ABSTRACT
Histidine kinases (HKs) are among the most prominent sensing proteins studied in the kingdom Fungi. Their distribution and biological functions in early diverging fungi (EDF), however, remain elusive. We have taken advantage of recent genomic resources to elucidate whether relationships between the occurrence of specific HKs in some EDF and their respective habitat/lifestyle could be established. This led to the unexpected discovery of fungal HKs that share a high degree of similarity with receptors for plant hormones (ethylene and cytokinin). Importantly, these phytohormone receptor homologs are found not only in EDF that behave as plant root symbionts or endophytes but also in EDF species that colonize decaying plant material. We hypothesize that these particular sensing proteins promoted the interaction of EDF with plants, leading to the conquest of land by these ancestral fungi.
OPINION/HYPOTHESIS
Histidine kinases (HKs) are prominent sensing proteins present in bacteria, amoebae, plants, and fungi. When activated, for instance following the perception of an external stimulus, HKs initiate more or less complex phosphorylation cascades, ranging from two-component systems (prevailing in bacteria) to multistep phosphorelays (in plants and fungi) that lead to an adapted response (1).
In bacteria, it is now well documented that HKs regulate a large panel of fundamental processes, including nutrient acquisition, various metabolic activities, adaptation to changes in the environment, developmental pathways, virulence, antibiotic resistance, and many others (2). In plants, HKs act as osmosensors by regulating responses to drought, salt stress, and stomatal closure, but more importantly they have been implicated in the perception of two major phytohormones, cytokinins and ethylene (3). HKs are also widespread in the kingdom Fungi, and to date in Dikarya (i.e., Ascomycota and Basidiomycota) they have been reported to be involved in stress adaptation, red light perception, morphogenesis, and virulence (4). With the exception of a few recent insights into the distribution of these sensing proteins in Mucoromycotina (5), no extensive analysis of HKs has been conducted to date across the so-called early diverging fungal (EDF) lineages, which comprise a large portion of the phylogenetic diversity of the kingdom Fungi though just a small proportion of described species (6–8).
EDF are currently categorized into 10 lineages (depicted in Fig. 1) (9). From an ecological point of view, the habitats of EDF are broadly diversified, ranging from marine or freshwater environments to soils, with more or less tight associations with plants. Many other EDF lifestyles require strong interactions with hosts, as illustrated by Glomeromycotina, which live exclusively as obligate symbionts of vascular plants, bryophytes, and cyanobacteria, whereas several EDF have been identified as insect or vertebrate pathogens (6–8). In this way, to survive in a wide range of ecological niches or to fine-tune interactions with their respective hosts, EDF have likely developed a variety of cell signaling strategies that allow them to perceive and to cope appropriately with a broad range of external cues. Since HKs remain among the most important sensing proteins in Dikarya (1), we were primarily interested for this study in exploring the structure and distribution of HKs in EDF and in elucidating whether relationships could be established between the occurrence of particular HKs in some species/groups and their respective habitats and lifestyles.
FIG 1 .
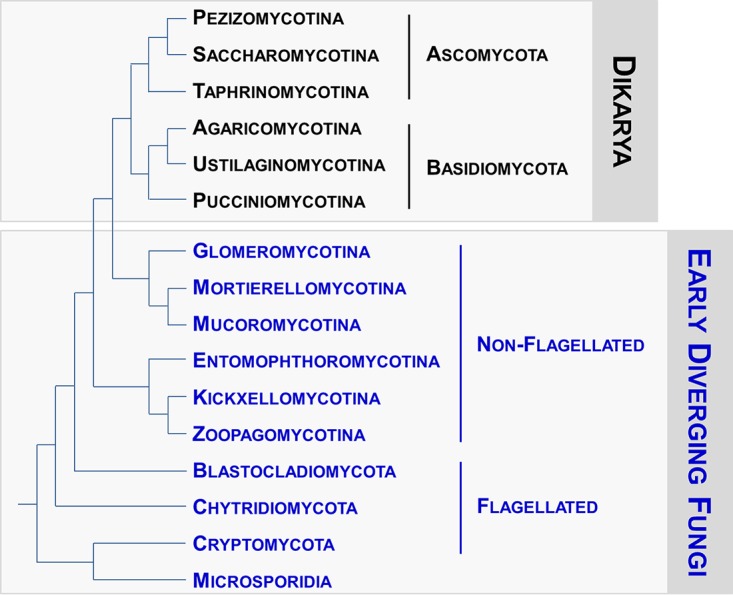
Phylogeny of the kingdom Fungi. The 10 early diverging fungal lineages are indicated in blue. The topologies resemble the current understanding of the relationships of the fungal groups, according to reference 9.
EXPLORING THE STRUCTURAL DIVERSITY AND DISTRIBUTION OF HKs AMONG THE EDF LINEAGES
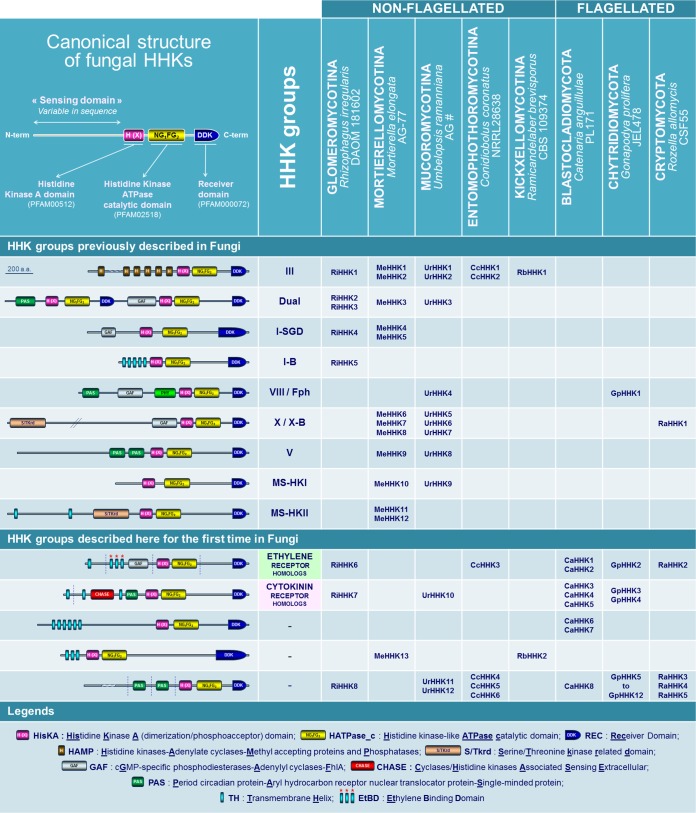
The basic structure of fungal HKs is now well established (Fig. 2, top left panel). In contrast to most bacterial HKs, the histidine kinase A (HisKA) and histidine kinase ATPase catalytic (HATPase_c) domains are fused to the C-terminal receiver domain (REC); thus, importantly, fungal HKs are generically defined as hybrid HKs (HHKs). It is worth noting that the variable N-terminal region, referred to as the sensing domain, displays a combination of motifs that drives the perception properties of the HHK (Fig. 2, top left panel) (10). Based on the sequence analysis of both histidine kinase A and sensing domains from more than 200 predicted proteins, fungal HHKs are currently categorized into 16 groups (4).
FIG 2 .
Compilation of HHKs in some early diverging fungi. Gene loci encoding HHKs in EDF genomes were identified following multiple tBLASTn and BLASTp searches against selected genomes of the Joint Genome Institute (JGI) MycoCosm database (6) (http://genome.jgi.doe.gov/programs/fungi/index.jsf). Conserved Domain Database (CDD) sequences for HisKA (PFAM00512), HATPase_c (PFAM02518), and REC (PFAM00072) were used in BLAST searches of each EDF genome. All hits producing E values below 10−4 were further analyzed. Functional domains were identified with CDD, and predictions of transmembrane-spanning regions were carried out using TMHMM v2.0. All sequences are compiled in Text S1 in the supplemental material.
Further description of methods used for the phylogenetic analysis and a compilation of nucleotide sequences used in the study. Download Text S1, DOCX file, 0.1 MB (118.7KB, docx) .
Copyright © 2017 Hérivaux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To gain insight into the structure and distribution of these proteins in the EDF lineages, we were primarily interested in compiling predicted sequences that bear the HHK canonical domains described in top left panel of Fig. 2 (10). For this purpose, we browsed the genome of a representative species from each EDF lineage (Fig. 1); the characteristics of these species are summarized in Table 1. Due to the lack of genomic resources for Zoopagomycotina, this lineage was not incorporated into the present analysis, nor were the Microsporidia, which do not contain HHK-encoding genes in their genome.
TABLE 1 .
Early diverging fungi whose genomes were examined in this study
| Speciesa | Strain | Fungal lineage | Lifestyle or habitatb |
|---|---|---|---|
| Preliminary exploration of HHK structures | |||
| Rhizophagus irregularis | DAOM 181602 | Glomeromycotina (nonflagellated) | Obligate endosymbiont of plant roots |
| Mortierella elongata | AG-77 | Mortierellomycotina (nonflagellated) | Saprotrophic, widely distributed in soils |
| Umbelopsis ramanniana | AG # | Mucoromycotina (nonflagellated) | Saprotrophic, facultative endophyte in woody roots |
| Conidiobolus coronatus | NRRL28638 | Entomophthoromycotina (nonflagellated) | Saprotrophic, rarely parasite of insects and mammals |
| Ramicandelaber brevisporus | CBS 109374 | Kickxellomycotina (nonflagellated) | Saprotrophic, widely distributed in soils |
| Catenaria anguillulae | PL171 | Blastocladiomycota (flagellated) | Saprotrophic, decaying plant materials, facultative parasite of plant pathogenic nematodes |
| Gonapodya prolifera | JEL478 | Chytridiomycota (flagellated) | Saprotrophic, decaying plant material |
| Rozella allomycis | CSF55 | Cryptomycota (flagellated) | Obligate parasite of Allomyces macrogynus |
| Secondary explorationb | |||
| Gigaspora rosea | DAOM194757 | Glomeromycotina (nonflagellated) | Obligate endosymbiont of plant roots |
| Basidiobolus meristosporus | CBS 931.73 | Entomophthoromycotina (nonflagellated) | Saprotrophic, decaying plant material |
| Zoophthora radicans | ARSEF 4784 | Entomophthoromycotina (nonflagellated) | Parasite of insects |
| Allomyces macrogynus | ATCC 38327 | Blastocladiomycota (flagellated) | Saprotrophic, decaying plant material |
| Spizellomyces punctatus | DAOM BR117 | Chytridiomycota (flagellated) | Saprotrophic, decaying plant material |
| Batrachochytrium dendrobatidis | JAM81 | Chytridiomycota (flagellated) | Parasite of amphibians |
| Rhizoclosmatium globosum | JEL800 | Chytridiomycota (flagellated) | Saprotrophic |
| Piromyces sp. | E2 | Neocallimastigomycetes (flagellated) | Mutualist in gut in variety of herbivores |
All genomes were compared with information from the Joint Genome Institute MycoCosm database (6) (http://genome.jgi.doe.gov/programs/fungi/index.jsf). See Fig. 2 for further information on the genomes browsed for the preliminary exploration of HHK structures; for information regarding the genomes browsed in the secondary exploration, see Fig. 3, 4, and 6.
A compilation of HHK structures deduced from EDF genomes is provided in Fig. 2. Above all, this compilation gives evidence that 9 out of the 16 fungal HHK groups previously identified in Dikarya are also present in EDF. This includes, notably, the osmosensing group III HHKs, the dual HHK group, which was initially thought to be restricted to Basidiomycota, and the red light sensing phytochromes (the VIII/Fph group) (4, 11). Interestingly, some unprecedented HHKs are also scattered among the different EDF lineages (Fig. 2), and it was particularly surprising that most of their sensing domains harbor hydrophobic transmembrane helices, distinguishing them from the majority of Dikarya-related HHKs (4).
FIRST DESCRIPTION OF PLANT HORMONE RECEPTOR HOMOLOGS IN EDF
The striking finding of this analysis is actually the discovery of fungal HHKs with a high degree of similarity with two groups of plant hormone receptors, i.e., ethylene and cytokinin receptors (Fig. 2). Both phytohormones are known to play crucial roles in plant development, and recent works have highlighted a cytokinin/ethylene interaction at diverse levels of biosynthetic and metabolic pathways (3). From a general point of view, ethylene and cytokinins are also well-documented as key signaling molecules in plant biotic interactions (with viruses, protists, bacteria, worms, insects, and fungi). Importantly, recent advances have revealed that in several plant-fungus systems, both plant- and microorganism-borne phytohormones have concerted effects that promote interactions (12–17).
SOME EDF GENOMES ENCODE ETHYLENE RECEPTOR HOMOLOGS
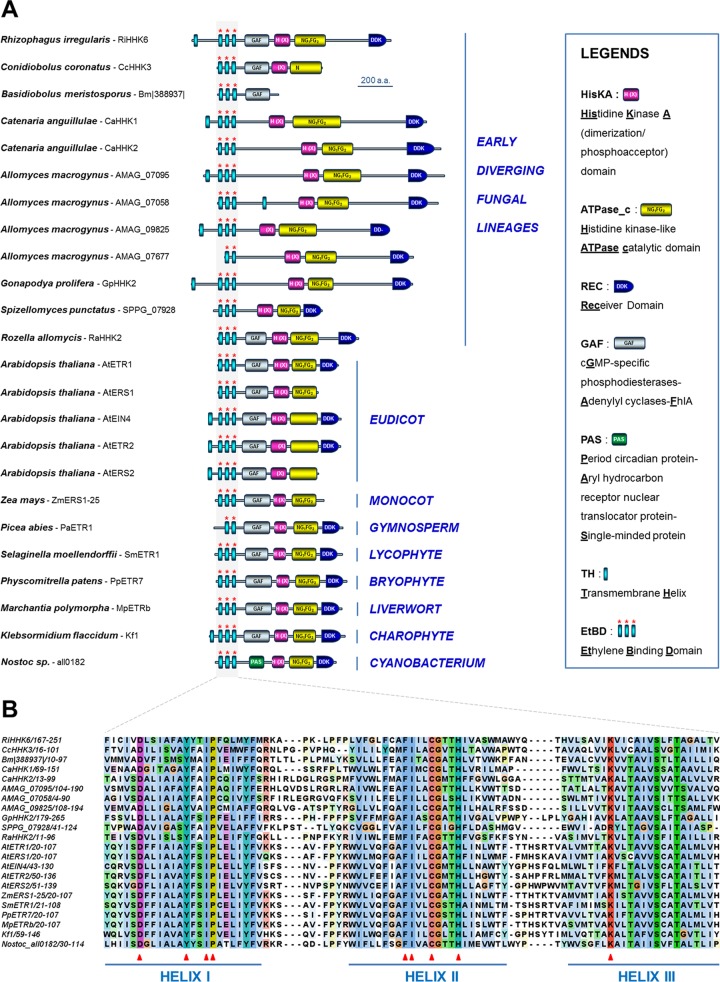
We first identified new fungal HHKs that share strong identities with plant ethylene receptors (Fig. 2 and 3A). The main feature that differentiates plant ethylene receptors from other bacterial or fungal HKs is the presence within the N-terminal sensing region of an ethylene binding domain consisting of a combination of three transmembrane helices that bear conserved amino acids essential for hormone perception (Fig. 3B) (18). We identified this particular N-terminal feature initially in the Rhizophagus (Glomeromycotina) RiHHK6 predicted protein (Fig. 2). Glomeromycotina form arbuscular mycorrhizae with plants (Table 1), and the importance of ethylene in the establishment of this type of symbiosis has been previously demonstrated (12, 13). Further BLAST analysis of more than 500 fungal genomes (using the Rhizophagus RiHHK6 ethylene binding domain as the query) led us to identify homologous sequences in several other EDF which are known to colonize plant materials (leaf litter, twigs, decaying fruits, soil) (Table 1), including Conidiobolus (CcHHK3), Catenaria (CaHHK1 and CaHHK2), Gonapodya (GpHHK2), Basidiobolus (Bm|388937|), and Spizellomyces (SPPG_07928) (Fig. 2 and 3A). We also noticed the presence of ethylene receptor homologs in Allomyces (AMAG_07677, AMAG_07095, AMAG_07058, and AMAG_09825) and Rozella (RaHHK2) (Fig. 2 and 3A). Since it is well-known that Rozella behaves as an obligate parasite of Allomyces (19), it could be hypothesized that ethylene orchestrates interactions between these two aquatic species. Alternatively, the presence of ethylene receptor homologs in these two flagellated fungi could be inherent to their location in the deepest branches of the tree and the inheritance of ethylene receptor homologs in the common ancestor of all fungi (9). All these new EDF proteins are depicted in Fig. 3A, along with several previously characterized ethylene receptors from plants, green algae, and cyanobacteria (20). As revealed by alignment of the ethylene binding domains from these proteins (Fig. 3B), all homologs to ethylene receptors identified in the genomes of the set of EDF display the three predicted transmembrane helices containing all of the crucial residues involved in ethylene perception (18). Plant ethylene receptors are currently categorized in two subfamilies: subfamily 1 includes members displaying an ethylene binding domain together with a GAF domain (cyclic GMP-specific phosphodiesterases/adenylyl cyclases/FhlA domain) in the sensing region, whereas subfamily 2 includes ethylene receptors containing a supplemental N-terminal transmembrane helix, compared to subfamily 1 members (21). As shown in Fig. 3A, both subfamilies are represented among EDF. In addition, it is now recognized that some members of the Arabidopsis ethylene receptor series lack several HHK functional domains, such as the C-terminal receiver domain (REC; AtERS1 and AtERS2) up to the histidine kinase ATPase catalytic (HATPase_c) domain (AtEIN4, AtERS2, and AtETR2) (21). Interestingly, such truncated features are also observed in some Entomophthoromycotina (Conidiobolus CcHHK3 and Basidiobolus Bm|388937|) (Fig. 3A).
FIG 3 .
Structures of homologs to ethylene receptors identified in early diverging fungi. (A) Diversity of structures found in various early diverging fungi. The structures of several previously characterized ethylene receptors from plant, green algae, and cyanobacteria are also provided (20). (B) Alignment of ethylene binding domains from homologs to ethylene receptors identified in early diverging fungi, along with others from plant, algae, and cyanobacteria. The ethylene binding domains consist of three transmembrane helices. Essential residues that have been reported to be involved in ethylene perception (18) are indicated with red arrows. AMAG_07677 and PaETR1 sequences were not included in this analysis (incomplete ethylene binding domain). The amino acid sequence alignment was formatted with the JALVIEW program.
SOME EDF GENOMES ENCODE CYTOKININ RECEPTOR HOMOLOGS
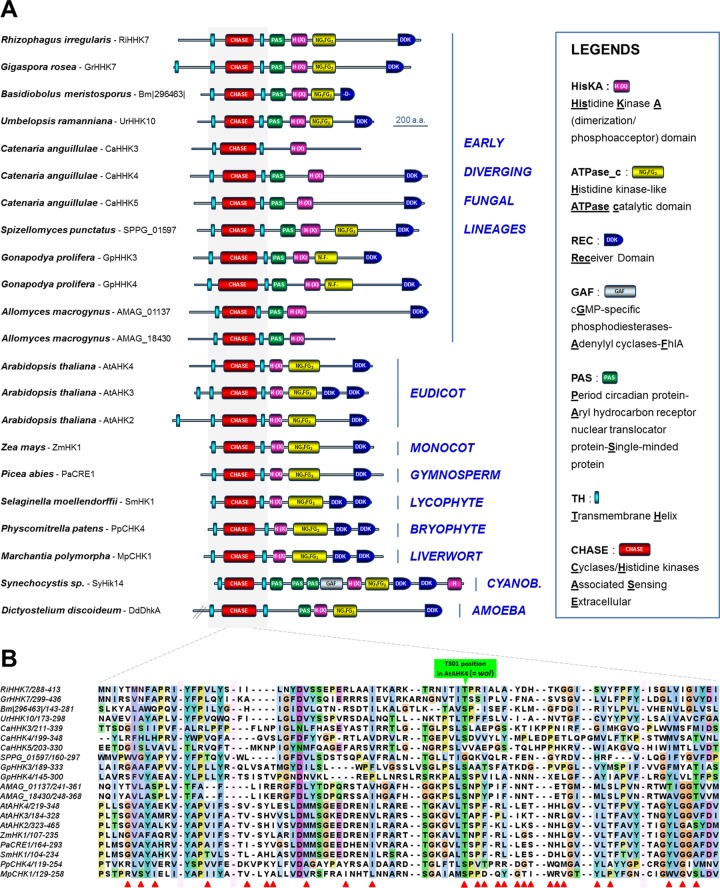
When exploring the structural diversity of HHKs in EDF, we were also surprised to observe unprecedented fungal HHKs that bear within their N terminus a CHASE domain (cyclases/histidine kinase-associated sensing extracellular domain) surrounded by two predicted transmembrane helices (Fig. 2). This remains indeed the characteristic feature of plant cytokinin receptors (22, 23). These CHASE domain-containing HHKs among the kingdom Fungi were initially identified in this study among the Glomeromycotina (Rhizophagus RiHHK7 and Gigaspora GrHHK7) (Fig. 2 and 4A). As for ethylene, the major importance of the host plant cytokinins in the development of arbuscular mycorrhizal symbiosis by Glomeromycotina is now well documented (15, 16). Further investigations (BLAST analysis using the Rhizophagus RiHHK7 CHASE sequence as the query) allowed us to identify homologous sequences in other EDF which, interestingly, are known to colonize decaying plant material (Table 1), including the genera Catenaria (CaHHK3, CaHHK4, and CaHHK5), Gonapodya (GpHHK3, GpHHK4), Basidiobolus (Bm|296463|), Allomyces (AMAG_01137, AMAG_18430), and Spizellomyces (SPPG_01597) (Fig. 4A). In Mucoromycotina, cytokinin receptor homologs are also found in the genus Umbelopsis (UrHHK10) (Fig. 4A) but not in the other 30 Mucoromycotina species for which the genome sequences are available. Umbelopsis species display a unique lifestyle compared to other Mucoromycotina (predominantly including saprotrophic or pathogenic species), as Umbelopsis spp. have been reported as endophytes in root xylem tissues (Table 1) (24). Figure 4A depicts this set of new fungal HHKs, along with the structure of several plant cytokinin receptors and CHASE domain-containing HHKs. As previously described for plant cytokinin receptors (22, 23), alignment of all these predicted protein sequences revealed that the CHASE domain remains highly degenerated (Fig. 4B). This suggests that EDF may sense numerous cytokinin derivatives, as observed in plants that typically contain a mixture of different biologically active cytokinin metabolites (25). Interestingly, the most important amino acid for hormone binding in the Arabidopsis AHK4 cytokinin receptor, i.e., threonine 301 (green arrow in Fig. 4B), is conserved or similar (serine) in almost all the CHASE domain-containing HHKs deduced from EDF genomes. Indeed, this residue was at the origin of the discovery of plant cytokinin receptors, since when mutated in AHK4, this led to the wooden leg (wol) mutant phenotype in Arabidopsis (an altered root morphology characteristic of the absence of cytokinin perception in plants) (26).
FIG 4 .
Structures of homologs to cytokinin receptors identified in early diverging fungi. (A) Diversity of structures found in various early diverging fungi. The structures of several previously characterized plant cytokinin receptors (23) and other CHASE domain-containing HHKs (which had not been demonstrated to date to act as cytokinin receptors) from Synechocystis sp. (Cyanobacteria) and Dictyostelium discoideum (Amoebae) are also provided. (B) Alignment of CHASE domains from homologs to cytokinin receptors identified in early diverging fungi, along with others from plant cytokinin receptors. The Dictyostelium and Synechocystis sequences were not included in this analysis because their subgroup was not supported by a high bootstrap value. Interruptions of the alignments are indicated by pink rectangles, and essential residues reported to be involved in cytokinin perception in A. thaliana AtAHK4 are indicated with red arrows (previously compiled in reference 23).
DECIPHERING THE PHYLOGENETIC RELATIONSHIPS OF EDF, PLANT, AND CYANOBACTERIAL ETHYLENE AND CYTOKININ RECEPTORS
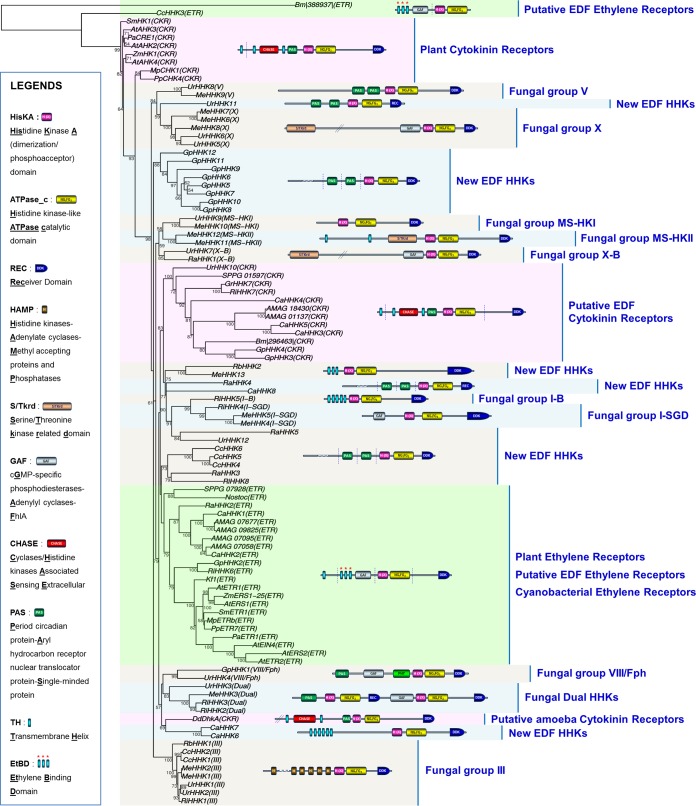
A cyanobacterial origin of plant ethylene receptors was previously suggested (27). Although it was recently proposed that cytokinin perception by plant HHKs through the CHASE domain might have emerged shortly before the conquest of land (23), the exact origin of these phytohormone receptors remains unclear. To gain insight into the phylogenetic relationships of EDF, plant, and cyanobacterial ethylene and cytokinin receptors, we generated a robust phylogenetic tree after multiple alignments of all the predicted sequences compiled in this analysis (Fig. 5). This revealed that cyanobacterial, plant, and EDF ethylene receptors tend to cluster, supporting a common origin (Fig. 5). Emergence of EDF cytokinin receptor homologs, as currently believed for plants (23), might have resulted from separate transfer and specialization of an ancestral, hitherto-unknown CHASE domain-containing prokaryotic HK, since plant and EDF cytokinin receptor homologs do not occur in the same cluster (Fig. 5).
FIG 5 .
Phylogeny estimation of HHK predicted protein sequences. Methods used to carry out this phylogenetic analysis are provided in Text S1 in the supplemental material. Early diverging fungi predicted HHK sequences were categorized following sequence analysis of both HisKA signatures and N-terminal sensing domains according to previous classifications (4, 10). Abbreviations: Ri, Rhizophagus irregularis (Glomeromycotina); Me, Mortierella elongata (Mortierellomycotina); Ur, Umbelopsis ramanniana (Mucoromycotina); Cc, Conidiobolus coronatus (Entomophthoromycotina); Rb, Ramicandelaber brevisporus (Kickxellomycotina); Ca, Catenaria anguillulae (Blastocladiomycota); Gp, Gonapodya prolifera (Chytridiomycota); Ra, Rozella allomycis (Cryptomycota); AMAG, Allomyces macrogynus (Blastocladiomycota); Dd, Dictyostelium discoideum (Amoebae); SPPG, Spizellomyces punctatus (Chytridiomycota); Bm, Basidiobolus meristoporus (Entomophthoromycotina); Gr, Gigaspora rosea (Glomeromycotina); At, Arabidopsis thaliana (dicots); Zm, Zea mays (monocots); Pa, Picea abies (gymnosperms); Sm, Selaginella moellendorffii (lycophytes); Pp, Physcomitrella patens (bryophytes); Mp, Marchantia polymorpha (liverworts); Kf, Klebsormidium flaccidum (charophytes).
PHYTOHORMONE RECEPTOR HOMOLOGS MAY HAVE PLAYED AN ESSENTIAL ROLE IN FUNGAL LAND COLONIZATION AND FUNGUS-PLANT INTERACTION PROCESSES
We have described here for the first time fungal HHKs that share a high degree of similarity with plant ethylene and cytokinin receptors. Importantly, these homologs to phytohormone receptors were found in large numbers in several flagellated genera (e.g., Gonapodya, Catenaria, Allomyces, and Spizellomyces), which are reported to colonize decaying plant material, where cytokinins and ethylene are omnipresent (25), and in a small number of some nonflagellated EDF they are known to behave as plant root symbionts or endophytes (e.g., Rhizophagus and Umbelopsis) (Fig. 3 and 4).
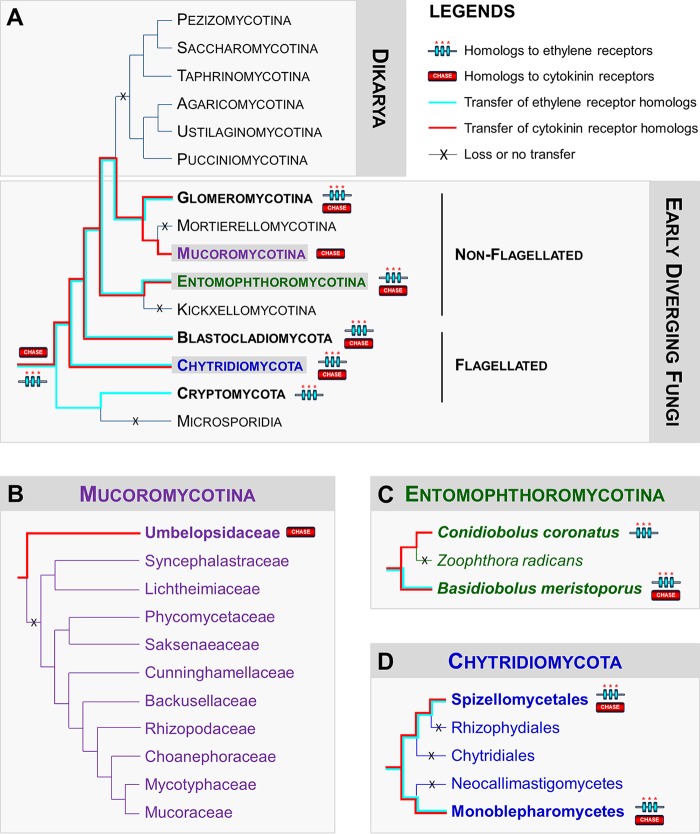
Strong arguments suggest that interacting with fungi was one of the major processes that promoted land colonization by plants (28, 29). In addition, as previously mentioned, ethylene and cytokinins are prominent phytohormones that orchestrate interactions in several plant-fungus symbiotic systems (12, 13, 15, 16). On the basis of our observations, some hypotheses may be therefore put forward to explain the presence and distribution of these ethylene and cytokinin receptor homologs in the EDF lineages (Fig. 6A). Ancestral aquatic fungi may have harbored a pool of archetypal ethylene receptors and CHASE domain-containing HK genes that may have been transferred horizontally from cyanobacteria or green algae (23, 27). These gene families have undergone rapid expansion in some flagellated EDF lineages, including, for instance, Chytridiomycota (e.g., Gonapodya and Spizellomyces) and Blastocladiomycota (e.g., Allomyces and Catenaria) (Fig. 6A and D). The presence of these phytohormone receptor homologs may have initially participated to potentiate EDF-EDF (e.g., ethylene receptors in Allomyces and its parasite, Rozella), EDF-cyanobacteria, and EDF-green algae communication (most cyanobacteria and green algae harbor such phytohormone homologs) (23, 27). Later, these same signaling pathways could have been coopted for EDF-plant interactions and coevolution (28). The presence of a unique member of both ethylene and cytokinin receptors in Glomeromycotina (e.g., Rhizophagus) (Fig. 6A), which currently represent the sole group of EDF that cannot be cultured without their plant partner, may reflect the optimization process that occurred in some EDF lineages to sense the host plant for symbiosis establishment. Once plants successfully colonized land, notably Glomeromycotina and the related Mucoromycotina (29), and developed more hospitable terrestrial habitats, such as soil, wood, and litter, ethylene and cytokinin receptor homologs were progressively lost in the genomes of fungi (absent in the Dikarya and most of the nonflagellated EDF lineages) that have shifted to other environmental niches. The striking truncated structures of ethylene receptor homologs found in Conidiobolus and Basidiobolus (Entomophthoromycotina) (Fig. 3A and 6C) and the intriguing presence of a unique cytokinin receptor homolog in Umbelopsis (Mucoromycotina) (Fig. 6B) and Basidiobolus (Fig. 6C) clearly illustrate this phenomenon of gene erosion, since these genera are considered basal within their respective lineages (9, 30). Moreover, we propose that EDF species that originally colonized plants and decaying vegetation lost their receptors at the same time they gained the ability to become pathogens of animals living in the same terrestrial ecological niche (e.g., Catenaria and Conidiobolus) (Fig. 6). Additional examples of plant host-to-animal host switches in the EDF will be necessary to confirm whether loss of these phytohormone receptor homologs is really linked to changes to non-plant-associated lifestyles.
FIG 6 .
Distribution of homologs to ethylene and cytokinin receptors in the early diverging fungal lineages. (A) Phylogeny of fungi. Homologs to phytohormone receptors were found in lineages which included flagellated early diverging fungal species (from Chytridiomycota, Blastocladiomycota, and Cryptomycota) and some nonflagellated species, which have been reported to colonize decaying plant material or to behave as symbionts or endophytes of plant roots (from Mucoromycotina, Glomeromycotina, and Entomophthoromycotina) (B) The particular case of Mucoromycotina. Homologs to cytokinin receptors are exclusively present in basal species from the Umbelopsidaceae clade (facultative endophytes of plant roots). (C) The case of Entomophthoromycotina. Homologs to ethylene and cytokinin receptors are detected in the basal lineages (e.g., Basidiobolus and Conidiobolus) that are capable of diverse ecologies, including colonizing decaying plant materials and parasitizing insects. As the lifestyle progresses to a strict reliance on insect parasitism (e.g., Zoophthora), the presence of the homologs is lost. (D) The case of Chytridiomycota. Both homologs to ethylene and cytokinin receptors are detected in species that colonize decaying plant materials (e.g., Gonapodya from Monoblepharomycetes and Spizellomyces from Spizellomycetales), but not in others (e.g., the amphibian pathogenic fungus Batrachochytrium, Rhizophydiales, the saprobe Rhizoclosmatium globosum, Chytridiales, or the mutualistic fungi of herbivore guts, Piromyces and Neocallimastigomycetes). The topologies resemble the current understanding of the relationships of the fungal groups according to information reported in reference (7–9 and 30).
CONCLUDING REMARKS
The discovery of unprecedented homologs to plant hormone receptors in EDF suggests the participation of these sensing proteins in fungus-plant interaction processes, which may have helped these early diversifying fungal lineages to colonize land. Work is under way to functionally characterize these receptors and to decipher their physiological roles in EDF via complementary biochemical, genetic, and modeling approaches.
ACKNOWLEDGMENTS
We thank the U.S. Department of Energy Joint Genome Institute and the following principal investigators from the 1000 Fungal Genomes Project (J. Spatafora, J. Stajich, A. Gryganskyi, S. Baker, and R. Vilgalys) for the permission to use EDF genomes before publication. We also acknowledge the Broad Institute Fungal Genome Initiative for making the genome sequences of A. macrogynus and S. punctuatus available.
Research in the laboratory of F.M. is funded by the Laboratory of Excellence Advanced Research on the Biology of Tree and Forest Ecosystems (ARBRE grant ANR 11 LABX 0002 01) and the U.S. DOE through the Oak Ridge National Laboratory Scientific Focus Area for Genomics Foundational Sciences (Plant Microbe Interfaces Project).
We are grateful to V. Puech-Pagès, P. M. Delaux, and G. Bécard (Université de Toulouse, UMR 5546, France) for their critical reading of the manuscript. We apologize to those whose work was not referenced due to citation restrictions.
Footnotes
Citation Hérivaux A, Dugé de Bernonville T, Roux C, Clastre M, Courdavault V, Gastebois A, Bouchara J-P, James TY, Latgé J-P, Martin F, Papon N. 2017. The identification of phytohormone receptor homologs in early diverging fungi suggests a role for plant sensing in land colonization by fungi. mBio 8:e01739-16. https://doi.org/10.1128/mBio.01739-16.
REFERENCES
- 1.Hérivaux A, So YS, Gastebois A, Latgé JP, Bouchara JP, Bahn YS, Papon N. 2016. Major sensing proteins in pathogenic fungi: the hybrid histidine kinase family. PLoS Pathog 12:e1005683. doi: 10.1371/journal.ppat.1005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bem AE, Velikova N, Pellicer MT, Baarlen Pv, Marina A, Wells JM. 2015. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol 10:213–224. doi: 10.1021/cb5007135. [DOI] [PubMed] [Google Scholar]
- 3.Zdarska M, Dobisová T, Gelová Z, Pernisová M, Dabravolski S, Hejátko J. 2015. Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. J Exp Bot 66:4913–4931. doi: 10.1093/jxb/erv261. [DOI] [PubMed] [Google Scholar]
- 4.Defosse TA, Sharma A, Mondal AK, Dugé de Bernonville T, Latgé JP, Calderone R, Giglioli-Guivarc’h N, Courdavault V, Clastre M, Papon N. 2015. Hybrid histidine kinases in pathogenic fungi. Mol Microbiol 95:914–924. doi: 10.1111/mmi.12911. [DOI] [PubMed] [Google Scholar]
- 5.Corrochano LM, Kuo A, Marcet-Houben M, Polaino S, Salamov A, Villalobos-Escobedo JM, Grimwood J, Álvarez MI, Avalos J, Bauer D, Benito EP, Benoit I, Burger G, Camino LP, Cánovas D, Cerdá-Olmedo E, Cheng JF, Domínguez A, Eliáš M, Eslava AP, Glaser F, Gutiérrez G, Heitman J, Henrissat B, Iturriaga EA, Lang BF, Lavín JL, Lee SC, Li W, Lindquist E, López-García S, Luque EM, Marcos AT, Martin J, McCluskey K, Medina HR, Miralles-Durán A, Miyazaki A, Muñoz-Torres E, Oguiza JA, Ohm RA, Olmedo M, Orejas M, Ortiz-Castellanos L, Pisabarro AG, Rodríguez-Romero J, Ruiz-Herrera J, Ruiz-Vázquez R, Sanz C, Schackwitz W, et al. 2016. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr Biol 26:1577–1584. doi: 10.1016/j.cub.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y, Wang S, Sekimoto S, Aerts AL, Choi C, Clum A, LaButti KM, Lindquist EA, Yee Ngan C, Ohm RA, Salamov AA, Grigoriev IV, Spatafora JW, Berbee ML. 2015. Phylogenomic analyses indicate that early fungi evolved digesting cell walls of algal ancestors of land plants. Genome Biol Evol 7:1590–1601. doi: 10.1093/gbe/evv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, et al. 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 9.Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, O’Donnell K, Roberson RW, Taylor TN, Uehling J, Vilgalys R, White MM, Stajich JE. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046. doi: 10.3852/16-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catlett NL, Yoder OC, Turgeon BG. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell 2:1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavín JL, Sarasola-Puente V, Ramírez L, Pisabarro AG, Oguiza JA. 2014. Dual-histidine kinases in basidiomycete fungi. C R Biol 337:111–116. doi: 10.1016/j.crvi.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Foo E, McAdam EL, Weller JL, Reid JB. 2016. Interactions between ethylene, gibberellins, and brassinosteroids in the development of rhizobial and mycorrhizal symbioses of pea. J Exp Bot 67:2413–2424. doi: 10.1093/jxb/erw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de los Santos RT, Vierheilig H, Ocampo JA, Garrido JM. 2011. Altered pattern of arbuscular mycorrhizal formation in tomato ethylene mutants. Plant Signal Behav 6:755–758. doi: 10.4161/psb.6.5.15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanclud E, Kisiala A, Emery NR, Chalvon V, Ducasse A, Romiti-Michel C, Gravot A, Kroj T, Morel JB. 2016. Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLoS Pathog 12:e1005457. doi: 10.1371/journal.ppat.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosme M, Ramireddy E, Franken P, Schmülling T, Wurst S. 2016. Shoot- and root-borne cytokinin influences arbuscular mycorrhizal symbiosis. Mycorrhiza 26:709–720. doi: 10.1007/s00572-016-0706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laffont C, Rey T, André O, Novero M, Kazmierczak T, Debellé F, Bonfante P, Jacquet C, Frugier F. 2015. The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS One 10:e0116819. doi: 10.1371/journal.pone.0116819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinsch J, Galuszka P, Tudzynski P. 2016. Functional characterization of the first filamentous fungal tRNA-isopentenyltransferase and its role in the virulence of Claviceps purpurea. New Phytol 211:980–992. doi: 10.1111/nph.13960. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Esch JJ, Shiu SH, Agula H, Binder BM, Chang C, Patterson SE, Bleecker AB. 2006. Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18:3429–3442. doi: 10.1105/tpc.106.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gleason FH, Lilje O, Marano AV, Sime-Ngando T, Sullivan BK, Kirchmair M, Neuhauser S. 2014. Ecological functions of zoosporic hyperparasites. Front Microbiol 5:244. doi: 10.3389/fmicb.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasumura Y, Pierik R, Fricker MD, Voesenek LA, Harberd NP. 2012. Studies of physcomitrella patens reveal that ethylene-mediated submergence responses arose relatively early in land-plant evolution. Plant J 72:947–959. doi: 10.1111/tpj.12005. [DOI] [PubMed] [Google Scholar]
- 21.Gallie DR. 2015. Appearance and elaboration of the ethylene receptor family during land plant evolution. Plant Mol Biol 87:521–539. doi: 10.1007/s11103-015-0296-z. [DOI] [PubMed] [Google Scholar]
- 22.Heyl A, Wulfetange K, Pils B, Nielsen N, Romanov GA, Schmülling T. 2007. Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain. BMC Evol Biol 7:62. doi: 10.1186/1471-2148-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruhn N, Halawa M, Snel B, Seidl MF, Heyl A. 2014. A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two-component signaling system of plants. Plant Physiol 165:227–237. doi: 10.1104/pp.113.228080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoff JA, Klopfenstein NB, McDonald GI, Tonn JR, Kim M-S, Zambino PJ, Hessburg PF, Rogers JD, Peever TL, Carris LM. 2004. Fungal endophytes in woody roots of Douglas-fir (Pseudotsuga menziesii) and ponderosa pine (Pinus ponderosa). Forest Pathol 34:255–271. doi: 10.1111/j.1439-0329.2004.00367.x. [DOI] [Google Scholar]
- 25.Spíchal L. 2012. Cytokinins—recent news and views of evolutionally old molecules. Functional Plant Biol 39:267–284. doi: 10.1071/FP11276. [DOI] [PubMed] [Google Scholar]
- 26.Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. 2000. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacey RF, Binder BM. 2016. Ethylene regulates the physiology of the cyanobacterium Synechocystis sp. PCC 6803 via an ethylene receptor. Plant Physiol 171:2798–2809. doi: 10.1104/pp.16.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brundrett MC. 2002. Coevolution of roots and mycorrhizas of land plants. New Phytol 154:275–304. doi: 10.1046/j.1469-8137.2002.00397.x. [DOI] [PubMed] [Google Scholar]
- 29.Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. 2015. Symbiotic options for the conquest of land. Trends Ecol Evol 30:477–486. doi: 10.1016/j.tree.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann K, Pawłowska J, Walther G, Wrzosek M, de Hoog GS, Benny GL, Kirk PM, Voigt K. 2013. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 30:57–76. doi: 10.3767/003158513X666259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R, Charron P, Duensing N, Frei dit Frey N, Gianinazzi-Pearson V, Gilbert LB, Handa Y, Herr JR, Hijri M, Koul R, Kawaguchi M, Krajinski F, Lammers PJ, Masclaux FG, Murat C, Morin E, Ndikumana S, Pagni M, Petitpierre D, Requena N, Rosikiewicz P, Riley R, Saito K, San Clemente H, Shapiro H, van Tuinen D, Bécard G, Bonfante P, Paszkowski U, Shachar-Hill YY, Tuskan GA, Young JP, Sanders IR, Henrissat B, Rensing SA, Grigoriev IV, Corradi N, Roux C, Martin F. 2013. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci U S A 110:20117–20122. doi: 10.1073/pnas.1313452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James TY, Pelin A, Bonen L, Ahrendt S, Sain D, Corradi N, Stajich JE. 2013. Shared signatures of parasitism and phylogenomics unite Cryptomycota and Microsporidia. Curr Biol 23:1548–1553. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- 33.Rosenblum EB, James TY, Zamudio KR, Poorten TJ, Ilut D, Rodriguez D, Eastman JM, Richards-Hrdlicka K, Joneson S, Jenkinson TS, Longcore JE, Parra Olea G, Toledo LF, Arellano ML, Medina EM, Restrepo S, Flechas SV, Berger L, Briggs CJ, Stajich JE. 2013. Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc Natl Acad Sci U S A 110:9385–9390. doi: 10.1073/pnas.1300130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang N, San Clemente H, Roy S, Bécard G, Zhao B, Roux C. 2016. A survey of the gene repertoire of Gigaspora rosea unravels conserved features among Glomeromycota for obligate biotrophy. Front Microbiol 7:233. doi: 10.3389/fmicb.2016.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russ C, Lang BF, Chen Z, Gujja S, Shea T, Zeng Q, Young S, Cuomo CA, Nusbaum C. 2016. Genome sequence of Spizellomyces punctatus. Genome Announc 4:e00849-16. doi: 10.1128/genomeA.00849-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Further description of methods used for the phylogenetic analysis and a compilation of nucleotide sequences used in the study. Download Text S1, DOCX file, 0.1 MB (118.7KB, docx) .
Copyright © 2017 Hérivaux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.