of SM change cholesterol dynamics? These questions and more are now within reach as we continue to understand the molecular mechanisms of substrate discrimination, and subsequently develop novel membrane technologies.
Acknowledgments
The authors would like to acknowledge the principle contributions of Dr. David Daleke, Dr. Wray Huestis, and the late Dr. Philippe Devaux for their pioneering work in this field. We apologize to those who conducted studies that were not discussed within this manuscript due to space limitations.
Biographies
TRG Bio:
Todd is a Professor in the Department of Biological Sciences at Vanderbilt University. He received his B.S. at Maryville College and Ph.D. at St. Louis University. Todd went on to do a postdoctoral fellowship with Scott Emr at CalTech and University of CA at San Diego before starting up his own lab at Vanderbilt University focused on membrane biology and vesicle trafficking.
BPR Bio:
Bart is a postdoctoral fellow in the Department of Biological Sciences at Vanderbilt University. He received his B.S. from Central Michigan University and his Ph.D. in Molecular Pharmacology at the University of Pittsburgh. He joined the Graham lab to gain training in the fields of lipid biochemistry and membrane biology.
Footnotes
Declaration of Interest
This work was supported by the National Institutes of Health (R01-GM107978 TRG; F32-GM116310 BPR).
ORCiDs: BPR: 0000-0002-9033-9942; TRG: n/a
References Cited
- Alder-Baerens N, Lisman Q, Luong L, Pomorski T, Holthuis JC. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol Biol Cell. 2006;17:1632–42. doi: 10.1091/mbc.E05-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara A, Mercer J. Viral apoptotic mimicry. Nat Rev Microbiol. 2015;13:461–9. doi: 10.1038/nrmicro3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JP, Vestergaard AL, Mikkelsen SA, Mogensen LS, Chalat M, Molday RS. P4-ATPases as Phospholipid Flippases-Structure, Function, and Enigmas. Front Physiol. 2016;7:275. doi: 10.3389/fphys.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arashiki N, Takakuwa Y, Mohandas N, Hale J, Yoshida K, Ogura H, Utsugisawa T, Ohga S, Miyano S, Ogawa S, Kojima S, Kanno H. ATP11C is a major flippase in human erythrocytes and its defect causes congenital hemolytic anemia. Haematologica. 2016;101:559–65. doi: 10.3324/haematol.2016.142273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- Baldridge RD, Graham TR. Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc Natl Acad Sci U S A. 2012;109:E290–8. doi: 10.1073/pnas.1115725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD, Graham TR. Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc Natl Acad Sci U S A. 2013;110:E358–67. doi: 10.1073/pnas.1216948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD, Xu P, Graham TR. Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J Biol Chem. 2013;288:19516–27. doi: 10.1074/jbc.M113.476911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bers DM, Patton CW, Nuccitelli R, Zucker R, Thomas D, Hanley MR. Ca2+ Buffers and [Ca2+]i Perturbation Techniques. In: Nuccitelli R, editor. A Practical Guide to the Study of Calcium in Living Cells. San Diego, CA: Academic Press Inc; 1994. pp. 4–84. [Google Scholar]
- Bevers EM, Comfurius P, Van Rijn JL, Hemker HC, Zwaal RF. Generation of prothrombin-converting activity and the exposure of phosphatidylserine at the outer surface of platelets. Eur J Biochem. 1982;122:429–36. doi: 10.1111/j.1432-1033.1982.tb05898.x. [DOI] [PubMed] [Google Scholar]
- Bevers EM, Williamson PL. Phospholipid scramblase: An update. Febs Letters. 2010;584:2724–2730. doi: 10.1016/j.febslet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol. 1999;147:1223–36. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–30. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Kwok MC, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem. 2009;284:32670–9. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Molday RS. Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J Biol Chem. 2011;286:17205–16. doi: 10.1074/jbc.M111.229419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Quazi F, Molday RS. Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim Biophys Acta. 2013;1831:555–74. doi: 10.1016/j.bbalip.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Vestergaard AL, Molday RS, Vilsen B, Andersen JP. Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc Natl Acad Sci U S A. 2012;109:1449–54. doi: 10.1073/pnas.1108862109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfurius P, Bevers EM, Zwaal RF. Interaction between phosphatidylserine and the isolated cytoskeleton of human blood platelets. Biochim Biophys Acta. 1989;983:212–6. doi: 10.1016/0005-2736(89)90236-8. [DOI] [PubMed] [Google Scholar]
- Contreras FX, Sanchez-Magraner L, Alonso A, Goni FM. Transbilayer (flip-flop) lipid motion and lipid scrambling in membranes. Febs Letters. 2010;584:1779–86. doi: 10.1016/j.febslet.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Cribier S, Morrot G, Neumann JM, Devaux PF. Lateral diffusion of erythrocyte phospholipids in model membranes comparison between inner and outer leaflet components. Eur Biophys J. 1990;18:33–41. doi: 10.1007/BF00185418. [DOI] [PubMed] [Google Scholar]
- Cribier S, Morrot G, Zachowski A. Dynamics of the membrane lipid phase. Prostaglandins Leukot Essent Fatty Acids. 1993;48:27–32. doi: 10.1016/0952-3278(93)90006-i. [DOI] [PubMed] [Google Scholar]
- Czogalla A, Grzybek M, Jones W, Coskun U. Validity and applicability of membrane model systems for studying interactions of peripheral membrane proteins with lipids. Biochim Biophys Acta. 2014;1841:1049–59. doi: 10.1016/j.bbalip.2013.12.012. [DOI] [PubMed] [Google Scholar]
- Daleke DL, Huestis WH. Incorporation and translocation of aminophospholipids in human erythrocytes. Biochemistry. 1985;24:5406–16. doi: 10.1021/bi00341a019. [DOI] [PubMed] [Google Scholar]
- Daleke DL, Huestis WH. Erythrocyte morphology reflects the transbilayer distribution of incorporated phospholipids. J Cell Biol. 1989;108:1375–85. doi: 10.1083/jcb.108.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux PF. Static and dynamic lipid asymmetry in cell membranes. Biochemistry. 1991;30:1163–73. doi: 10.1021/bi00219a001. [DOI] [PubMed] [Google Scholar]
- Ding J, Wu Z, Crider BP, Ma Y, Li X, Slaughter C, Gong L, Xie XS. Identification and functional expression of four isoforms of ATPase II, the putative aminophospholipid translocase. Effect of isoform variation on the ATPase activity and phospholipid specificity. J Biol Chem. 2000;275:23378–86. doi: 10.1074/jbc.M910319199. [DOI] [PubMed] [Google Scholar]
- Drummond DC, Daleke DL. Synthesis and characterization of N-acylated, pH-sensitive ‘caged’ aminophospholipids. Chem Phys Lipids. 1995;75:27–41. doi: 10.1016/0009-3084(94)02398-O. [DOI] [PubMed] [Google Scholar]
- Elvington SM, Bu F, Nichols JW. Fluorescent, acyl chain-labeled phosphatidylcholine analogs reveal novel transport pathways across the plasma membrane of yeast. J Biol Chem. 2005;280:40957–64. doi: 10.1074/jbc.M507926200. [DOI] [PubMed] [Google Scholar]
- Fadok VA, Voelker DR, Campbell PA, Bratton DL, Cohen JJ, Noble PW, Riches DW, Henson PM. The ability to recognize phosphatidylserine on apoptotic cells is an inducible function in murine bone marrow-derived macrophages. Chest. 1993;103:102S. doi: 10.1378/chest.103.2_supplement.102s. [DOI] [PubMed] [Google Scholar]
- Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148:2207–16. [PubMed] [Google Scholar]
- Fadok VA, Warner ML, Bratton DL, Henson PM. CD36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (alpha v beta 3) J Immunol. 1998;161:6250–7. [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–75. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann P, Herve P, Devaux PF. Transmembrane distribution and translocation of spin-labeled plasmalogens in human red blood cells. Chem Phys Lipids. 1993;66:225–30. doi: 10.1016/0009-3084(93)90010-z. [DOI] [PubMed] [Google Scholar]
- Fellmann P, Herve P, Pomorski T, Muller P, Geldwerth D, Herrmann A, Devaux PF. Transmembrane movement of diether phospholipids in human erythrocytes and human fibroblasts. Biochemistry. 2000;39:4994–5003. doi: 10.1021/bi992649q. [DOI] [PubMed] [Google Scholar]
- Fujii T, Sakata A, Nishimura S, Eto K, Nagata S. TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc Natl Acad Sci U S A. 2015;112:12800–5. doi: 10.1073/pnas.1516594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall WE, Geething NC, Hua Z, Ingram MF, Liu K, Chen SI, Graham TR. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr Biol. 2002;12:1623–7. doi: 10.1016/s0960-9822(02)01148-x. [DOI] [PubMed] [Google Scholar]
- Goren MA, Morizumi T, Menon I, Joseph JS, Dittman JS, Cherezov V, Stevens RC, Ernst OP, Menon AK. Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat Commun. 2014;5:5115. doi: 10.1038/ncomms6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Burd CG. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 2011;21:113–21. doi: 10.1016/j.tcb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon Y, Broccardo C, Chambenoit O, Luciani MF, Toti F, Chaslin S, Freyssinet JM, Devaux PF, Mcneish J, Marguet D, Chimini G. ABC1 promotes engulfment of apoptotic cells and transbilayer redistribution of phosphatidylserine. Nat Cell Biol. 2000;2:399–406. doi: 10.1038/35017029. [DOI] [PubMed] [Google Scholar]
- Han X. Lipidomics for studying metabolism. Nat Rev Endocrinol. 2016 doi: 10.1038/nrendo.2016.98. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Hankins HM, Baldridge RD, Xu P, Graham TR. Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic. 2015a;16:35–47. doi: 10.1111/tra.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR. Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell. 2015b;26:4674–85. doi: 10.1091/mbc.E15-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann PR, Decathelineau AM, Ogden CA, Leverrier Y, Bratton DL, Daleke DL, Ridley AJ, Fadok VA, Henson PM. Phosphatidylserine (PS) induces PS receptor-mediated macropinocytosis and promotes clearance of apoptotic cells. J Cell Biol. 2001;155:649–59. doi: 10.1083/jcb.200108080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- Kato U, Emoto K, Fredriksson C, Nakamura H, Ohta A, Kobayashi T, Murakami-Murofushi K, Umeda M. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J Biol Chem. 2002;277:37855–62. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- Kay JG, Grinstein S. Phosphatidylserine-mediated cellular signaling. Adv Exp Med Biol. 2013;991:177–93. doi: 10.1007/978-94-007-6331-9_10. [DOI] [PubMed] [Google Scholar]
- Larijani B, Poccia DL. Effects of phosphoinositides and their derivatives on membrane morphology and function. Curr Top Microbiol Immunol. 2012;362:99–110. doi: 10.1007/978-94-007-5025-8_5. [DOI] [PubMed] [Google Scholar]
- Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, Taguchi T, Arai H. Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 2015;34:669–88. doi: 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir G, Williamson P, Puts CF, Holthuis JC. Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J Biol Chem. 2009;284:17956–67. doi: 10.1074/jbc.M109.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Marques RL, Poulsen LR, Bailly A, Geisler M, Pomorski TG, Palmgren MG. Structure and mechanism of ATP-dependent phospholipid transporters. Biochim Biophys Acta. 2015;1850:461–75. doi: 10.1016/j.bbagen.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Lopez-Marques RL, Theorin L, Palmgren MG, Pomorski TG. P4-ATPases: lipid flippases in cell membranes. Pflugers Arch. 2014;466:1227–40. doi: 10.1007/s00424-013-1363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno S, Takakuwa Y, Mohandas N. Identification of a functional role for lipid asymmetry in biological membranes: Phosphatidylserine-skeletal protein interactions modulate membrane stability. Proc Natl Acad Sci U S A. 2002;99:1943–8. doi: 10.1073/pnas.042688399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montigny C, Lyons J, Champeil P, Nissen P, Lenoir G. On the molecular mechanism of flippase- and scramblase-mediated phospholipid transport. Biochim Biophys Acta. 2016;1861:767–83. doi: 10.1016/j.bbalip.2015.12.020. [DOI] [PubMed] [Google Scholar]
- Morrot G, Cribier S, Devaux PF, Geldwerth D, Davoust J, Bureau JF, Fellmann P, Herve P, Frilley B. Asymmetric lateral mobility of phospholipids in the human erythrocyte membrane. Proc Natl Acad Sci U S A. 1986;83:6863–7. doi: 10.1073/pnas.83.18.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrot G, Herve P, Zachowski A, Fellmann P, Devaux PF. Aminophospholipid translocase of human erythrocytes: phospholipid substrate specificity and effect of cholesterol. Biochemistry. 1989;28:3456–62. doi: 10.1021/bi00434a046. [DOI] [PubMed] [Google Scholar]
- Nagata S, Suzuki J, Segawa K, Fujii T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016;23:952–61. doi: 10.1038/cdd.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Takatsu H, Miyano R, Takada N, Nakayama K, Shin HW. Phospholipid Flippase ATP10A Translocates Phosphatidylcholine and Is Involved in Plasma Membrane Dynamics. J Biol Chem. 2015;290:15004–17. doi: 10.1074/jbc.M115.655191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Wang J, Hua Z, Graham TR. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc Natl Acad Sci U S A. 2004;101:10614–9. doi: 10.1073/pnas.0404146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Alonso A, Basanez G, Goni FM, Gulik A, Vargas R, Luzzati V. Topological properties of two cubic phases of a phospholipid:cholesterol:diacylglycerol aqueous system and their possible implications in the phospholipase C-induced liposome fusion. Febs Letters. 1995;368:143–7. doi: 10.1016/0014-5793(95)00631-i. [DOI] [PubMed] [Google Scholar]
- Onat OE, Gulsuner S, Bilguvar K, Nazli Basak A, Topaloglu H, Tan M, Tan U, Gunel M, Ozcelik T. Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur J Hum Genet. 2013;21:281–5. doi: 10.1038/ejhg.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P. P-type ATPases. Annu Rev Biophys. 2011;40:243–66. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- Panatala R, Hennrich H, Holthuis JC. Inner workings and biological impact of phospholipid flippases. J Cell Sci. 2015;128:2021–32. doi: 10.1242/jcs.102715. [DOI] [PubMed] [Google Scholar]
- Paterson JK, Renkema K, Burden L, Halleck MS, Schlegel RA, Williamson P, Daleke DL. Lipid specific activation of the murine P4-ATPase Atp8a1 (ATPase II) Biochemistry. 2006;45:5367–76. doi: 10.1021/bi052359b. [DOI] [PubMed] [Google Scholar]
- Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–85. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl A, Devaux PF, Herrmann A. Function of prokaryotic and eukaryotic ABC proteins in lipid transport. Biochim Biophys Acta. 2005;1733:29–52. doi: 10.1016/j.bbalip.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Pomorski T, Lombardi R, Riezman H, Devaux PF, Van Meer G, Holthuis JC. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell. 2003;14:1240–54. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14:166–80. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AS, Zhang A, Park SJ, Farzan M, Zong M, Choe H. Virion-associated phosphatidylethanolamine promotes TIM1-mediated infection by Ebola, dengue, and West Nile viruses. Proc Natl Acad Sci U S A. 2015;112:14682–7. doi: 10.1073/pnas.1508095112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR, Voelker DR. Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J Biol Chem. 2006;281:36588–96. doi: 10.1074/jbc.M608851200. [DOI] [PubMed] [Google Scholar]
- Riekhof WR, Wu J, Gijon MA, Zarini S, Murphy RC, Voelker DR. Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J Biol Chem. 2007;282:36853–61. doi: 10.1074/jbc.M706718200. [DOI] [PubMed] [Google Scholar]
- Roland BP, Graham TR. Directed evolution of a sphingomyelin flippase reveals mechanism of substrate backbone discrimination by a P4-ATPase. Proc Natl Acad Sci U S A. 2016;113:E4460–6. doi: 10.1073/pnas.1525730113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:3418–32. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344:1164–8. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- Segawa K, Nagata S. An Apoptotic ‘Eat Me’ Signal: Phosphatidylserine Exposure. Trends Cell Biol. 2015;25:639–50. doi: 10.1016/j.tcb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Seigneuret M, Devaux PF. ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: relation to shape changes. Proc Natl Acad Sci U S A. 1984;81:3751–5. doi: 10.1073/pnas.81.12.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneuret M, Zachowski A, Hermann A, Devaux PF. Asymmetric lipid fluidity in human erythrocyte membrane: new spin-label evidence. Biochemistry. 1984;23:4271–5. doi: 10.1021/bi00314a002. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci U S A. 1974;71:4457–61. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms BA, Zamponi GW. Neuronal voltage-gated calcium channels: structure, function, and dysfunction. Neuron. 2014;82:24–45. doi: 10.1016/j.neuron.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Smriti, Nemergut EC, Daleke DL. ATP-dependent transport of phosphatidylserine analogues in human erythrocytes. Biochemistry. 2007;46:2249–59. doi: 10.1021/bi061333x. [DOI] [PubMed] [Google Scholar]
- Stone A, Chau C, Eaton C, Foran E, Kapur M, Prevatt E, Belkin N, Kerr D, Kohlin T, Williamson P. Biochemical characterization of P4-ATPase mutations identified in patients with progressive familial intrahepatic cholestasis. J Biol Chem. 2012;287:41139–51. doi: 10.1074/jbc.M112.413039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341:403–6. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- Takar M, Wu Y, Graham TR. The Essential Neo1 Protein from Budding Yeast Plays a Role in Establishing Aminophospholipid Asymmetry of the Plasma Membrane. J Biol Chem. 2016;291:15727–39. doi: 10.1074/jbc.M115.686253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Tanaka G, Segawa K, Suzuki J, Nagata S, Nakayama K, Shin HW. Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J Biol Chem. 2014;289:33543–56. doi: 10.1074/jbc.M114.593012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim Biophys Acta. 2009;1793:941–6. doi: 10.1016/j.bbamcr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Iwasawa S, Ogawa H, Hirata A, Tsueda J, Inesi G. Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature. 2013;495:260–4. doi: 10.1038/nature11899. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Mizutani T. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 2004;430:529–35. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–55. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–11. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- Van Den Eijnde SM, Van Den Hoff MJ, Reutelingsperger CP, Van Heerde WL, Henfling ME, Vermeij-Keers C, Schutte B, Borgers M, Ramaekers FC. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J Cell Sci. 2001;114:3631–42. doi: 10.1242/jcs.114.20.3631. [DOI] [PubMed] [Google Scholar]
- Van Geffen JP, Swieringa F, Heemskerk JW. Platelets and coagulation in thrombus formation: aberrations in the Scott syndrome. Thromb Res. 2016;141(Suppl 2):S12–6. doi: 10.1016/S0049-3848(16)30355-3. [DOI] [PubMed] [Google Scholar]
- Van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard AL, Coleman JA, Lemmin T, Mikkelsen SA, Molday LL, Vilsen B, Molday RS, Dal Peraro M, Andersen JP. Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc Natl Acad Sci U S A. 2014;111:E1334–43. doi: 10.1073/pnas.1321165111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–9. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–47. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XS, Stone DK, Racker E. Purification of a vanadate-sensitive ATPase from clathrin-coated vesicles of bovine brain. J Biol Chem. 1989;264:1710–4. [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR. Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol. 2013;202:875–86. doi: 10.1083/jcb.201305094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabas M, Teh CE, Frankenreiter S, Lal D, Roots CM, Whittle B, Andrews DT, Zhang Y, Teoh NC, Sprent J, Tze LE, Kucharska EM, Kofler J, Farell GC, Broer S, Goodnow CC, Enders A. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat Immunol. 2011;12:441–9. doi: 10.1038/ni.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–51. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- Zachowski A, Favre E, Cribier S, Herve P, Devaux PF. Outside-inside translocation of aminophospholipids in the human erythrocyte membrane is mediated by a specific enzyme. Biochemistry. 1986;25:2585–90. doi: 10.1021/bi00357a046. [DOI] [PubMed] [Google Scholar]
- Zachowski A, Fellman P, Devaux PF. Absence of transbilayer diffusion of spin-labeled sphingomyelin on human erythrocytes. Comparison with the diffusion of several spin-labeled glycerophospholipids. Biochim Biophys Acta. 1985;815:510–4. doi: 10.1016/0005-2736(85)90380-3. [DOI] [PubMed] [Google Scholar]
- Zhang P, Toyoshima C, Yonekura K, Green NM, Stokes DL. Structure of the calcium pump from sarcoplasmic reticulum at 8-A resolution. Nature. 1998;392:835–9. doi: 10.1038/33959. [DOI] [PubMed] [Google Scholar]
- Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci U S A. 2009;106:16586–91. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]