Abstract
While bacteria were long thought to rely primarily on transcriptional control, it is now well established that they also use numerous small RNAs to regulate mRNA translation and stability. There has recently been a surge in studies, including one by Waters et al (2017) in this issue of The EMBO Journal, that have used clever variations of the RNA‐seq technique to comprehensively map small RNA–target networks.
Subject Categories: Methods & Resources; Microbiology, Virology & Host Pathogen Interaction; RNA Biology
A major recent shift in our view of gene expression is that even the simplest organisms—bacteria—extensively control their genes post‐transcriptionally using small noncoding RNAs (sRNAs). Of the >200 sRNAs expressed in Escherichia coli or Salmonella, many regulate target mRNAs via base pairing to alter translation or transcript stability; and those that act via the global RNA‐binding protein Hfq typically target multiple mRNAs. Understanding the components of such RNA regulons is not only important to dissect the regulatory circuits underlying bacterial physiology and virulence, but also to fathom where and why organisms favor regulatory RNA over proteins.
Global transcript profiling by RNA‐seq has been amply used to map bacterial regulons, but sRNA‐mediated control seldom causes strong alterations in transcript abundance and observed changes may not always be a direct consequence of sRNA activity. As a result, there has recently been a surge in variations of the RNA‐seq technique to map bacterial RNA regulons more directly.
Co‐immunoprecipitation of cellular RNA followed by deep sequencing (RIP‐seq) has yielded semi‐quantitative snapshots of sRNA and mRNA association patterns with Hfq in different growth phases of Salmonella (Chao et al, 2012). These patterns were refined by the inclusion of UV cross‐linking in vivo (Tree et al, 2014; Holmqvist et al, 2016), showing more precisely where Hfq recognizes its RNA targets. Other recent work used RNA‐seq to capture the target suites of individual sRNAs. In MAPS, potential mRNA targets were co‐purified and sequenced with in vivo expressed aptamer‐tagged E. coli sRNAs (Lalaouna et al, 2015). GRIL‐seq works similarly but additionally seeks to seal the typically short sRNA–mRNA duplexes in vivo by expressing an RNA ligase prior to RNA capture (Han et al, 2016). However, while these approaches stepwise built better inventories and often discovered novel RNA constituents of Hfq networks (Chao et al, 2012; Tree et al, 2014; Lalaouna et al, 2015), they were unable to report on all sRNAs at once.
Two papers, one in this issue of The EMBO Journal (Waters et al, 2017) and the other in Molecular Cell (Melamed et al, 2016), now provide deep sequencing‐based global sRNA interactome maps in E. coli; they constitute major leaps forward in imaging RNA networks in bacteria. Both studies ligate sRNA–mRNA pairs after purification with central proteins, either Hfq itself (Melamed et al, 2016) or the major endoribonuclease RNase E (Waters et al, 2017) which degrades mRNAs upon recognition by cognate sRNAs (Fig 1).
Figure 1. How CLASH and RIL‐seq work.
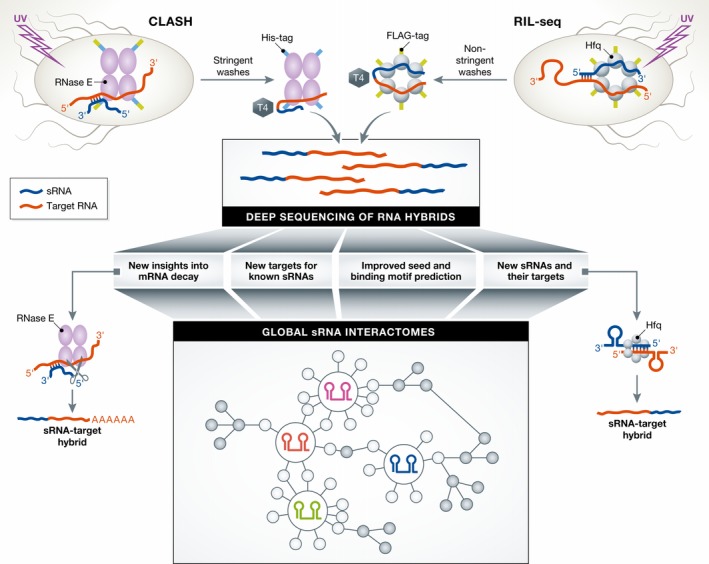
CLASH: Following in vivo cross‐linking, RNase E is pulled down using a FLAG‐tag, the bound RNA is trimmed, and the complex is purified under denaturing conditions using a His‐tag. Hybrid RNAs are created by ligation of the interacting RNAs with T4 ligase. RIL‐seq: In vivo cross‐linked Hfq is pulled down using a FLAG‐tag, followed by RNA trimming and ligation of the interacting RNAs with T4 ligase. Subsequently, the resulting hybrid RNAs of either protocol are sequenced, allowing in silico analysis of sRNA–target interactions. Bottom left: CLASH reveals new insights into mRNA decay. After binding 3′ of the sRNA–mRNA seed region, RNase E cleaves the mRNA, an oligo(A) stretch is added to the 3′ end, which finally allows decay by 3′ exonucleases. Bottom right: RIL‐seq reveals new sRNAs along with their targets. Bottom middle: Sequencing of RNA hybrids reveals global sRNA interactomes. sRNAs (colored) directly interact with targets (white nodes), which then interact with further targets (gray nodes).
Waters et al (2017) build on previous work from the Tollervey group who originally developed cross‐linking, ligation, and sequencing of hybrids (CLASH) to discover new snoRNA–rRNA interactions on snoRNA‐related yeast proteins (Kudla et al, 2011). An earlier attempt to apply this proximity ligation method to Hfq in enterohemorrhagic E. coli (EHEC) yielded few RNA hybrids (Tree et al, 2014); however, encouraged by the observation that RNase E recognizes the short seed helix formed between an sRNA and its target (Bandyra et al, 2012), the authors now focused on ligation to RNase E, which much elevated the proportion of RNA hybrids (Waters et al, 2017). Since these hybrids are significantly enriched in pairs of known sRNA seed regions and co‐regulated targets, they likely represent bona fide sRNA–mRNA interactions. What recruits RNase E to these many duplexes cannot be directly concluded from the data. However, plenty of coincidences with Hfq sites suggest a model whereby RNase E is recruited 3′ to where Hfq binds in mRNAs. RNase E would then cleave the target a few nucleotides downstream, followed by polyadenylation and full degradation (Waters et al, 2017; Fig 1).
Some of the most interesting sRNAs in pathogens like EHEC come from horizontally acquired virulence regions. However, lacking recognizable homologs in other species, these sRNAs rarely reveal their seed regions in sequence alignments, which complicates in silico target predictions. Waters et al (2017) demonstrate the power of CLASH for unveiling functions of pathogen sRNAs by capturing hybrids of the EHEC‐specific sRNA Esr41 with multiple mRNAs. These targets encode iron uptake and storage proteins, and in agreement with this, knockout of Esr41 confers a fitness advantage to EHEC in iron‐limited medium. Conversely, overexpressed Esr41 renders bacteria insensitive to colicin, which aligns with a CLASH prediction that Esr41 represses the mRNA of siderophore receptor CirA whereby this toxin anchors.
The study by Melamed et al (2016) revisits the idea of obtaining sRNA–mRNA pairs directly from Hfq, expressed with a single FLAG epitope tag in nonpathogenic E. coli. RIL‐seq recorded an aggregate of ~2,800 putative RNA interactions on Hfq, ten times the established E. coli sRNA interactome (Melamed et al, 2016). The overlap with known sRNA–mRNA pairs was remarkably high (56%), also considering that not all E. coli genes are expressed under the three growth conditions sampled. Other observations support the reliability of RIL‐seq, too. With a seed mutant of the multi‐target sRNA GcvB, almost all GcvB–mRNA hybrids are lost. Moreover, the mRNA sequences in the obtained hybrids often carry motifs complementary to the seed of the respective sRNA partner.
The E. coli sRNA interactome revealed by RIL‐seq looks very dynamic, with substantial re‐wiring occurring upon changes in cellular conditions. Overall, sRNA antagonists emerge as major theme in the Hfq network; RIL‐seq not only captured the known RNA sponges of the ChiX and RyhB sRNAs but also discovered one for Spot42. Other new regulations include the acid stress response of E. coli where RIL‐seq discovers trans‐encoded mRNA targets of ArrS and GadY, previously known as cis‐regulatory antisense RNAs only (Melamed et al, 2016; Fig 1).
For methods to be widely applicable, they must be able to robustly assess the same phenomenon in different species. Intriguingly, CLASH and RIL‐seq recover a comparable number of sRNA–mRNA interactions in related conditions and similar mRNA motifs complementary to sRNA seeds for several model sRNAs, all of this despite using two different bait proteins and considering that EHEC and E. coli K12 share only ~80% genomic content. Both studies bolster a view that Hfq‐dependent sRNAs primarily sequester the 5′ region of mRNAs to repress translation. Yet, hundreds of sRNA hybrids with mRNA fragments outside translation initiation regions offer plenty of leads for potentially new molecular mechanisms of negative or positive post‐transcriptional regulation.
While CLASH and RIL‐seq provide global snapshots of bacterial sRNA interactomes (Melamed et al, 2016; Waters et al, 2017; Fig 1), these approaches must now be made more quantitative. For example, does the frequency of hybrids recovered for a given sRNA or mRNA fully reflect its cellular activity or regulation, respectively? How do the RNase E interactions with sRNA–mRNA pairs determined by CLASH (Waters et al, 2017) compare to global in vivo profiling of RNase E cleavage by RNA‐seq (Chao et al, 2017)? With these methods at hand, we can dig deeper into the intricate details of sRNA and target loading onto Hfq (Schu et al, 2015). Moreover, following progress on profiling sRNA activities in host–pathogen interactions (Westermann et al, 2016), CLASH and RIL‐seq may enable us to capture sRNA–mRNA interactomes in intracellular pathogens replicating inside eukaryotic host cells. Finally, Hfq just got a sibling: The ProQ protein has been discovered as a second global sRNA chaperone in E. coli and Salmonella (Smirnov et al, 2016), and these new interactome methods lend themselves to accelerate sRNA target discovery in this unexplored domain of post‐transcriptional control in bacteria.
See also: SA Waters et al (February 2017)
References
- Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF (2012) The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 47: 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J (2012) An atlas of Hfq‐bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31: 4005–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Li L, Girodat D, Förstner KU, Said N, Corcoran C, Śmiga M, Papenfort K, Reinhardt R, Wieden H‐J, Luisi BF, Vogel J (2017) In vivo cleavage map illuminates the central role of RNase E in coding and noncoding RNA pathways. Mol Cell 65: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Tjaden B, Lory S (2016) GRIL‐Seq: a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation. Nat Microbiol 2: 16239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J (2016) Global RNA recognition patterns of post‐transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo . EMBO J 35: 991–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D (2011) Cross‐linking, ligation, and sequencing of hybrids reveals RNA‐RNA interactions in yeast. Proc Natl Acad Sci USA 108: 10010–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaouna D, Carrier M‐C, Semsey S, Brouard J‐S, Wang J, Wade JT, Massé E (2015) A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell 58: 393–405 [DOI] [PubMed] [Google Scholar]
- Melamed S, Peer A, Faigenbaum‐Romm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H (2016) Global mapping of small RNA‐target interactions in bacteria. Mol Cell 63: 884–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu DJ, Zhang A, Gottesman S, Storz G (2015) Alternative Hfq‐sRNA interaction modes dictate alternative mRNA recognition. EMBO J 34: 2557–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J (2016) Grad‐seq guides the discovery of ProQ as a major small RNA‐binding protein. Proc Natl Acad Sci USA 113: 11591–11596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL (2014) Identification of bacteriophage‐encoded anti‐sRNAs in pathogenic Escherichia coli . Mol Cell 55: 199–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters SA, McAteer SP, Kudla G, Pang I, Deshpande NP, Amos TG, Leong KW, Wilkins MR, Strugnell R, Gally DL, Tollervey D, Tree JJ (2017) Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J 36: 374–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J (2016) Dual RNA‐seq unveils noncoding RNA functions in host‐pathogen interactions. Nature 529: 496–501 [DOI] [PubMed] [Google Scholar]


