Abstract
We report the first imported case of Rift Valley fever (RVF) in China. The patient returned from Angola, a non-epidemic country, with an infection of a new reassortant from different lineages of Rift Valley fever viruses (RVFVs). The patient developed multiorgan dysfunction and gradually recovered with continuous renal replacement therapy and a short regimen of methylprednisolone treatment. The disordered cytokines and chemokines in the plasma of the patient revealed hypercytokinemia, but the levels of protective cytokines were low upon admission and fluctuated as the disease improved. Whole-genome sequencing and phylogenetic analysis revealed that the imported strain was a reassortant comprising the L and M genes from lineage E and the S gene from lineage A. This case highlights that RVFV had undergone genetic reassortment, which could potentially alter its biological properties, cause large outbreaks and pose a serious threat to global public health as well as the livestock breeding industry.
Keywords: hypercytokinemia, imported case, reassortment, Rift Valley fever virus
INTRODUCTION
The first clinical report of Rift Valley fever (RVF) in humans was made in an area near Lake Naivasha of the Rift Valley province in Kenya in 1930.1 RVF epizootics and epidemics in livestock and humans have periodically occurred and were geographically restricted to sub-Saharan Africa, but since 2000, this disease has spread to the Arabian Peninsula.2 Rift Valley fever virus (RVFV) infection is correlated with several risk factors, including contact with sick animals or contaminated products or exposure to virus-carrying mosquitoes.3 Sero-epidemiology revealed anti-RVFV IgG antibodies among livestock and human in countries such as Djibouti where RVF outbreaks have never been reported in either humans or animals,4 suggesting the presence of subclinical virus circulation in non-epidemic areas.
Although RVFV has been described in an Angolan returning from South Africa,5 and circulating RVFV was reported among animals and humans in other Central African countries such as Central African Republic,6 no epizootic or epidemic occurrences have been reported in Angola. Herein, we describe the first case of imported RVFV infection from Angola to China. The longitudinal observation of the clinical manifestations and pro-inflammatory immune mediators of this severe case were reported, and our phylogenetic analysis revealed the virus to be a novel reassortant between lineages E and A.
MATERIALS AND METHODS
Laboratory diagnosis
On day 7 after initial presentation of symptoms in Angola, the patient returned to Beijing, and total RNA was extracted from 140 μL of blood and saliva samples using the QIAmp Viral RNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Using a conventional real-time PCR (RT-PCR) by using Agpath-ID One-Step RT-PCR kit (Thermo, Carlsbad, CA, USA) assay, yellow fever, malaria and chikungunya fever were excluded. Dengue fever, haemorrhagic fever with renal syndrome and viral hepatitis A to E were excluded based on the results of enzyme-linked immunosorbent assays (ELISAs, the antigen and antibody detection kits were from Panbio, Windsor, Australia and Wantai, Beijing, China). The serum sample and saliva sample were both positive for RVFV based on the results of the Agpath-ID One-step RT-PCR kit (ABI, San Francisco, CA, USA), which targets the M segment of RVFV (forward primer: 5′-AGG AAC AAT GGA CTC TGG T-3′, reverse primer: 5′-TTC TTA CTA CCA TGT CCT CC-3′, probe: 5′-AGC TTT GAT ATC TCT CAG TGC CCC A-3′).
Inflammatory mediator tests
We collected plasma samples from the patient daily since hospitalization and measured the levels of different cytokines and chemokines (Supplementary Table S1) using the Bio-Plex Pro Human Cytokine Array 27-Plex Group I and 21-Plex Group II Kits on a Luminex200 Multiplexing Instrument (Merck Millipore, Darmstadt, Germany) following the manufacturers' instructions. The raw data were analysed using xPONENT 3.1 software (Merck Millipore). Plasma from seven healthy individuals were used as controls.
Virus genome sequencing and analysing
RNA extracted from blood was used for genome sequencing. Whole genome sequencing was performed using an Ion Torrent PGM Platform (Thermo Fisher Scientific, San Francisco, CA, USA). No readouts corresponding to other haemorrhagic fever viruses were found. All of the RVFV nucleotide sequences available from GenBank were downloaded, and only those that were nearly full length (>85%) were used. This left three datasets corresponding to the L (n=107), M (n=115) and S (n=171) gene segments, which were aligned using Muscle7 and then manually adjusted. Phylogenetic analysis was performed on the three single gene segments and the four coding gene regions using RAxML.8, 9
The NCBI accession numbers for the L, M and S segments of RVFV are KX611605, KX611606 and KX611607, respectively.
RESULTS
Patient history
The 45-year-old patient presented a fever (38.3 °C) associated with chills, malaise, headache, myalgia and large-joint arthralgia on day 1 of disease onset (16 July 2016) in Luanda, Angola. The patient was empirically treated for yellow fever with oral acetaminophen, an intravenous infusion of 5% glucose and a sodium chloride injection at a local hospital. Three days later, his temperature returned to normal, but he developed worsening symptoms of fatigue, malaise, nausea, vomiting, anorexia and severe oliguria. On day 5, he presented midepigastric discomfort and jaundice, and the laboratory tests suggested liver and kidney injury. On the morning of day 7, the patient returned to Beijing and was admitted to Beijing Ditan Hospital.
The patient worked as a forklift worker in Luanda, Angola since February 2014, and lived in the rural district of Luanda. The patient was frequently bitten by mosquitoes. There was no history of contact with livestock or humans with fever, and the patient did not travel out of Luanda since February 2014. Based on the epidemiological data, the most likely source of infection was a mosquito bite.
Findings on admission
Upon admission to our hospital on day 7, the patient's vital signs were normal (temperature, 37 °C; heart rate, 90 beats per min; respiration rate, 20 per min; and blood pressure, 120/70 mm Hg). He was awake, alert and fully oriented. Physical examination revealed scleral icterus, no splenomegaly or hepatomegaly and no joint tenderness or swelling. Rash and haemorrhagic tendency were absent.
The laboratory tests on day 7 showed renal failure and severe liver damage, with creatinine levels of 1005 μmol/L, blood urea nitrogen levels of 35 mmol/L, total bilirubin levels of 83.8 μmol/L, alanine aminotransferase of 5910 IU/L, aspartate amino transferase of 7570 IU/L and prothrombin levels of 74% (Table 1). Additional abnormal laboratory values included lactate dehydrogenase (1880 U/L), creatine kinase (6680 U/L), and myohaemoglobin (1200 ng/mL; Table 1). The patient was transferred to the intensive care unit because of his presentation of multiorgan dysfunction, including acute kidney injury, acute hepatitis, acute myocardial injury, pancreatitis and rhabdomyolysis.
Table 1. Clinical variables and laboratory values during the course of the patient's illnessa.
| Clinical and laboratory findings | Day7b | Day 8 | Day 9 | Day 10 | Day 11 | Day 12 | Day 13 | Day 17 | Day 20 | Day 29 | Normal range |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical variables | |||||||||||
| Temperature (°C) | 37.0 | 36.8 | 36.7 | 37.2 | 36.9 | 36.7 | 37.8 | 37.7 | 38.0 | 37.0 | 36–37 |
| Mean artery blood pressure (mm Hg) | 77 | 81 | 89 | 103 | 82 | 102 | 105 | 110 | 97 | 94 | 70–105 |
| Urinary output (mL/24 h) | 0 | 0 | 0 | 200 | 300 | 500 | 500 | 1800 | 3500 | 2800 | 1500–2500 |
| Laboratory values | |||||||||||
| WBC ( × 109/L) | 6.77 | 5.91 | 8.19 | 8.61 | 9.09 | 7.07 | 6.54 | 6.27 | 6.97 | 6.85 | 4.00–10.00 |
| Neutrophils ( × 109/L) | 4.84 | 4.53 | 5.93 | 6.06 | 6.20 | 4.53 | 4.19 | 4.28 | 4.50 | 5.10 | 2.00–8.00 |
| Platelets ( × 109/L) | 99 | 104 | 70 | 79.4 | 85.4 | 63 | 54 | 129 | 202 | 280 | 100–300 |
| Haemoglobin (g/L) | 155.4 | 137.4 | 129.4 | 130.4 | 129.0 | 130.0 | 115.0 | 97.0 | 77.0 | 75.0 | 120.0–160.0 |
| ALT (U/L) | 5910 | 5394 | 3052 | 1968 | 857 | 622 | 398 | — | 67 | 194 | 9–50 |
| AST (U/L) | 7570 | 6180 | 1819 | 612 | 125 | 70 | 87 | — | 46 | 90 | 15–40 |
| TBIL (μmol/L) | 83.8 | 87.3 | 87.4 | 59.7 | 46.1 | 45.0 | 68.2 | — | 19.0 | 16.6 | 0–18.8 |
| DBIL (μmol/L) | 71.8 | 73.4 | 75.9 | 52.4 | 37.6 | 37.7 | 55.3 | — | 14.4 | 11.1 | 0–6.8 |
| LDH (U/L) | 1880 | 2754 | 1580 | 807 | 523 | — | 362 | — | — | — | 80–285 |
| MYO (ng/mL) | 1200 | 1020 | — | — | 197 | 102 | — | — | — | — | 0–140 |
| CK (U/L) | 6680 | 6332 | — | 819 | 74 | — | 44 | — | — | — | 38–174 |
| CK-Mb (U/L) | 86 | 77 | — | 26 | 15 | — | 7 | — | — | — | <25 |
| Creatinine (μmol/L) | 1005 | 1097 | 540 | 275 | 217 | 274 | 244 | 338 | 233 | 139 | 59–104 |
| BUN (mmol/L) | 35.2 | 37.9 | 19.7 | 10.4 | 10.6 | 13.0 | 10.9 | 8.7 | 16.6 | 8.0 | 1.7–8.3 |
| PT (s) | 14.0 | 14.1 | 10.3 | 10.3 | 10.7 | 10.8 | 12.0 | 11.3 | — | 10.2 | 9.4–12.5 |
| AMY (U/L) | 132 | — | 284 | 275 | 192 | — | — | — | — | — | 35–135 |
| LPS (U/L) | 58.0 | — | 468.0 | 341.0 | 67.0 | — | — | — | — | — | 5.6–51.0 |
| CRP (mg/L) | 18.5 | 17.7 | — | — | 16.1 | 28.5 | — | — | — | 0–5.0 | |
| PCT (ng/mL) | 71.80 | — | — | — | — | — | — | 6.80 | — | — | <0.05 |
Abbreviations: alanine aminotransferase, ALT; amylase, AMY; aspartate aminotransferase, AST; blood urea nitrogen, BUN; creatine kinase, CK; C-reactive protein, CRP; direct bilirubin, DBIL; lactate dehydrogenase, LDH; lipase, LPS; myohaemoglobin, MYO; procalcitonin, PCT; prothrombin time, PT; total bilirubin, TBIL; white blood cell, WBC.
Unavailable information for the tests is denoted as '—'.
Days after disease onset.
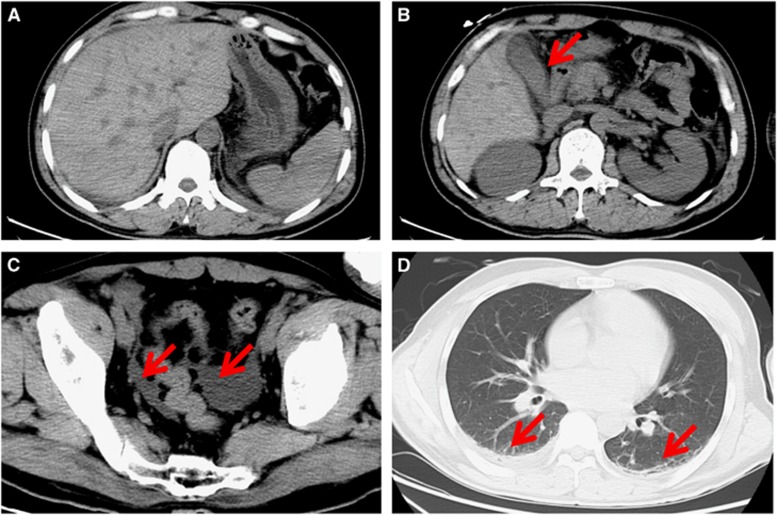
Computed tomography scanning showed pneumonia in the double upper and lower lobes of the lung, pleural effusion, cholecystitis and a small amount of ascites; however, the head CT scan was normal (Figure 1). An ultrasonic cardiogram showed that the left ventricular ejection fraction was 60% and that the heart structure had no obvious abnormalities.
Figure 1.
Computed tomography images of the patient on day 7 after clinical presentation of infection. (A) Normal morphology and density of the liver, spleen, pancreas and kidney. (B) Gallbladder wall thickening (red arrow). (C) Fluid in the lower abdominal and pelvic cavities (red arrows). (D) A reduced amount of lower pulmonary effusion and pleural effusion (red arrows).
RVFV nucleic acids were detectable in the serum and saliva samples with Ct values of 28.7 and 31.0, respectively, on day 7.
Clinical course and management
The patient was treated with continuous renal replacement therapy to maintain fluid volume and electrolyte balance. Nausea and pancreatitis limited oral intake, and parenteral nutrition of three litres was given every day with a zero net-volume balance during the first week. For the first three days after admission to the hospital, the patient was also treated with intravenous glycyrrhizinate and reduced glutathione for acute liver injury as well as with methylprednisolone (80 mg/day) to reduce the inflammatory response.
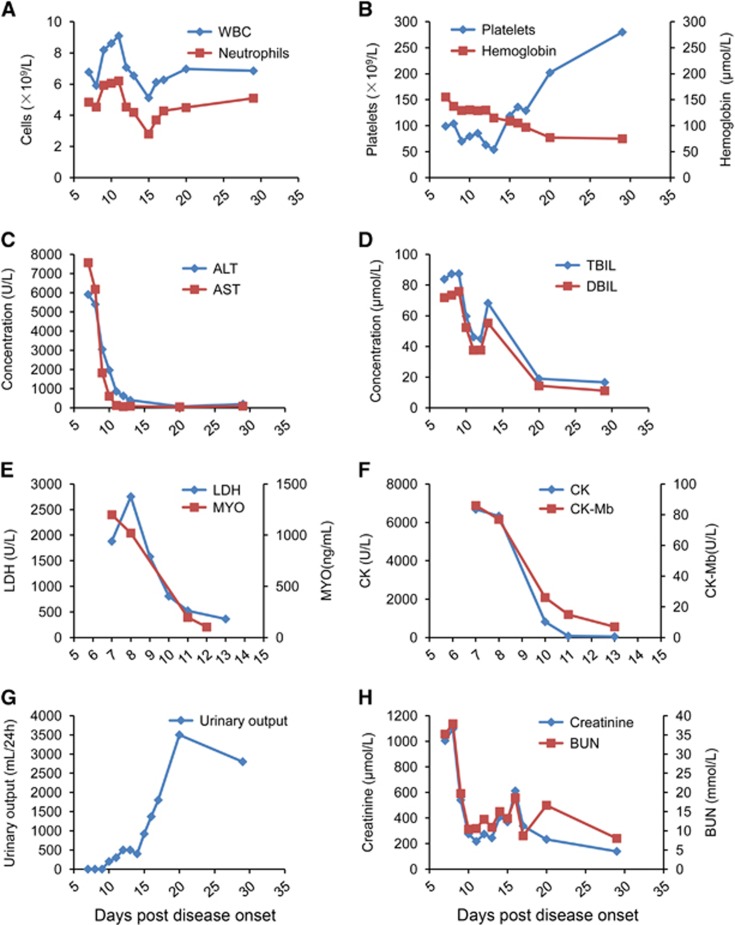
On day 9, the nausea and vomiting improved, but the anorexia and weakness persisted without further complications. On day 13, his symptoms were ameliorated, and the renal failure and liver injury had improved (Figure 2). He underwent intermittent haemodialysis once every two days starting on day 13. On day 17, the thrombocytopenia improved, and urine output significantly increased (Figure 2). On day 20, the serum creatinine level spontaneously began to decline; thus, the venous catheter was removed. On day 29, his renal function almost recovered, with only the serum aminotransferase levels still slightly elevated (Figure 2). His visual field testing and neurological exam were normal throughout the duration of his hospital stay. Clinical signs and laboratory tests were observed continuously and evaluated daily (Table 1). The patient made a full recovery and was discharged on day 51 (5 September 2016).
Figure 2.
The longitudinal variation of the clinical and laboratory values. The clinical and laboratory variables during the patient's disease progression are shown. (A) White blood cells (WBC) and neutrophils. (B) Platelets and hemoglobin. (C) Alanine aminotransferase (ALT) and aspartate aminotransferase (AST). (D) Total bilirubin (TBIL) and direct bilirubin (DBIL). (E) Lactate dehydrogenase (LDH) myohaemoglobin (MYO). (F) Creatine kinase (CK) and CK-Mb isoenzyme. (G) Urinary output per day. (H) Creatinine and blood urea nitrogen (BUN).
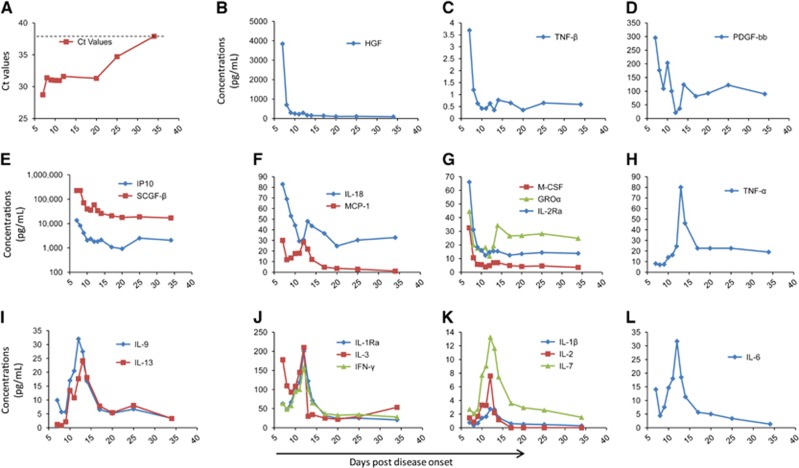
To evaluate the viremia of the patient, we used the Ct values from the RT-PCR results of the patient's blood samples; these values are inversely correlated with the viral RNA shedding. The viremia was ameliorated over the first several days with RVFV nucleic acid Ct values of 28.7 on day 7 to 31.4 on day 8. This Ct value was maintained over the following days. From day 20 to day 35, the viremia showed continuous abatement in the patient, and on day 35, the Ct value reached the detection cut-off of the RT-PCR kit for RVFV (38.0, Figure 3A), reflecting the end of the viremia.
Figure 3.
Viremia and hypercytokinemia in the RVF patient. (A) The Ct values are based on the RVFV-specific RT-PCR tests by using the blood collected daily from the patient. (B–L). The longitudinal test of the cytokine and chemokine levels in the plasma from the patient. The daily cytokines and chemokines values are listed in Supplementary Table S1. hepatocyte growth factor, HGF; tumour necrosis factor, TNF; platelet-derived growth factor, PDGF; interferon gamma-induced protein 10, IP10; stem cell growth factor, SCGF-β interleukin, IL; monocyte chemoattractant protein, MCP; macrophage colony-stimulating factor, M-CSF; growth-regulated oncogene, GRO; IL-1 receptor antagonist, IL-1Ra; interferon-gamma, IFN-γ. The concentration of IP10 on day 7 was greater than the highest concentration in the standard curve. Thus, in the 3E, we used the highest point of standard curve (13518.8 pg/mL) as the IP10 concentration on day 7.
Inflammatory mediators
In general, hypercytokinemia was observed on day 7 after disease onset. Overall, in plasma collected from the patient on day 7, 21 cytokines and chemokines were higher than the upper end of the 95% confidence interval (CI) of the means of the corresponding controls (Supplementary Table S1). Notably, the elevation of the cytokines and chemokines HGF, TNF-β, IP10, SCGF-b, IL-18, MCP-1, M-CSF, GROa and IL-2Ra on day 7 were higher than the upper end of the 95% CI of the healthy controls and showed an obviously rapid decrease after treatment initiation in the following days (Figures 3B, 3C, 3E and 3G). These data confirmed the presence of hypercytokinemia, also known as a ‘cytokine storm', which may have contributed to the immunopathogenesis in the patient. In contrast, the levels of cytokines such as IL-1β, IL-1Ra, IL-2, IL-3, IL-6, IL-7, IL-9, IL-13, IFN-γ and TNF-α, which are correlated with protective immune responses to viruses, were gradually increased in conjunction with the improvement of the symptoms (Figures 3H–3L). The IL-9, IL-13 and TNF-α levels on day 7 were even below the lower end of the 95% CI of healthy controls but increased afterwards (Supplementary Table S1). Starting at day 14, the levels of these cytokines gradually reduced concomitant with the amelioration of viremia in the patient, which may reveal the role of the adaptive immune process. Interestingly, PDGF-bb, which has a significant role in blood vessel formation, showed a generally decreasing trend in the following days after admission, indicating the reduction of the expression of vessel damage factors (Figure 3D).
Phylogenetic analysis
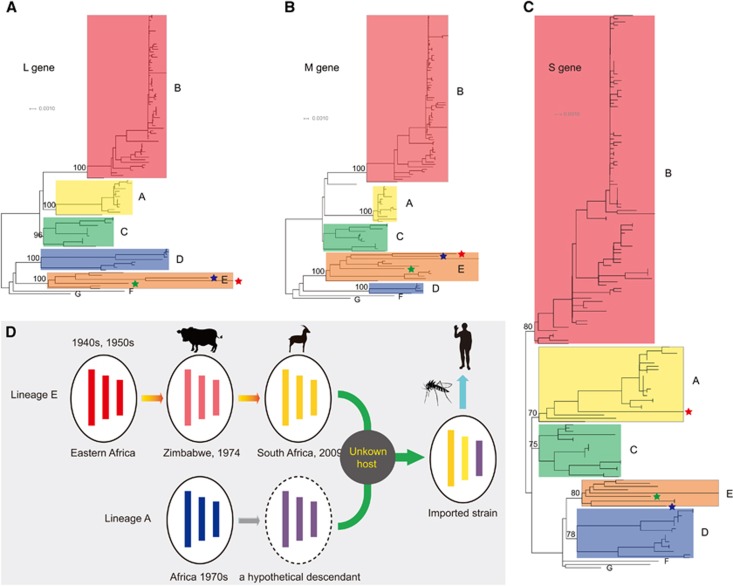
Consistent with a previous report,10 our phylogenetic trees estimated from the L, M and S gene sequences could also be classified into seven independent lineages (A–G, Figures 4A–4C) despite the inclusion of more newly sequenced strains. In the phylogenetic trees constructed using the L and M gene segments (Figures 4A and 4B, Supplementary Figures S1 and S2), the imported strain fell within lineage E and was closely related to a South African strain (Kakamas) isolated from sheep from 2009. Lineage E also includes several viruses isolated from eastern Africa in the 1940s and 1950s as well as one bovine strain from Zimbabwe in 1974. In the S gene tree, the 2009 South African strain still fell within lineage E close to the bovine strain from Zimbabwe in 1974 (Figure 4C and Supplementary Figure S3). However, the imported strain in China fell within lineage A and was clustered with several strains from Egypt, Madagascar, Zimbabwe and Central Africa from the 1970s, one South African strain from 1981 and one Namibian strain from 2004 (Figure 4C and Supplementary Figure S3).10 We then estimated the mean distances of the S gene sequences. In accordance with the phylogenetic analysis, the mean distance between the imported strain and lineage A was 2.5%±0.4%, while that between the imported strain and lineage E was 3.5%±0.4%. We further detected potential recombination events using the complete S gene segment, the non-structural gene region and the nucleocapsid gene region using the Recombination Detection Program (RDP),11 respectively, and no positive recombination signals were observed. Therefore, all of the evidence suggested that the imported strain might be a reassortant with the L and M genes from lineage E and the S gene from lineage A.
Figure 4.
Phylogenetic analysis of the worldwide RVFV gene sequences and the proposed model for the potential source of the first case of imported RVF in China. The phylogenetic trees constructed using the L, M and S gene sequences of the worldwide RVFVs are displayed in A–C, respectively. In these panels, the red star represents the first imported RVF case in China, the blue star represents the South African strain from sheep from 2009, and the green star represents the bovine strain from Zimbabwe in 1974. (D) The proposed model for the potential source of the first imported RVF case in China based on phylogenetic evidence.
DISCUSSION
RVFV can cause symptoms with different severities. Most people with RVF have either no symptoms or a mild illness associated with fever and liver abnormalities. A small percentage of patients develop severe indications including liver failure, renal failure, thrombocytopenia, encephalitis, haemorrhage and miscarriage.12, 13, 14, 15, 16 For the first case of imported RVF in China, we used high-volume continuous renal replacement therapy, which treated the renal failure, and methylprednisolone, which reduced the inflammation in the patient.17
In previous studies,18, 19 the pro-inflammatory mediators IL-1α, IL-1Ra, IL-6, IL-8, IL-10, MIG and IP10 were significantly increased in fatal cases. In our study, 21 cytokines and chemokines in the plasma of the patient were measured upon admission and were higher than the levels in healthy controls, which further demonstrated the contribution of dysregulated inflammatory responses in host individuals to RVFV pathogenesis. The non-structural protein NSs, which is encoded by the S segment of RVFV, has been implicated as the primary virulence factor. NSs can counteract the antiviral effects of the host's type I interferon response.20, 21 In this patient on day 7 of disease onset, the levels of cytokines related to the protective immune response (Figures 3H–3L) were equal to or lower than normal levels but gradually increased on day 9 concomitant with the improvement of the disease symptoms. Whether the disordered cytokines and chemokines are associated with the novel genetic constellation of this virus warrants further investigation.
Genetic reassortment of RVFV has been previously reported, and in the present study, we describe a novel RVFV reassortant between lineages E and A. Combined with the phylogenetic evidence and epidemiological data, we propose a model to illustrate the potential source of the imported strain (Figure 4D). RVFV was regarded as originating from eastern Africa and was transmitted to southern Africa in the 1950s.2 The descendants of these eastern African strains, such as the Zimbabwe strain (2373/74) and the South African strain (Kakamas), continued to circulate in southern Africa. In the 1970s, another RVFV lineage (lineage A) with a genetic constellation that differed from that of the southern African lineage swept through several African countries and was also transmitted to southern Africa no later than the 1970s. Genetic reassortment might have occurred between descendants of these two lineages in an unknown host somewhere in southern Africa, which would have given rise to the imported strain described here.
It should be noted that the imported strain contains a reassorted S gene segment from lineage A. As mentioned above, the S gene of RVFV is an important virulence factor. Because many strains from lineage A are highly pathogenic and lethal to WF rats10 and infected more than 200 000 humans in the 1977–1978 Egyptian outbreak (including 598 fatalities),2 we believe that the potential influence of the reassorted S gene on the biological properties of the imported strain requires further study. In addition, whether this reassortment was responsible for the severe clinical outcomes of this patient should be pursued as well.
The patient returned from Angola, a non-epidemic country of RVFV. The subclinical infection results in long-term RVFV circulation in a wide range of hosts throughout Africa, thus increasing the possibility of the accumulation of genetic mutations in the virus via genetic recombination, genetic reassortment and random point mutations. These ‘neglected' viral variants might result in large outbreaks and potentially pose a threat to public health, as we have learned from the 2014–2016 Ebola outbreak in western Africa,22 the 2016 Zika virus outbreak in South America and the Caribbean countries23 and the 2015 Middle East respiratory syndrome virus outbreak in eastern Asia.24, 25 Therefore, vaccines and antiviral agents should be developed in order to control these ‘neglected' tropical diseases.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC, Grant NO 81590761, 81301483, 3150050174, 81373141, 81401312, 81502857 and 31401270), the Megaproject for Infectious Disease Research of China (Grant NO: 2016ZX10004222-003), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (Grant NO: ZJLX201602) and Beijing Municipal Science & Technology Commission Support (Grant NO: Z161100000116049). Weifeng Shi was supported by the ‘Taishan Scholar' project of Shandong Province. George F Gao is a primary principal investigator of the NSFC Innovative Research Group (Grant NO: 81321063).
Footnotes
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
Supplementary Material
References
- Daubney R, Hudson JR, Garnham P. Enzootic hepatitis or Rift Valley fever: an undescribed virus disease of sheep cattle and man from east Africa. J Pathol Bacteriol 1931; 34: 545–579. [Google Scholar]
- Nanyingi MO, Munyua P, Kiama SG et al. A systematic review of Rift Valley Fever epidemiology 1931-2014. Infect Ecol Epidemiol 2015; 5: 28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyangu AS, Gould LH, Sharif SK et al. Risk factors for severe Rift Valley fever infection in Kenya, 2007. Am J Trop Med Hygien 2010; 83: 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andayi F, Charrel RN, Kieffer A et al. A sero-epidemiological study of arboviral fevers in Djibouti, Horn of Africa. PLoS Negl Trop Dis 2014; 8: e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobbelaar AA, Weyer J, Leman PA et al. Molecular epidemiology of Rift Valley fever virus. Emerg Infect Dis 2011; 17: 2270–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakoune E, Kamgang B, Berthet N et al. Rift Valley fever virus circulating among ruminants, mosquitoes and humans in the Central African Republic. PLoS Negl Trop Dis 2016; 10: e0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res 2004; 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Bird BH, Khristova ML, Rollin PE et al. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J Virol 2007; 81: 2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics 2000; 16: 562–563. [DOI] [PubMed] [Google Scholar]
- El Imam M, El Sabiq M, Omran M et al. Acute renal failure associated with the Rift Valley fever: a single center study. Saudi J Kidn Dis Transpl 2009; 20: 1047–1052. [PubMed] [Google Scholar]
- Kahlon SS, Peters CJ, Leduc J et al. Severe Rift Valley fever may present with a characteristic clinical syndrome. Am J Trop Med Hygien 2010; 82: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanain AM, Noureldien W, Karsany MS et al. Rift Valley Fever among febrile patients at New Halfa hospital, eastern Sudan. Virol J 2010; 7: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam AA, Karsany MS, Adam I. Manifestations of severe Rift Valley fever in Sudan. InternJ Infect Dis 2010; 14: e179–e180. [DOI] [PubMed] [Google Scholar]
- Baudin M, Jumaa AM, Jomma HJ et al. Association of Rift Valley fever virus infection with miscarriage in Sudanese women: a cross-sectional study. Lancet Glob Health 2016; 4: e864–e871. [DOI] [PubMed] [Google Scholar]
- Mansfield KL, Banyard AC, McElhinney L et al. Rift Valley fever virus: A review of diagnosis and vaccination, and implications for emergence in Europe. Vaccine 2015; 33: 5520–5531. [DOI] [PubMed] [Google Scholar]
- Jansen van Vuren P, Shalekoff S, Grobbelaar AA et al. Serum levels of inflammatory cytokines in Rift Valley fever patients are indicative of severe disease. Virol J 2015; 12: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy AK, Nichol ST. Rift Valley fever virus inhibits a pro-inflammatory response in experimentally infected human monocyte derived macrophages and a pro-inflammatory cytokine response may be associated with patient survival during natural infection. Virology 2012; 422: 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Mansuroglu Z, Leger P et al. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog 2008; 4: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A. Targeting TFIIH to inhibit host cell transcription by Rift Valley Fever Virus. Mol Cell 2004; 13: 456–458. [DOI] [PubMed] [Google Scholar]
- Wong G, Gao GF, Qiu X. Can Ebola virus become endemic in the human population? Protein Cell 2016; 7: 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen W, Wong G et al. Highly diversified Zika viruses imported to China. 2016Protein Cell 2016; 7: 461–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu D, Shi W et al. Origin and possible genetic recombination of the middle east respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. mBio 2015; 6: e01280–01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Kim YJ, Lemey P et al. The recent ancestry of Middle East respiratory syndrome coronavirus in Korea has been shaped by recombination. Sci Rep 2016; 6: 18825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.