Abstract
People with autism spectrum disorder (ASD) and other neurodevelopmental disorders (NDDs) are behaviorally and medically heterogeneous. The combination of polygenicity and gene pleiotropy - the influence of one gene on distinct phenotypes - raises questions of how specific genes and their protein products interact to contribute to NDDs. A preponderance of evidence supports developmental and pathophysiological roles for the MET receptor tyrosine kinase, a multi-functional receptor that mediates distinct biological responses depending upon cell context. MET influences neuron architecture and synapse maturation in the forebrain, and regulates homeostasis in gastrointestinal and immune systems, both commonly disrupted in NDDs. Peak expression of synapse-enriched MET is conserved across rodent and primate forebrain, yet regional differences in primate neocortex are pronounced, with enrichment in circuits that participate in social information processing. A functional risk allele in the MET promoter, enriched in subgroups of children with ASD, reduces transcription and disrupts socially-relevant neural circuits structurally and functionally. In mice, circuit-specific deletion of Met causes distinct atypical behaviors. MET activation increases dendritic complexity and nascent synapse number, but synapse maturation requires reductions in MET. MET mediates its specific biological effects through different intracellular signaling pathways, and has a complex protein interactome that is enriched in ASD and other NDD candidates. The interactome is co-regulated in developing human neocortex. We suggest that a gene as pleiotropic and highly regulated as MET, together with its interactome, is biologically relevant in normal and pathophysiological contexts, impacting central and peripheral phenotypes that contribute to NDD risk and clinical symptoms.
Keywords: forebrain, synapse development, protein interactome, symptom heterogeneity, immune system, gastrointestinal system
Autism spectrum disorder (ASD) is a neurodevelopmental disorder (NDD) affecting 1-2% of children (1). Diagnosis of ASD is based on two behaviorally-defined criteria: 1) dysfunction in social interaction and communication, and 2) restricted, repetitive behavior, including hyper- or hypo-reactivity to sensory input (2). Described more than 70 years ago by Kanner (3), the nature and severity of clinical presentation is highly heterogeneous. There has been more recent attention to another layer of heterogeneity, the medical and psychiatric co-occurring disturbances expressed by most individuals with ASD. Indeed, multiple comorbidities have been described, including gastrointestinal disturbances (GID), immunological dysfunction, sleep abnormalities, epilepsy, motor deficits, generalized anxiety disorder, attention deficit hyperactivity disorder and aggression (4–6).
While a diagnosis of ASD is based on clinical observation using categorical measures, there is a growing consensus that understanding the genetic contribution and underlying pathophysiology of complex NDDs will be better served using a dimensional approach, with each dimension, or endophenotype, ranging along a continuum from typical to atypical (7, 8). This approach recognizes that 1) distinct neural circuitry underlies different phenotypes – for example, language, social interaction and behavioral inflexibility, 2) alterations in a single dimension will cross categorical diagnoses – for example, impairments in social interactions are observed in many DSM-V disorders, and 3) co-occurring psychiatric conditions or shared traits across conditions probably arise from shared genetic and environmental burdens. In such a scheme, co-occurring medical conditions that likely share disruptions in common biological pathways define subpopulations of individuals with ASD, facilitating the identification of specific genetic and environmental contributions to the etiology of the disorder. Further, a focus solely on identifying genome-wide disease risk (usually small risk effect size) will miss identifying factors that contribute to biological dimensions that may be vulnerable in particular categorically-defined disorders. A similar approach has benefited several fields of medicine, recently emphasized in studies of psychiatric disorders (9).
The concept of pleiotropy and studies of the underlying complex biology of ASD heterogeneity begs the question of how specific genes may contribute. This review focuses a biological lens on basic and clinical studies of the MET receptor tyrosine kinase, which is an important regulator of development and cellular homeostasis in organs in which it is expressed (10). MET is a Category 2 risk gene on Simons Foundation Autism Research Initiative Gene (SFARIgene, https://gene.sfari.org/autdb/GS_Home.do). A functional promoter variant reduces MET transcription and is enriched in ASD. This has led to extensive, interdisciplinary studies, reviewed here (supplemental Table S1), demonstrating the biological significance of dysregulation of MET expression in the context of typical and atypical neurodevelopment. We also describe MET function in peripheral systems relevant to ASD, particularly GI and immune systems. We also review studies that place MET functionally in the broader context of specific gene and protein networks implicated in NDDs.
Genetic studies: association of the MET tyrosine kinase receptor with ASD
ASD is a polygenic disorder. Multiple rare de novo and inherited variations that are enriched in the ASD population have been identified (11–13). These represent a modest fraction of ASD diagnoses, with common heritable variations representing the largest component of genetic risk (14). Our laboratory identified genetic association, in multiplex families, of a common promoter variant, rs1858830, in the gene encoding the MET receptor tyrosine kinase with ASD (15). The association of this and other variants has been replicated across independent cohorts (16–19). There are, however, two important facts to emphasize in our and others genetic findings. First, the promoter variant individually has a small effect on ASD risk. Second, the promoter variant, like the majority of genetic findings in ASD, does not reach genome-wide significance across populations selected solely on an ASD diagnosis. Rather, the MET contribution to ASD risk likely occurs in unique subgroups of children, and moreover, the variant alone is not sufficient to cause ASD. Thus, the common promoter variant is likely one of a number of genetic and specific environmental factors that result in ASD. The variant is nonetheless functional. The ‘G’-to-‘C’ single nucleotide polymorphism results in a striking 50% decrease in promoter activity (20). Given that the promoter variant may be more biologically influential in ASD subpopulations, how might one identify clinically relevant subgroups across the autism spectrum? Attempts to do this with ASD diagnostic tools have not succeeded (21). We have used a different approach, examining subgroups for genetic risk by focusing on biomedical phenotypes (Figure 1). The strategy takes advantage of the pleiotropic nature of MET, which, in addition to its role in brain development, has demonstrated functions in systems vulnerable in ASD, including GI (22, 23) and immune (24, 25). The MET ‘C’ allele is enriched in children with ASD and co-occurring GID (26) and is associated in mothers who express ASD-associated antibodies that react with fetal brain proteins (27). In those individuals with ASD from families with GID, the MET ‘C’ allele also is associated with more disrupted social communication (28). This observation is consistent with the strong link between the most common types of GID in ASD and increased social impairment and lack of expressive language (29). Further, siblings in a family pedigree with a rare, functional mutation in MET that generates haploinsufficiency have either ASD or social-communication deficits (30). Finally, a subgroup of children who are homozygous for the MET ‘C’ allele and whose mothers had been exposed to high levels of air pollution during pregnancy are at an increased risk of ASD (31).
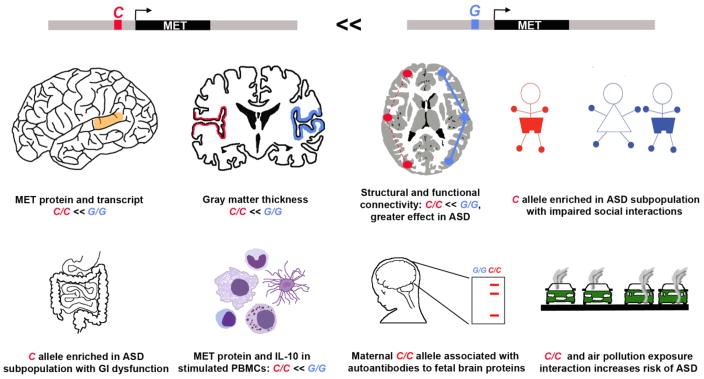
Figure 1. The MET promoter variant ‘C’- allele is associated with children with ASD in multiplex families and influences development and function of human brain and peripheral organ systems.
A common ‘G’-to-‘C’ single nucleotide polymorphism in the MET promoter results in a 50% decrease in promoter activity. Because the ‘C’ variant is biologically functional, it is associated with specific phenotypes in typical individuals and those diagnosed with ASD. From Top Left to Lower Right: In the typical population, reduced levels of MET protein in the temporal cortex correlate with the C/C genotype. People with ASD have reduced MET, irrespective of genotype. Human neuroimaging studies demonstrate reduced gray matter thickness in MET-expressing cortical regions and altered functional and structural connectivity in C/C individuals independent of diagnosis. The phenotype effects are greater in people with ASD. Examination of ASD subgroups based on behavioral and biomedical phenotypes shows an enrichment of the C/C allele in children with ASD whose social communication is more impaired, and in children with ASD who have co-occurring gastrointestinal (GI) dysfunction. In the immune system, levels of MET in activated peripheral blood mononuclear cells (PBMCs), critical components of the immune system, isolated from individuals with a C/C genotype is approximately half that from those with G/G. In addition, the C/C genotype is enriched in mothers who produce ASD-associated maternal antibodies that recognize brain proteins expressed only in the fetus. Finally, there is an increased risk of ASD in children with the C/C allele whose mothers had been exposed to high levels of air pollution during pregnancy, suggesting an interaction between genetic and environmental burdens that influence ASD risk.
Regulation of MET expression: biological significance
Because gene dysregulation is central to human disease (32), including ASD and psychiatric disorders (33), and cell type-specific regulation of gene expression likely defines the most vulnerable circuits in ASD (34), we have focused on defining precisely spatial, temporal and evolutionary regulation of MET expression in the developing brain (35–38). Spatial and temporal mapping of transcript and protein in the developing rodent forebrain revealed low levels of MET expression prenatally. Met is not expressed in progenitor and migratory zones of neocortex and subcortical structures (36, 38), a pattern distinct from stem and migrating cell expression during peripheral organ development (39). Over the first postnatal week, there is a dramatic increase in MET expression in discrete forebrain regions, including neocortex, hippocampus and subcortical limbic regions, with levels remaining elevated through the second postnatal week (36, 40). This corresponds to the early and peak periods of process outgrowth and synaptogenesis. In these regions, MET is expressed in excitatory projection neurons, with no in vivo evidence of expression in interneurons, astrocytes or oligodendrocytes, consistent with the failure to alter interneuron development in Metfx/fx/Dlx5/6Cre mice (38). This latter point is important, as MET expression may occur ectopically in some cells placed in culture or after injury. This occurs in cultured ganglion eminence (GE), which gives rise to interneurons. The cells that generate interneurons respond to the MET receptor ligand, hepatocyte growth factor (HGF), in vitro, but in contrast to our initial interpretation (41), this signaling would not operate for cortical interneurons in vivo because they lack Met. There also is no evidence for MET expression in prenatal human GE, dorsal pallial progenitors or any migrating neurons (35), as well as in our own and GENSAT MetGFP mice (42). We note, however, a report that Metfx/fx/Dlx5/6Cre mice on a different background exhibit neocortical interneuron decreases (43), though without direct evidence of GE or interneuron Met expression in vivo using in situ hybridization co-labeling. At later stages of cortical development, ligand-activated pMET expression rapidly declines during synapse refinement (36, 40).
In developing rhesus monkey, three striking findings emerged: 1) the temporal pattern of MET expression is conserved, with peak expression also occurring during the period of rapid process outgrowth and synapse formation (37); 2) subcortical and hippocampal expression patterns are similar between rodent and primate; and 3) there are substantial differences in neocortical regional expression. Specifically, whereas MET is expressed across all neocortical regions in the rodent, it is largely limited to temporal, posterior parietal and occipital regions in developing monkey and human neocortex (35, 37). The conserved temporal expression pattern of MET in forebrain likely reflects conserved functions during synapse and circuit development. The enrichment in regions that include sensory processing of social information highlights the circuits most vulnerable to dysregulated MET expression, noteworthy because of recent human imaging findings (44, 45).
The effects of MET dysregulation have been extensively studied in cancer, where overexpression or gain-of-function mutations in MET result in increased receptor signaling, leading to uncontrolled cell proliferation and metastasis (46). In contrast, the MET ‘C’ allele causes a 50% reduction in gene transcription (15). This allele is present in typical and ASD populations, thus prompting the question as to how the MET ‘C’ allele shapes typical neurodevelopmental processes to influence disorder risk (Figure 1). For example, there is a significant effect of the MET ‘C’ allele on MET expression in typically developing subjects (20). Such reductions in protein expression likely impact development and function. The is evident in typically-developing children and adolescents, with a reduction in the thickness of MET-expressing areas of the neocortex that is related to the dose of the MET ‘C’ allele (44). These cortical regions contribute to circuits underlying social behavior, including face processing. Consistent with this, a functional neuroimaging study revealed that, in typically-developing subjects, those with the MET ‘C’ allele exhibit altered activity patterns in response to emotional faces (45). Further, resting state and structural connectivity between temporal and parietal cortices is disrupted.
What about MET expression and function in ASD? Compared to age-matched control subjects, MET transcript and protein are reduced approximately 2-fold in the temporal cortex of individuals with ASD, irrespective of MET promoter genotype (20). This decrease suggests that, in addition to the promoter allele, other genetic or environmental factors may contribute to reduced MET expression in ASD. MET reduction in temporal cortex in ASD was replicated in an independent transcriptome analysis (47). This reduction likely underlies the disruption in ASD of the typical regional differences in MET expression, which is normally high in temporal and low in frontal cortices. In a functional context, the neuroimaging study described above for the typical population (45) revealed alterations in circuit structure and function in subjects with ASD that also correlate with the ‘C’ allele. Two important findings emerged: 1) the MET ‘C’ allele has a more pronounced quantitative impact on circuits in individuals with ASD compared to the measures in controls of the same genotype; 2) the effect size on atypical brain circuitry is greater in typically-developing C/C individuals than in ASD G/G individuals. While these findings may seem counterintuitive because typically developing individuals do not meet criteria for ASD, this is consistent with the well-known overlap of specific dimensional measures between clinically and non-clinically diagnosed individuals (45, 48). Moreover, this emphasizes the fact that for genetic factors like MET, which contribute small to moderate ASD risk, there are likely other genetic and environmental factors that are necessary to reach criteria for a clinical diagnosis (14, 33, 49). Thus, the functional promoter variant can influence neurobiological phenotypes, but does not cause ASD.
The differential circuit expression of MET led to our hypothesis that the behavioral readout of dysregulated MET signaling would be complex. Three Cre-LoxP mouse lines revealed that the specific behaviors disrupted depend on the neuronal population targeted by Cre deletion and, in some instances, even gene dosage (50, 51). Global deletion of Met from all neural cells was achieved using Nestincre. These mice exhibit disruptions in contextual fear conditioning (50). In the absence of MET signaling, only fear memory is disrupted, whereas reduction of MET signaling (heterozygous mice) additionally alters fear learning. In contrast, fear conditioning is normal when using Emx1cre to delete Met in neocortical and hippocampal neurons (50). These mice display hypoactivity and blunted spontaneous alternation, with heterozygous mice performance comparable to wild types. Neither line shows differences in sociability and social novelty preference. This contrasts Met deletion from a subset of dorsal raphe neurons using Pet1cre; these mice exhibit deficits in social approach, but normal baseline activity (51). The lack of phenotypic overlap of the 3 lines suggests that circuit-specific manipulations of Met can generate both unique dysfunction and adaptive capacities that may contribute to distinct phenotypes.
MET in the forebrain: synapse formation and plasticity
Human genetic studies and animal models indicate that disruption of synapse formation and/or stabilization is a major target in the etiology of ASD (52, 53). MET function falls within this framework, as accumulating evidence demonstrates a key role for the receptor in modulating forebrain dendritic growth and synapse formation. MET expression is enriched spatially and temporally during forebrain development to impact these neurodevelopmental processes. Further, MET protein expression is robust in neuropil, with an enrichment in pre- and post-synaptic compartments, and developing axon tracts, reflecting active transport to the synapse (36, 54). Strikingly, localization of endogenously-activated MET (pMET) in the developing forebrain reveals enriched receptor activation in the neuropil, with an absence from axon tracts (55).
MET signaling modulates dendritic growth and synapse formation of glutamatergic neocortical and hippocampal neurons in vitro (Figure 2). In the short-term, HGF increases dendritic length and branching (40, 55–58), the expression and clustering of postsynaptic proteins, including NR2B and GluR1 (59, 60), and synapse density (55). Like many receptor tyrosine kinases, ligand-induced activation of MET engages the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) and phosphatidylinositol-3 kinase (PI3K)/Akt pathways (61–63). In hippocampal neurons, activation of Akt is required for HGF-induced dendritic growth, although the role of the MAPK/ERK pathway was not examined (57). In neocortical neurons, however, inhibition of the PI3K/Akt pathway has no effect on HGF-induced dendritic growth; rather, this response is abolished in the presence of inhibitors of the MAPK/ERK pathway (55, 58), reflecting cell-specific intracellular mediation of the same outcome to MET activation even in closely-related neuronal populations. In contrast, similar treatments demonstrate that the PI3K/Akt pathway is required for a rapid increase in synapse density following MET activation, with no role for MAPK/ERK in synaptogenesis (55). Based on all findings, mediating different growth effects via distinct intracellular pathways has potential pathophysiological implications for developmental disruption of MAPK/Akt-PI3K balance (64).
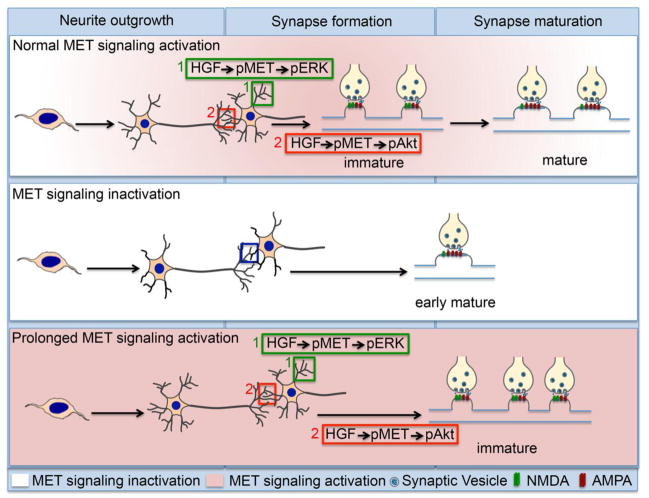
Figure 2. Functional outcomes of normal, decreased or increased MET receptor signaling during forebrain development.
During normal development (top panel), MET signaling is absent (white shading) during progenitor cell proliferation and neuron migration. MET signaling increases and is maintained at high levels (pink shading) during the early and peak periods of process outgrowth and synapse formation. At this time, activation of MET by its ligand, hepatocyte growth factor (HGF), results in an increase in the phosphorylated receptor (pMET), which activates the MAPK/ERK pathway to increase dendritic growth. In the same neurons, pMET also activates the PI3K/Akt pathway to promote immature synapse formation, denoted by the NMDA (green) and AMPA (red) glutamate receptor ratio in the postsynaptic membrane. During the period of synaptic refinement, MET signaling rapidly declines (white shading), and is accompanied by a maturation (increased AMPA:NMDA ratio) of the nascent synapses. In the absence of MET signaling (middle panel), there is a reduction in spine density, but an increase in spine head volume that is accompanied by a premature onset of synapse maturation (early AMPA:NMDA ratio increase). In contrast, prolonged MET signaling (bottom panel) increases dendritic growth and promotes nascent synapse formation, but delays synapse maturation.
In vitro and animal models have addressed the morphological and functional outcomes following long-term alterations in MET signaling. Increases or decreases in MET expression in vitro have opposing effects on spine development in CA1 hippocampal neurons (40). Specifically, overexpression of MET results in increased density of immature-looking dendritic protrusions and a decrease in spine head volume, whereas reduced MET expression leads to decreased mature spine density but a concomitant increase in spine head volume. The Metfx/fx/Emx1cre mouse has complex alterations in neocortical dendritic architecture (65), including an increase in spine volume, but no change in spine density. Increases in spine volume have been associated with increases in excitatory synaptic transmission and functional maturation (66, 67). Likewise, in hippocampal CA1 neurons, there is altered dendritic branching and increases in spine head volume, but these are accompanied by a decrease in spine density (40, 68). Interestingly, medium spiny neurons in the striatum also exhibit altered dendritic growth patterns and increases in spine head volume. These neurons receive MET-positive afferents from the neocortex, but do not themselves express MET, suggesting that MET signaling may also influence neurodevelopmental outcomes in a non-cell autonomous fashion (65).
MET function regulation of developing cortical circuits was determined using different electrophysiological methods in Metfx/fx/Emx1cre mice (40, 68, 69). In frontal cortex of both homozygous null and heterozygous mice, caged glutamate activation reveals a 2.3-fold increase in excitatory drive from layer 2/3 onto discrete populations of layer 5 neurons (69). This phenotype is restricted to contralaterally-projecting intratelencephalic corticostriatal neurons. There are no input changes in layer 5 pyramidal tract corticopontine neurons, emphasizing again that contribution to ASD risk may be instantiated in the findings of circuit-specific disruption following dysregulation of MET expression in humans and rodents. The local hyperconnectivity also is reminiscent of that reported in ASD using functional neuroimaging (70, 71). In the mouse hippocampal CA1 region, single-neuron analyses during the second postnatal week reveals excitatory synaptic changes. Premature maturation is evident in three measures, including a significantly larger ratio of stimulus-evoked AMPA to NMDA currents, a more rapid replacement of the GluN2B subunit of the NMDA receptor by GluN2A, and fewer silent synapses (40). This early maturation contrasts with the repression of the number and maturational state of glutamatergic synapses following prolonged activation of MET signaling in vitro (68). Interestingly, changes in trafficking and localization of postsynaptic proteins to the synapse, rather than altered levels of expression, underlie the physiological phenotype. Measures of glutamatergic maturational state become normalized by puberty, indicating that altered phenotypes expressed in the adult are likely developmentally mediated through a complex process of altered maturation due to MET receptor dysfunction (Figure 2).
MET, the periphery and ASD: GI and immune functions
MET signaling is pleiotropic in multiple peripheral organ systems (22–25, 72–74). Here, we describe the role of MET in two systems known to be dysregulated in subpopulations of individuals with ASD: GI and immune. Findings that the MET ‘C’ allele is enriched in children with ASD and co-occurring GID (26) stimulated a focus on the GI system. MET is expressed by intestinal epithelial cells, as well as enteric nervous system precursors and a subpopulation of differentiated myenteric neurons (22, 75). Rodent models of GI injury support a key role for MET signaling in intestinal mucosal wound repair and remodeling. Following injury, treatment with HGF increases MET activation in and improves the regeneration of colonic epithelium (76). Unexpectedly, mice in which Met is deleted from enteric neurons, but is still expressed in intestinal epithelium, also have an increased susceptibility to mucosal injury (22). Recent evidence suggests vagal innervation may modulate intestinal mucosal immune homeostasis by a mechanism known as the cholinergic anti-inflammatory pathway (reviewed in (77)), such that vagotomized mice also display increased susceptibility to experimentally-induced colitis (78, 79). In this context, mapping studies in rodent and monkey reveal highly enriched MET expression in neurons comprising brainstem autonomic nuclei contributing central efferents to the vagus nerve (80). Thus, MET signaling can contribute to GI homeostasis and injury responses in multiple ways, via epithelial, and peripheral and central neural mechanisms.
The role of MET in mediating aspects of immune responses also is well-delineated. It is not known, however, whether MET immune-mediated function relates to the reported enrichment of autoimmune disorders, allergies and asthma in the families of children with ASD (81), although these disturbances have been postulated as contributing factors to ASD symptoms (for review (82)). From a functional perspective, how might MET signaling be involved? Within the immune system, MET expression is low in inactive cells, but increases following activation in several cell classes, including monocytes/macrophages, dendritic cells and B cells (24, 83, 84). Normally, MET negatively regulates immune responsiveness in mice, attenuating inflammatory responses in a variety of contexts (85, 86). Thus, the reduced levels of MET and interleukin-10 expression in activated immune cells from individuals homozygous for the MET ‘C’ allele (27) may underlie immune comorbidities in at least some patients with ASD, as well as their family members. There is one direct link between disrupted MET signaling and immune function in mothers of children with ASD (27). Specifically, the MET ‘C’ allele is highly enriched with the presence of ASD-associated maternal antibodies that recognize a specific pattern of fetal brain proteins; these antibodies are functionally active in terms of brain disruptions (87, 88). The relationship with GI and immune dysfunction is well-delineated in ASD and, as noted above, MET signaling clearly is involved in both systems. The genetic, clinical and biological correlations are intriguing, but meaningful convergence of these data will require additional studies.
Beyond ASD: Broader implications for MET-related networks in NDDs
Thus far, we have considered MET in the context of normal neurodevelopmental function, and in ASD brain and periphery-related disturbances. MET may be more broadly involved in other NDDs. The weakest evidence comes from a haplotype block association, of small effect size, with schizophrenia (89). More intriguing data are from studies of Rett syndrome (RTT), typically defined genetically by mutations in the transcription factor MECP2. MeCP2 binds directly to the MET promoter in human neural progenitor cells in vitro (90). There is enhanced binding and transcription in the presence of the MET ‘C’ allele and RTT-specific mutations in MECP2 reduce MET transcription. There are two important findings in this study (90): 1) in females with RTT, MET transcript is reduced dramatically in postmortem temporal cortex compared to sex-matched controls, with a much greater effect size than in ASD - this contrasts with ASD, where reduced MET expression is observed only in males, and 2) unlike ASD, the MET ‘C’ allele is not enriched in RTT. These data indicate that dysregulated MET expression, potentially through distinct disease- and sex-related mechanisms, may contribute to the pathophysiology of multiple NDDs, not just ASD.
The broader NDD involvement by MET may reflect circuit-specific networks of proteins with which the receptor interacts to mediate neurodevelopmental processes at risk in NDDs. The nature of the MET protein network is likely dependent on cell and subcellular context, as well as activation state. Further, disruption of discrete components of the network may modulate the balance between activation of the MAPK/ERK and PI3K/Akt pathways, thus influencing functional outcome. Mutations in components or regulators of these two signaling pathways have been associated with ASD (64, 91) as well as syndromic or rare mutation NDDs (13, 92–100). Few studies to date, however, have examined specific protein networks in a neurodevelopmental context. To address this knowledge gap, we recently characterized the MET interactome in murine neocortical synaptosomes, isolated at the peak of synaptogenesis, using co-immunoprecipitation/mass spectrometry (101). Seventy-two interacting proteins were identified, which could include those that bind directly or indirectly to MET within the protein complex. Of these, nine (12.5%) – including MET - are associated with NDDs, based on high confidence criteria set by genetic consortia. Specifically, genes encoding five proteins in the MET interactome are associated with ASD, two with bipolar disorder and three with schizophrenia; no proteins encoded by genes associated with attention deficit hyperactivity disorder or major depressive disorder are represented in the MET interactome. The 9 NDD-associated candidates, together with their secondary protein interactors, form an interactome network, in which NDD-associated candidates are highly connected with each other at the center (Figure 3). A more refined enrichment analysis, using proteins determined experimentally to be expressed in a mouse neocortical synaptosome fraction (102) as a background comparison, reveals a very important relationship - only NDD- and ASD-associated proteins are enriched in the synaptic MET interactome. Furthermore, based on analysis of the human neocortical transcriptome (in BrainSpan), MET and its interactome proteins are co-regulated temporally and spatially from 0–24 postnatal months, compared to all other synaptic proteins. The MET synaptic protein network is complex, but this enrichment for NDD partners points towards a pronounced modulatory role for MET signaling in balancing synapse development along with other contributors to typical and atypical maturation in specific circuits.
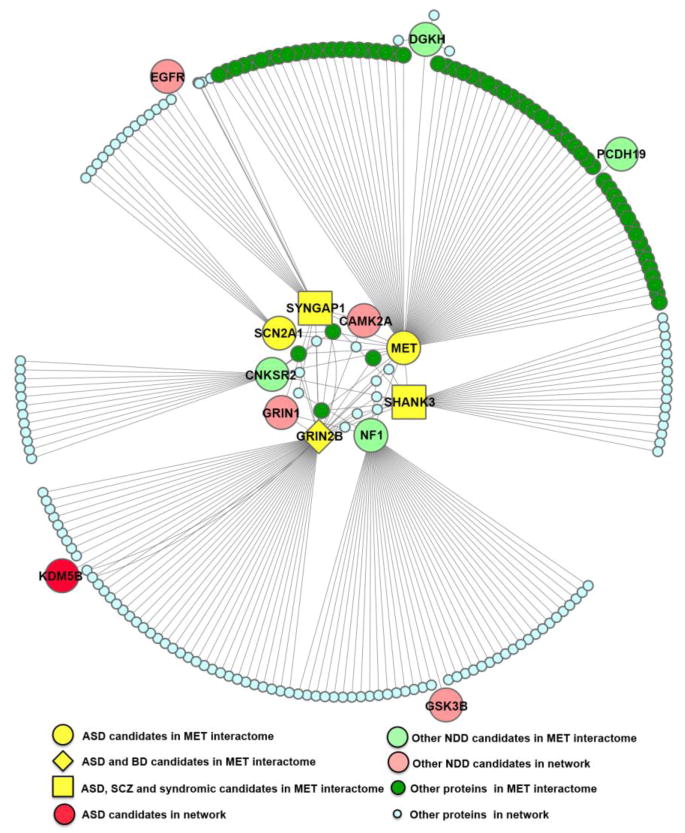
Figure 3. The MET interactome in the developing neocortical synapse is in a protein network enriched in NDD-associated candidates.
A protein-protein interaction network was generated using MET and the 72 MET-interacting candidates identified by immunoprecipitation and mass spectrometry analysis. Of the 72 proteins, there are 9 NDD-associated candidates (large yellow, pale green and salmon shapes), including MET. Secondary interactors of the NDD-associated candidates, identified in GeneMania (a human protein-protein interaction database), are shown as pale blue and dark green small circles, with several high confidence NDD candidates at the periphery of the network (larger circles). Note that NDD-associated candidates are highly connected with each other at the center of the network.
Not the center of the universe – but clearly part of it
The preponderance of data obtained over the past 10 years from model systems, clinical and neuroimaging studies supports the inclusion of the MET receptor tyrosine kinase as an important element when considering mechanisms of action in synapse development relevant to ASD and other NDDs. MET pleiotropy in systems causal to ASD co-occurring medical conditions provides further links. The dynamic spatial and temporal regulation of MET expression across evolution, in addition to co-regulation of key proteins with which MET interacts at the synapse, speaks to an involvement in influencing the quality of circuit development. This is not to overstate that dysregulated MET signaling will have dramatic systemic impact, but rather a greater influence on modulating the developmental timing, and ultimately the quality of circuit function. We suggest that MET may be biological relevant in a number of pathophysiological contexts, both centrally and peripherally. Why else would MET be one of only 32 genes (joining the serotonin transporter) across the entire genome overlapping in selection between two highly social species - humans and domesticated dogs (103)?
Supplementary Material
Acknowledgments
This work was supported by National Institute of Mental Health Grant MH067842, the Simms/Mann Chair in Developmental Neurogenetics and the Keck Chair in Neurogenetics.
Footnotes
Financial Disclosures
P Levitt is on the Scientific Advisory Board for Pediatric Biosciences.
K Eagleson and Z Xie report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Christensen DL, Baio J, Braun KV, Bilder D, Charles J, Constantino JN, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR Surveill Summ. 2016;65:1–23. doi: 10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Psychiatric AA. Diagnostic and statistical manual of mental disorders. 5. Arlington VA: American Psychiatric Association; 2013. [Google Scholar]
- 3.Kanner L. Autistic disturbances of affective contact. Nervous Child. 1943;2:217–250. [PubMed] [Google Scholar]
- 4.Levy SE, Giarelli E, Lee LC, Schieve LA, Kirby RS, Cunniff C, et al. Autism spectrum disorder and co-occurring developmental, psychiatric, and medical conditions among children in multiple populations of the United States. J Dev Behav Pediatr. 2010;31:267–275. doi: 10.1097/DBP.0b013e3181d5d03b. [DOI] [PubMed] [Google Scholar]
- 5.Peters B, Williams KC, Gorrindo P, Rosenberg D, Lee EB, Levitt P, et al. Rigid-compulsive behaviors are associated with mixed bowel symptoms in autism spectrum disorder. J Autism Dev Disord. 2014;44:1425–1432. doi: 10.1007/s10803-013-2009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salazar F, Baird G, Chandler S, Tseng E, O'Sullivan T, Howlin P, et al. Co-occurring Psychiatric Disorders in Preschool and Elementary School-Aged Children with Autism Spectrum Disorder. J Autism Dev Disord. 2015;45:2283–2294. doi: 10.1007/s10803-015-2361-5. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 8.Casey BJ, Oliveri ME, Insel T. A neurodevelopmental perspective on the research domain criteria (RDoC) framework. Biol Psychiatry. 2014;76:350–353. doi: 10.1016/j.biopsych.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC) toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 10.Organ SL, Tsao M-S. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3:S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corominas R, Yang X, Lin GN, Kang S, Shen Y, Ghamsari L, et al. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun. 2014;5:3650. doi: 10.1038/ncomms4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014 doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of Genes and Cellular Pathways Dysregulated in Autism Spectrum Disorders. The American Journal of Human Genetics. 2014:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson PB, Boccuto L, Skinner C, Collins JS, Neri G, Gurrieri F, et al. Further evidence that the rs1858830 C variant in the promoter region of the MET gene is associated with autistic disorder. Autism Res. 2009;2:232–236. doi: 10.1002/aur.87. [DOI] [PubMed] [Google Scholar]
- 17.Sousa I, Clark TG, Toma C, Kobayashi K, Choma M, Holt R, et al. MET and autism susceptibility: family and case-control studies. Eur J Hum Genet. 2009;17:749–758. doi: 10.1038/ejhg.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thanseem I, Nakamura K, Miyachi T, Toyota T, Yamada S, Tsujii M, et al. Further evidence for the role of MET in autism susceptibility. Neurosci Res. 2010;68:137–141. doi: 10.1016/j.neures.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Xu Y, Wang J, Zhou H, Liu X, Ayub Q, et al. Replication of the association of a MET variant with autism in a Chinese Han population. PLoS One. 2011;6:e27428. doi: 10.1371/journal.pone.0027428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell DB, D'Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62:243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- 21.Chaste P, Devlin B. A genomewide association study of autism using the Simons Simplex Collection: Does reducign phenotypic heterogeneity in autism increase genetic homogeneity? Biological Psychiatry. 2014:1–4. doi: 10.1016/j.biopsych.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avetisyan M, Wang H, Schill EM, Bery S, Grider JR, Hassell JA, et al. Hepatocyte Growth Factor and MET Support Mouse Enteric Nervous System Development, the Peristaltic Response, and Intestinal Epithelial Proliferation in Response to Injury. Journal of Neuroscience. 2015;35:11543–11558. doi: 10.1523/JNEUROSCI.5267-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura S, Kondo S, Shinomura Y, Isozaki K, Kanayama S, Higashimoto Y, et al. Expression of hepatocyte growth factor and c-met in ulcerative colitis. Inflammation research : official journal of the European Histamine Research Society [et al] 2000;49:320–324. doi: 10.1007/PL00000212. [DOI] [PubMed] [Google Scholar]
- 24.Beilmann M, Odenthal M, Jung W, Vande Woude GF, Dienes HP, Schirmacher P. Neoexpression of the c-met/hepatocyte growth factor-scatter factor receptor gene in activated monocytes. Blood. 1997;90:4450–4458. [PubMed] [Google Scholar]
- 25.McCall-Culbreath KD, Li Z, Zutter MM. Crosstalk between the alpha2beta1 integrin and c-met/HGF-R regulates innate immunity. Blood. 2008;111:3562–3570. doi: 10.1182/blood-2007-08-107664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM, et al. Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions. Pediatrics. 2009;123:1018–1024. doi: 10.1542/peds.2008-0819. [DOI] [PubMed] [Google Scholar]
- 27.Heuer L, Braunschweig D, Ashwood P, Van de Water J, Campbell DB. Association of a MET genetic variant with autism-associated maternal autoantibodies to fetal brain proteins and cytokine expression. Transl Psychiatry. 2011;1:e48. doi: 10.1038/tp.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell DB, Warren D, Sutcliffe JS, Lee EB, Levitt P. Association of MET with social and communication phenotypes in individuals with autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:438–446. doi: 10.1002/ajmg.b.30998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorrindo P, Williams KC, Lee EB, Walker LS, McGrew SG, Levitt P. Gastrointestinal dysfunction in autism: parental report, clinical evaluation, and associated factors. Autism Res. 2012;5:101–108. doi: 10.1002/aur.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambert N, Wermenbol V, Pichon B, Acosta S, van den Ameele J, Perazzolo C, et al. A familial heterozygous null mutation of MET in autism spectrum disorder. Autism Res. 2014;7:617–622. doi: 10.1002/aur.1396. [DOI] [PubMed] [Google Scholar]
- 31.Volk HE, Kerin T, Lurmann F, Hertz-Picciotto I, McConnell R, Campbell DB. Autism spectrum disorder: interaction of air pollution with the MET receptor tyrosine kinase gene. Epidemiology. 2014;25:44–47. doi: 10.1097/EDE.0000000000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349:1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willsey AJ, State MW. Autism spectrum disorders: from genes to neurobiology. Current Opinion in Neurobiology. 2015;30:92–99. doi: 10.1016/j.conb.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukamel Z, Konopka G, Wexler E, Osborn GE, Dong H, Bergman MY, et al. Regulation of MET by FOXP2, genes implicated in higher cognitive dysfunction and autism risk. J Neurosci. 2011;31:11437–11442. doi: 10.1523/JNEUROSCI.0181-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Judson MC, Bergman MY, Campbell DB, Eagleson KL, Levitt P. Dynamic gene and protein expression patterns of the autism-associated met receptor tyrosine kinase in the developing mouse forebrain. J Comp Neurol. 2009;513:511–531. doi: 10.1002/cne.21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Judson MC, Amaral DG, Levitt P. Conserved subcortical and divergent cortical expression of proteins encoded by orthologs of the autism risk gene MET. Cereb Cortex. 2011;21:1613–1626. doi: 10.1093/cercor/bhq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eagleson KL, Campbell DB, Thompson BL, Bergman MY, Levitt P. The autism risk genes MET and PLAUR differentially impact cortical development. Autism Res. 2011;4:68–83. doi: 10.1002/aur.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 40.Qiu S, Lu Z, Levitt P. MET receptor tyrosine kinase controls dendritic complexity, spine morphogenesis, and glutamatergic synapse maturation in the hippocampus. J Neurosci. 2014;34:16166–16179. doi: 10.1523/JNEUROSCI.2580-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell EM, Mars WM, Levitt P. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 2001;30:79–89. doi: 10.1016/s0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 42.Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 43.Martins GJ, Shahrokh M, Powell EM. Genetic disruption of Met signaling impairs GABAergic striatal development and cognition. Neuroscience. 2011;176:199–209. doi: 10.1016/j.neuroscience.2010.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedrick A, Lee Y, Wallace GL, Greenstein D, Clasen L, Giedd JN, et al. Autism Risk Gene MET Variation and Cortical Thickness in Typically Developing Children and Adolescents. Autism Res. 2012 doi: 10.1002/aur.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudie JD, Hernandez LM, Brown JA, Beck-Pancer D, Colich NL, Gorrindo P, et al. Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron. 2012;75:904–915. doi: 10.1016/j.neuron.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu GC-Y, Chung LWK. RANK-mediated signaling network and cancer metastasis. Cancer Metastasis Rev. 2014 doi: 10.1007/s10555-013-9488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson EB, Pourcain BS, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J, et al. Nat Genet. Nature Publishing Group; 2016. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population; pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson BL, Levitt P. Complete or partial reduction of the Met receptor tyrosine kinase in distinct circuits differentially impacts mouse behavior. J Neurodev Disord. 2015;7:35. doi: 10.1186/s11689-015-9131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okaty BW, Freret ME, Rood BD, Brust RD, Hennessy ML, de Bairos D, et al. Multi-Scale Molecular Deconstruction of the Serotonin Neuron System. Neuron. 2015;88:774–791. doi: 10.1016/j.neuron.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas MSC, Davis R, Karmiloff-Smith A, Knowland VCP, Charman T. The over-pruning hypothesis of autism. Developmental Science. 2015:n/a–n/a. doi: 10.1111/desc.12303. [DOI] [PubMed] [Google Scholar]
- 53.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nature Publishing Group. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 54.Eagleson KL, Milner TA, Xie Z, Levitt P. Synaptic and extrasynaptic location of the receptor tyrosine kinase Met during postnatal development in the mouse neocortex and hippocampus. J Comp Neurol. 2013 doi: 10.1002/cne.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eagleson KL, Lane CJ, McFadyen-Ketchum L, Solak S, Wu HH, Levitt P. Distinct intracellular signaling mediates C-MET regulation of dendritic growth and synaptogenesis. Dev Neurobiol. 2016 doi: 10.1002/dneu.22382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutierrez H, Dolcet X, Tolcos M, Davies A. HGF regulates the development of cortical pyramidal dendrites. Development. 2004;131:3717–3726. doi: 10.1242/dev.01209. [DOI] [PubMed] [Google Scholar]
- 57.Lim CS, Walikonis RS. Hepatocyte growth factor and c-Met promote dendritic maturation during hippocampal neuron differentiation via the Akt pathway. Cell Signal. 2008;20:825–835. doi: 10.1016/j.cellsig.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finsterwald C, Martin JL. Cellular mechanisms underlying the regulation of dendritic development by hepatocyte growth factor. Eur J Neurosci. 2011;34:1053–1061. doi: 10.1111/j.1460-9568.2011.07839.x. [DOI] [PubMed] [Google Scholar]
- 59.Tyndall SJ, Walikonis RS. The receptor tyrosine kinase Met and its ligand hepatocyte growth factor are clustered at excitatory synapses and can enhance clustering of synaptic proteins. Cell Cycle. 2006;5:1560–1568. doi: 10.4161/cc.5.14.2918. [DOI] [PubMed] [Google Scholar]
- 60.Nakano M, Takagi N, Takagi K, Funakoshi H, Matsumoto K, Nakamura T, et al. Hepatocyte growth factor promotes the number of PSD-95 clusters in young hippocampal neurons. Exp Neurol. 2007;207:195–202. doi: 10.1016/j.expneurol.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 61.Borowiak M, Garratt AN, Wustefeld T, Strehle M, Trautwein C, Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci U S A. 2004;101:10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brusevold IJ, Aasrum M, Bryne M, Christoffersen T. Migration induced by epidermal and hepatocyte growth factors in oral squamous carcinoma cells in vitro: role of MEK/ERK, p38 and PI-3 kinase/Akt. J Oral Pathol Med. 2012;41:547–558. doi: 10.1111/j.1600-0714.2012.01139.x. [DOI] [PubMed] [Google Scholar]
- 63.Chang CC, Chiu JJ, Chen SL, Huang HC, Chiu HF, Lin BH, et al. Activation of HGF/c-Met signaling by ultrafine carbon particles and its contribution to alveolar type II cell proliferation. Am J Physiol Lung Cell Mol Physiol. 2012;302:L755–763. doi: 10.1152/ajplung.00350.2011. [DOI] [PubMed] [Google Scholar]
- 64.Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest. 2009;119:747–754. doi: 10.1172/JCI37934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Judson MC, Eagleson KL, Wang L, Levitt P. Evidence of cell-nonautonomous changes in dendrite and dendritic spine morphology in the met-signaling-deficient mouse forebrain. J Comp Neurol. 2010;518:4463–4478. doi: 10.1002/cne.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 68.Peng Y, Lu Z, Li G, Piechowicz M, Anderson M, Uddin Y, et al. The autism-associated MET receptor tyrosine kinase engages early neuronal growth mechanism and controls glutamatergic circuits development in the forebrain. Mol Psychiatry. 2016 doi: 10.1038/mp.2015.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu S, Anderson CT, Levitt P, Shepherd GM. Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated Met receptor tyrosine kinase. J Neurosci. 2011;31:5855–5864. doi: 10.1523/JNEUROSCI.6569-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2:56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Courchesne E, Pierce K. Why the frontal cortex in autism might be talking only to itself: local over-connectivity but long-distance disconnection. Curr Opin Neurobiol. 2005;15:225–230. doi: 10.1016/j.conb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 72.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, et al. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 73.Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, et al. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 74.Ishibe S, Karihaloo A, Ma H, Zhang J, Marlier A, Mitobe M, et al. Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development. 2009;136:337–345. doi: 10.1242/dev.024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Renzo MF, Narsimhan RP, Olivero M, Bretti S, Giordano S, Medico E, et al. Expression of the Met/HGF receptor in normal and neoplastic human tissues. Oncogene. 1991;6:1997–2003. [PubMed] [Google Scholar]
- 76.Tahara Y, Ido A, Yamamoto S, Miyata Y, Uto H, Hori T, et al. Hepatocyte growth factor facilitates colonic mucosal repair in experimental ulcerative colitis in rats. J Pharmacol Exp Ther. 2003;307:146–151. doi: 10.1124/jpet.103.054106. [DOI] [PubMed] [Google Scholar]
- 77.Matteoli G, Boeckxstaens GE. The vagal innervation of the gut and immune homeostasis. Gut. 2013;62:1214–1222. doi: 10.1136/gutjnl-2012-302550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O'Mahony C, van der Kleij H, Bienenstock J, Shanahan F, O'Mahony L. Loss of vagal anti-inflammatory effect: in vivo visualization and adoptive transfer. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1118–1126. doi: 10.1152/ajpregu.90904.2008. [DOI] [PubMed] [Google Scholar]
- 79.Ghia JE, Blennerhassett P, Collins SM. Vagus nerve integrity and experimental colitis. Am J Physiol Gastrointest Liver Physiol. 2007;293:G560–567. doi: 10.1152/ajpgi.00098.2007. [DOI] [PubMed] [Google Scholar]
- 80.Wu HH, Levitt P. Prenatal expression of MET receptor tyrosine kinase in the fetal mouse dorsal raphe nuclei and the visceral motor/sensory brainstem. Dev Neurosci. 2013;35:1–16. doi: 10.1159/000346367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aldinger KA, Lane CJ, Veenstra-VanderWeele J, Levitt P. Patterns of Risk for Multiple Co-Occurring Medical Conditions Replicate Across Distinct Cohorts of Children with Autism Spectrum Disorder. Autism Res. 2015;8:771–781. doi: 10.1002/aur.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Estes ML, McAllister AK. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci. 2015;16:469–486. doi: 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Q, DeFrances MC, Zarnegar R. Induction of met proto-oncogene (hepatocyte growth factor receptor) expression during human monocyte-macrophage differentiation. Cell Growth Differ. 1996;7:821–832. [PubMed] [Google Scholar]
- 84.Ilangumaran S, Villalobos-Hernandez A, Bobbala D, Ramanathan S. The hepatocyte growth factor (HGF)-MET receptor tyrosine kinase signaling pathway: Diverse roles in modulating immune cell functions. Cytokine. 2016;82:125–139. doi: 10.1016/j.cyto.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 85.Futamatsu H, Suzuki J, Mizuno S, Koga N, Adachi S, Kosuge H, et al. Hepatocyte growth factor ameliorates the progression of experimental autoimmune myocarditis: a potential role for induction of T helper 2 cytokines. Circ Res. 2005;96:823–830. doi: 10.1161/01.RES.0000163016.52653.2e. [DOI] [PubMed] [Google Scholar]
- 86.Ito W, Kanehiro A, Matsumoto K, Hirano A, Ono K, Maruyama H, et al. Hepatocyte growth factor attenuates airway hyperresponsiveness, inflammation, and remodeling. Am J Respir Cell Mol Biol. 2005;32:268–280. doi: 10.1165/rcmb.2004-0058OC. [DOI] [PubMed] [Google Scholar]
- 87.Piras IS, Haapanen L, Napolioni V, Sacco R, Van de Water J, Persico AM. Anti-brain antibodies are associated with more severe cognitive and behavioral profiles in Italian children with Autism Spectrum Disorder. Brain Behavior and Immunity. 2014 doi: 10.1016/j.bbi.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bauman MD, Iosif AM, Ashwood P, BRAUNSCHWEIG D, Lee A, Schumann CM, et al. Maternal antibodies from mothers of children with autism alter brain growth and social behavior development in the rhesus monkey. Translational Psychiatry. 2013;3:e278–212. doi: 10.1038/tp.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burdick KE, DeRosse P, Kane JM, Lencz T, Malhotra AK. Association of genetic variation in the MET proto-oncogene with schizophrenia and general cognitive ability. Am J Psychiatry. 2010;167:436–443. doi: 10.1176/appi.ajp.2009.09050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Plummer JT, Evgrafov OV, Bergman MY, Friez M, Haiman CA, Levitt P, et al. Transcriptional regulation of the MET receptor tyrosine kinase gene by MeCP2 and sex-specific expression in autism and Rett syndrome. Transl Psychiatry. 2013;3:e316. doi: 10.1038/tp.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13:247. doi: 10.1186/gb-2012-13-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marui T, Hashimoto O, Nanba E, Kato C, Tochigi M, Umekage T, et al. Association between the neurofibromatosis-1 (NF1) locus and autism in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2004;131b:43–47. doi: 10.1002/ajmg.b.20119. [DOI] [PubMed] [Google Scholar]
- 93.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 97.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 98.Varga EA, Pastore M, Prior T, Herman GE, McBride KL. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med. 2009;11:111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 99.Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- 100.Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, et al. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22:9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xie Z, Li J, Baker J, Eagleson KL, Coba MP, Levitt P. Receptor Tyrosine Kinase MET Interactome and Neurodevelopmental Disorder Partners at the Developing Synapse. Biol Psychiatry. 2016 doi: 10.1016/j.biopsych.2016.02.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moczulska KE, Pichler P, Schutzbier M, Schleiffer A, Rumpel S, Mechtler K. Deep and precise quantification of the mouse synaptosomal proteome reveals substantial remodeling during postnatal maturation. J Proteome Res. 2014;13:4310–4324. doi: 10.1021/pr500456t. [DOI] [PubMed] [Google Scholar]
- 103.Wang GD, Zhai W, Yang HC, Fan RX, Cao X, Zhong L, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nature Communications. 2013;4:1860. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.