Abstract
The functional role of the ubiquitin‐proteasome pathway during maternal‐to‐zygotic transition (MZT) remains to be elucidated. Here we show that the E3 ubiquitin ligase, Rnf114, is highly expressed in mouse oocytes and that knockdown of Rnf114 inhibits development beyond the two‐cell stage. To study the underlying mechanism, we identify its candidate substrates using a 9,000‐protein microarray and validate them using an in vitro ubiquitination system. We show that five substrates could be degraded by RNF114‐mediated ubiquitination, including TAB1. Furthermore, the degradation of TAB1 in mouse early embryos is required for MZT, most likely because it activates the NF‐κB pathway. Taken together, our study uncovers that RNF114‐mediated ubiquitination and degradation of TAB1 activate the NF‐κB pathway during MZT, and thus directly link maternal clearance to early embryo development.
Keywords: maternal‐to‐zygotic transition, NF‐κB pathway, RNF114, TAB1, two‐cell‐stage arrest
Subject Categories: Development & Differentiation; Post-translational Modifications, Proteolysis & Proteomics
Introduction
The earliest stages of embryogenesis are controlled by maternal factors, which include large amounts of mRNA and protein that are accumulated and stored in the cytoplasm of mature oocytes. After fertilization, these maternal factors need to be eliminated to support maternal‐to‐zygotic transition (MZT) and subsequent zygotic genome activation (ZGA) 1, to facilitate the transition from terminally differentiated eggs to pluripotent blastocysts. This drastic cellular renovation process is regulated at both translational and posttranslational levels 2. Previous studies demonstrated that maternal proteins and mRNAs are degraded by massive autophagy after fertilization in mammals 3. The ubiquitin‐proteasome system, another major eukaryotic protein degradation system, also plays a critical role during these stages, including pronuclear remodeling, activation of the embryonic genome, and mitotic progression in the early embryo 4, 5, 6. Several E3 ubiquitin ligases, such as Cullin 1, Cullin 4A, and G2E3, are essential for early embryo development in mammals 7, 8, 9. However, the involvement of the ubiquitin‐proteasome pathway in maternal clearance and its relationship to MZT are still unclear.
Recently, large‐scale proteomic profile studies of mouse oocyte at different developmental stages, including the germinal vesicle (GV) stage, the metaphase II (MII) stage, and the fertilized oocytes (zygotes), revealed that the ubiquitin‐proteasome pathway may play a very important role during the developmental transition 10, 11. RNF114 (Ring finger 114) protein was identified as one of the predominantly expressed proteins in the later stages of mouse oocytes 11. RNF114, also known as zinc finger protein 313 (ZNF313), belongs to the family of really interesting new gene (RING) domain E3 ubiquitin ligases. Apart from the RING domain, RNF114 was found to contain two C2H2 (Cys(2)‐His(2))‐type zinc fingers, as well as a UIM (ubiquitin‐interacting motif) 12, 13. The ubiquitin ligases typically determine the specificity for substrate ubiquitylation 14. Ubiquitin‐mediated degradation of key regulatory short‐lived proteins plays an important role in basic cellular processes such as cell cycle, cell division, cell death 15, the biogenesis of subcellular organelles, and the differentiation of cells or tissues 16, 17.
To explore the roles and mechanisms of the ubiquitin‐proteasome pathway in maternal clearance, here we studied the functional role of RNF114 during MZT. We found that the knockdown of Rnf114 results in decrease in early embryo developmental competence. By an unbiased screening on a protein microarray with more than 9,000 proteins, we identified 13 potential RNF114 substrates and found that the elimination of one of them, TAB1, is necessary for MZT likely due to the requirement for the activation of the NF‐κB pathway. Our results uncover a novel functional role of RNF114 during MZT, and provide direct coupling evidence about maternal clearance and early embryo development.
Results and Discussion
RNF114 is predominately expressed in the oocytes and early embryonic development
We previously found that RNF114 protein was one of the highly expressed E3 ligases at the later stages in mouse oocytes 11. Here we confirmed that while Rnf114 mRNA was ubiquitously expressed in various tissues and organs, it was predominately expressed in oocytes, followed by lung and uterus (Fig 1A). Immunohistochemistry of RNF114 in the ovary also confirmed that RNF114 was mainly expressed in the oocytes (Fig 1B). We then explored the expression pattern of RNF114 from germinal vesicle (GV) to blastocyst stages by immunofluorescence with anti‐RNF114 antibody. The results revealed that RNF114 was distributed in the cytoplasm from GV oocytes to the blastocyst stages (Fig EV1). Taken together, these results demonstrate that Rnf114 is highly expressed in oocytes and in early embryonic development.
Figure 1. Expression of Rnf114 during mouse early embryonic development and effect of Rnf114 knockdown on early embryonic development.
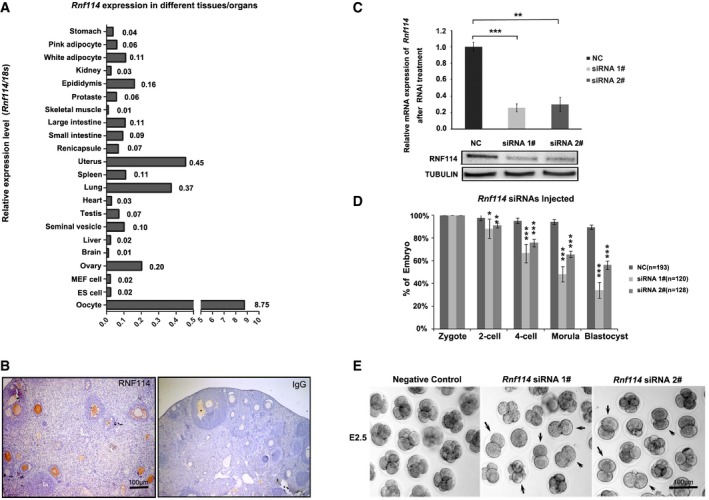
- Quantitative analysis of Rnf114 mRNA by real‐time PCR in various mouse tissues and organs shows that Rnf114 is predominately expressed in oocytes.
- Immunohistochemical analysis of RNF114 in mouse ovary shows that RNF114 is mainly expressed in the oocytes. Scale bar = 100 μm.
- Knockdown efficiency of Rnf114 in the two‐cell‐stage embryos derived from Rnf114 siRNA #1‐ or siRNA #2‐injected zygotes was verified by real‐time PCR and Western blotting. NC indicates negative control injected with non‐silencing siRNA. Three independent experiment replicates were performed for real‐time PCR, and error bars represent s.e.m.; **P < 0.01, ***P < 0.001 in unpaired two‐tailed t‐test.
- RNF114 depletion impaired the early embryonic development. Rnf114 siRNA #1‐ or siRNA #2‐injected zygotes were cultured and quantified (siRNA1#, n = 120; siRNA2#, n = 128; three independent experiments); NC indicates negative control (n = 193; six independent experiments). The error bars represent s.d., *P < 0.05, **P < 0.01, ***P < 0.001 in unpaired two‐tailed t‐test.
- Representative images to show early embryonic development defects. The embryos arrested at two‐cell stage are indicated by arrows. Scale bar = 100 μm.
Source data are available online for this figure.
Figure EV1. The expression of RNF114 during mouse early embryonic development.
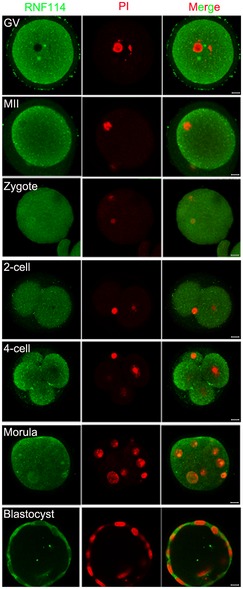
Immunofluorescence staining of RNF114 showed that the positive signals of RNF114 (green) are located in the cytoplasm from GV to blastocyst stages. PI nuclear staining (red). The results show that RNF114 was constantly expressed in the cytoplasm from GV oocytes to the blastocyst stages. Scale bar = 10 μm.
Knockdown of Rnf114 impairs early embryonic development
To assess the functional role of RNF114, zygotes were microinjected with specific siRNAs against Rnf114. The knockdown efficiency was confirmed by qRT–PCR and Western blotting at 24 h after microinjection. We observed an obvious decrease of Rnf114 at both mRNA and protein expression levels in Rnf114 siRNA #1 or #2 injection groups compared with the control group injected with non‐silencing siRNA (Fig 1C). We found that while almost all of the control embryos could normally develop into the four‐cell stage (non‐silencing siRNA, 95%, n = 193; six independent experiments), a considerable percent of the embryos injected with Rnf114 siRNAs could not develop to the four‐cell stage (siRNA1#, 33%, n = 120; siRNA2#, 24%, n = 128; three independent experiments) (Fig 1D and E). These results suggest that RNF114 plays an important role during early embryonic development and might be required for MZT.
Substrate screening of RNF114 using ProtoArray
As an E3 ligase 18, RNF114 might regulate MZT by targeting specific substrates. Therefore, the identification of its substrates could provide insight into its mechanisms and physiological functions during MZT. The ProtoArray® Human Protein Microarrays v5.0, which contains over 9,000 immobilized human proteins, provides a useful platform to identify potential substrates of E3 ligases 19, 20. To set up the in vitro ubiquitination system, we expressed and purified recombinant wild‐type and mutant human RNF114 in E. coli, and the critical animo acids (Cys44 and Cys49 in the C3HC4‐type RING domain) of the mutant RNF114 were changed to tryptophan. In the presence of E1, Ubc4, ATP, and FLAG‐tagged ubiquitin (FLAG‐ub), recombinant RNF114 could efficiently catalyze the formation of high molecular weight polyubiquitin chains (Fig 2A), while the mutant RNF114 lacked ubiquitination activity (Fig 2B and C), confirming that this RNF114 mutant could serve as a negative control in the substrate screening procedure.
Figure 2. RNF114 substrate screening by human protein microarrays.
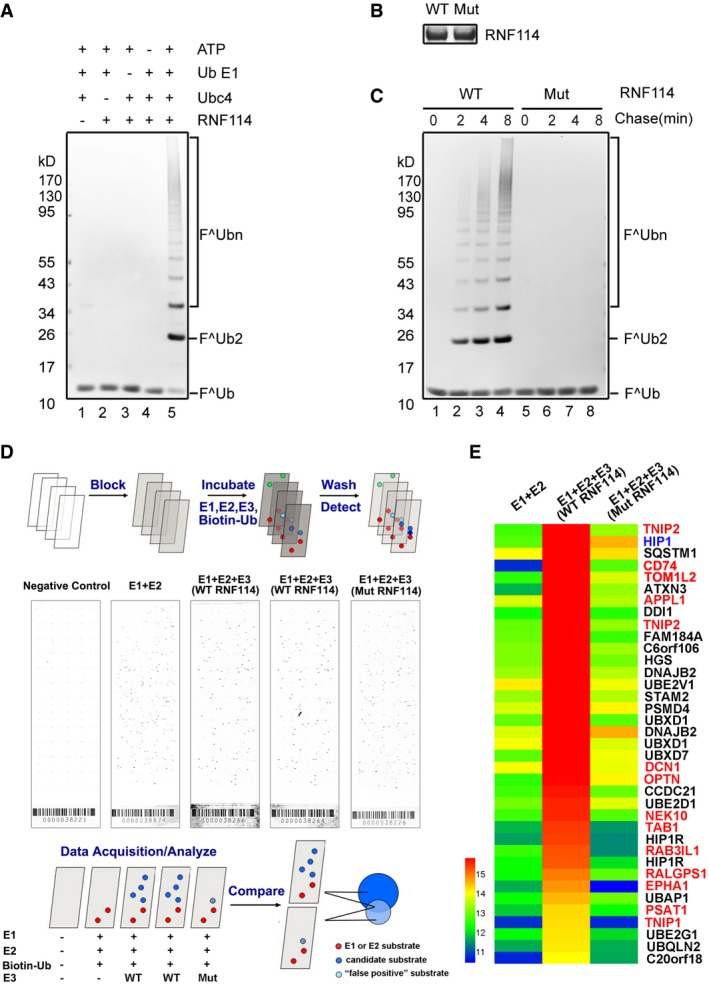
-
AIn vitro ubiquitination assay. Purified His‐RNF114 was subjected to ubiquitination assays with the indicated components, and the samples were analyzed by immunoblotting with anti‐FLAG (Ub) antibody. Lanes 1–4 indicate negative controls. The results show that recombinant RNF114 efficiently catalyzed the formation of polyubiquitin chains.
-
B, CPurified wild‐type and RING‐mutated His‐RNF114 were stained by Coomassie blue (B). Purified wild‐type and RING‐mutated His‐RNF114 were subjected to ubiquitination assays together with E1, Ubc4, FLAG‐Ub, and ATP. The results show that mutant RNF114 lacked ubiquitination activity (C).
-
DScreening for proteins ubiquitinated by RNF114 on a proteome array. Putative substrates of RNF114 were identified on Ubiquitin Ligase Substrate Identification on ProtoArray® Human Protein Microarrays v5.0. Ubiquitination reactions were performed on quintuplicate arrays using the indicated components, the five sub‐array are, respectively, negative controls, E1 + E2 (no E3), WT RNF114 (two arrays, duplicate), and mutated RNF114.
-
EA heat map comparing hits between E1 + E2, E1 + E2 + WT RNF114, and E1 + E2 + mutated RNF114 reactions. Candidate substrates of RNF114 selected for further characterization are indicated in red font; HIP1, indicated by blue font, was considered to be a false positive because it was also detected in the mutated RNF114 reaction. Black font indicates proteins with ubiquitin‐binding domains such as UIM, UBA, or CUE or proteins without clearly reported functions, which were excluded from further investigation.
To screen potential substrates of RNF114, E1, Ubc4 (E2), and E3 (RNF114 and mutant RNF114) together with biotin–ubiquitin were profiled on ProtoArray® Human Protein Microarrays. A brief schematic is shown in Fig 2D. The reaction was run in parallel with a negative control array probed with buffer alone and a control in which only E3 was omitted. After incubation with Alexa Fluor®647‐conjugated streptavidin, all arrays were scanned with an Axon 4000B fluorescent scanner (Molecular Devices). A small percentage (less than 0.5%) of the proteins exhibited significant signals in the negative control assay due to interaction with the detection reagent and were eliminated from the analysis. Possible substrates were evaluated by their Z‐score and signal used values within the array and were then compared with the control assay in which E3 was omitted. According to the threshold criteria described in Materials and Methods, 37 proteins were identified to exhibit significant ubiquitination signals with wild‐type RNF114. One of them (HIP1) might be a false positive as it was also detected when mutated RNF114 was used (Table EV1). Because ubiquitin or ubiquitin chains could charge on to some ubiquitin‐conjugating enzymes and can also be recognized by proteins that contain ubiquitin‐binding domains, such as UIM (ubiquitin‐interacting motif), UBA (ubiquitin associated), or CUE (coupling of ubiquitin conjugation to endoplasmic reticulum degradation) domains, proteins with these domains/motifs were excluded from further investigation. Furthermore, proteins without clearly reported functions were also not considered. Thirteen proteins—CD74, TAB1, TNIP1, TOM1L2, OPTN, TNIP2, EPHA1, NEK10, RALGPS1, RAB3IL1, PSAT1, APPL1, and DCN1—were selected as candidate substrates of RNF114 for further characterization (Fig 2E and Table EV1).
Experimental validation of RNF114 substrates
All seven lysines in ubiquitin can be used to build polyubiquitin chains with different topologies 21, and polyubiquitin chains with different linkages have distinct structural and functional relevance. Well‐accepted doctrine substrates modified with Lys48 or Lys11 polyubiquitin chains can be targeted to the proteasome for degradation, but this does not apply to other linkages 22, 23. To investigate the linkage specificity of RNF114‐mediated polyubiquitination, wild‐type ubiquitin and mutants with a single lysine changed to arginine (K6R, K11R, K27R, K29R, K33R, K48R, and K63R) or mutants in which only a single lysine was present with all others changed to arginine (K6, K11, K27, K29, K33, K48, and K63) were used in in vitro ubiquitination assays. As shown in Fig EV2, Lys11, Lys27, and Lys48 were necessary for RNF114‐mediated polyubiquitin chain assembly and Lys11 and Lys27 ubiquitin derivatives were both sufficient for high molecular weight ubiquitin chain formation, indicating that RNF114 assembles heterotypic polyubiquitination chains consisting of K11, K27, and K48 linkages. These data suggest that RNF114 ubiquitination of target substrates might promote their degradation.
Figure EV2. Linkage specificity of RNF114‐mediated polyubiquitin chains.
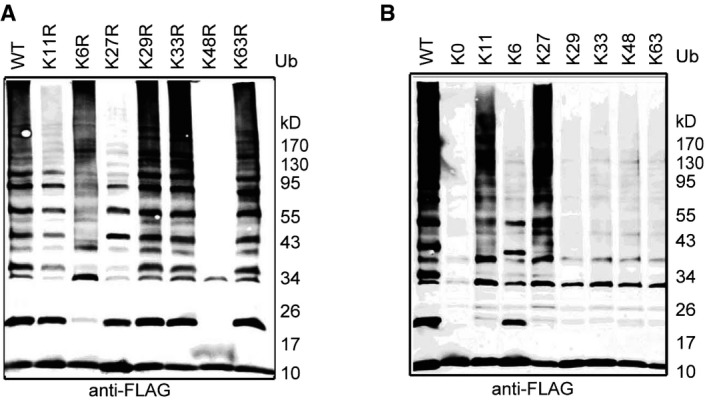
-
A, BUbiquitin mutants (K63R, K48R, K33R, K29R, K27R, K11R, K6R, or K11 only, K6 only, K27 only, K29 only, K33 only, K48 only, K63 only, and K0) were used for ubiquitin chain assembly. K0 indicates that all seven internal lysine residues were mutated to arginine. In the absence of K6, K29, K33, or K63 residues (K6R, K29R, K33R, or K63R), the mutated ubiquitins could still be used to assemble polyubiquitin chains, while in the absence of K11, K27, or K48 (K11R, K27R, or K48R), the mutated ubiquitins could not be used to assemble polyubiquitin chains by RNF114. K11‐only or K27‐only ubiquitin could be used to assemble polyubiquitin chains by RNF114.
We next established the eukaryotic expression vector containing the open reading frames of the 13 potential RNF114 substrate genes and a N‐terminal FLAG tag. Ten FLAG‐tagged candidate substrates were successfully detected by Western blotting after transfection into HEK293T cells (Fig EV3). The candidate substrate constructs were then co‐transfected into HEK293T cells with RGS‐His‐tagged wild‐type or mutant RNF114. EGFP (enhanced green fluorescent protein) was used as a transfection control. Compared with those co‐transfected with mutant RNF114, the protein expression levels of CD74, TAB1, OPTN, TNIP1, APPL1, PSAT1, and RALGPS1 were clearly reduced in the presence of wild‐type RNF114, while no changes were observed in the remaining three proteins (Fig 3A). These results suggest that RNF114 might regulate the stability of these seven proteins by promoting their ubiquitination and degradation.
Figure EV3. Eukaryotic expression of the potential substrate proteins.
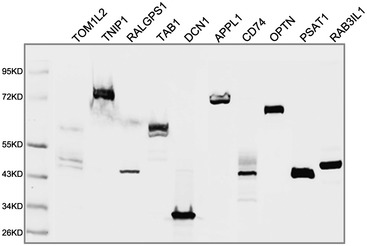
Ten FLAG‐tagged candidate substrates were successfully expressed by HEK293T cells and detected by Western blotting.
Figure 3. Verification of RNF114 substrates.
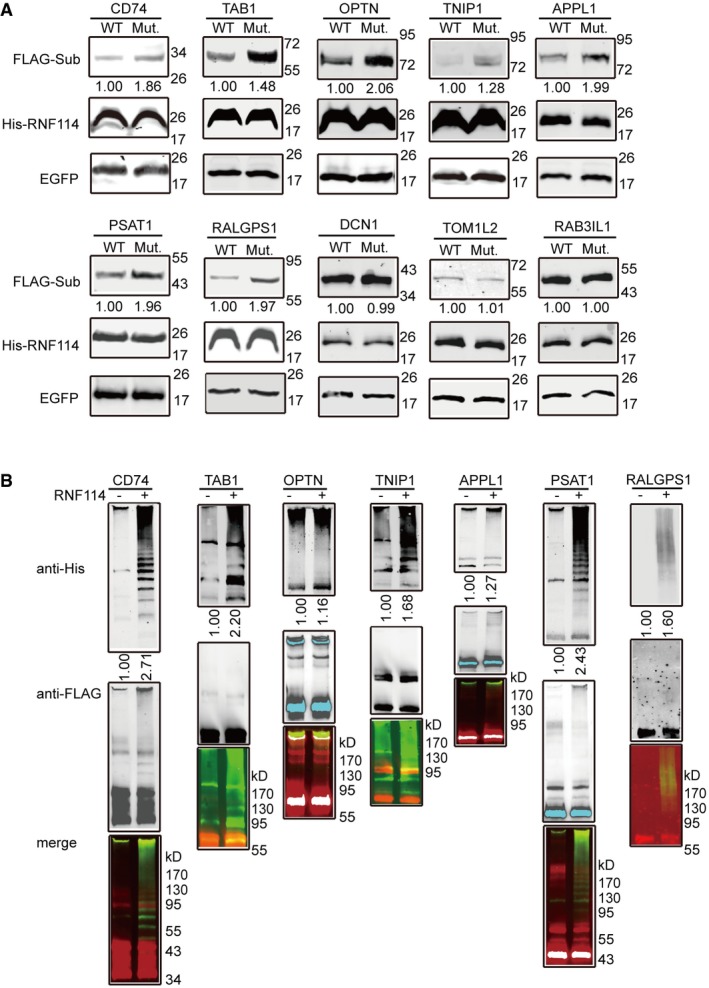
- RNF114 substrate validation. HEK293 cells were co‐transfected with constructs expressing FLAG‐tagged substrates, wild‐type or mutated His‐RNF114, and EGFP, and Western blot was performed with anti‐FLAG and anti‐His antibodies. The results show that the protein levels of CD74, TAB1, OPTN, TNIP1, APPL1, PSAT1, and RALGPS1 were reduced in the presence of wild‐type RNF114 compared to co‐transfecting with mutant RNF114.
- RNF114‐dependent in vitro ubiquitination of substrates. Substrate ubiquitination assays were performed with purified RNF114, the reaction products were immunoblotted using anti‐His antibody to detect the ubiquitin (upper panel) or anti‐FLAG antibody detecting the substrate protein (middle panel), and the lower panel shows the merged signals. The results show that clear polyubiquitin chains were formed in the systems containing recombinant CD74, TAB1, TNIP1, PSAT1, or RALGPS1.
To test the above possibility, we next performed in vitro ubiquitination assays using anti‐FLAG immunoprecipitates from HEK293T cells transfected with the candidate substrate proteins with or without the presence of RNF114. His‐ubiquitin was detected by Western blotting using the anti‐His tag antibody. The results showed that CD74, TAB1, TNIP1, PAST1, and RALGPS1 could be efficiently modified with ubiquitin in a RNF114‐dependent manner (Fig 3B) and suggest that these five proteins may be bona fide substrates of RNF114.
TAB1 accumulation impairs the early embryo developmental competence
Next we examined the potential role of these substrates in RNF114 function during MZT. Because knockdown Rnf114 results in the decrease of embryos develop beyond two‐cell stage, the overexpression of its substrate might also cause a similar phenotype if it is a critical substrate of RNF114 in this process. We microinjected in vitro‐transcribed mRNAs of myc‐Cd74, myc‐Tab1, myc‐Tnip1, myc‐Psat1, and myc‐Ralgps1 into zygotes, and the efficient overexpression of exogenous proteins was confirmed by Western blotting using anti‐myc tag antibody (Fig EV4). After 48–50 h of culture, the zygotes in the control groups developed normally, and approximately 97% reached the four‐cell stage. Zygotes injected with Ralgps1, Cd74, Tnip1, or Psat1 mRNA showed little changes. However, zygotes injected with exogenous Tab1 mRNA showed obvious developmental defects, which were similar to the phenotype caused by Rnf114 knockdown, with 27% of the zygotes arrested at the two‐cell stage (Fig 4A). Western blot analysis showed that during early embryo development, the expression of TAB1 decreased sharply, only weak expression could be detected at two‐cell and four‐cell stages, while when Rnf114 was knocked down by siRNAs, abnormal accumulation of TAB1 was observed (Fig 4B). In support of the two‐cell stage degradation, we found that the endogenous TAB1 could be ubiquitinated at this stage (Fig EV5); furthermore, more Ub signals on TAB1 were detected in the two‐cell‐stage embryos than in the zygotes (Fig 4C), suggesting endogenous TAB1 ubiquitination mainly occurs at two‐cell stage. To investigate whether the degradation of TAB1 was mediated by RNF114, Tab1‐GFP mRNA was injected into zygotes in the condition with or without knockdown RNF114. As expected, the GFP fluorescence signal gradually declined during the early embryo development. However, in the embryos injected with Rnf114 siRNAs, TAB1‐GFP levels remained approximately constant (Fig 4D), suggesting the elimination of TAB1 is indeed dependent on RNF114. Furthermore, when specific siRNA against Tab1 was microinjected into oocytes at GV stage together with Rnf114 siRNA 1#, the two‐cell‐stage arrest defect caused by RNF114 knockdown was partly rescued (Fig 4E). Together, these results suggest that TAB1 might be the major substrate of RNF114 during MZT.
Figure EV4. Overexpression of substrate proteins.
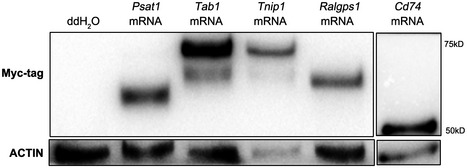
RNase‐free ddH2O (control group) or in vitro‐transcribed mRNAs of myc‐Cd74, myc‐Tab1, myc‐Tnip1, myc‐Psat1, and myc‐Ralgps1 were microinjected into zygotes, and then, the expression of exogenous protein was detected by Western blotting using anti‐myc tag antibody.
Figure 4. RNF114 mainly targets TAB1 for degradation during MZT .
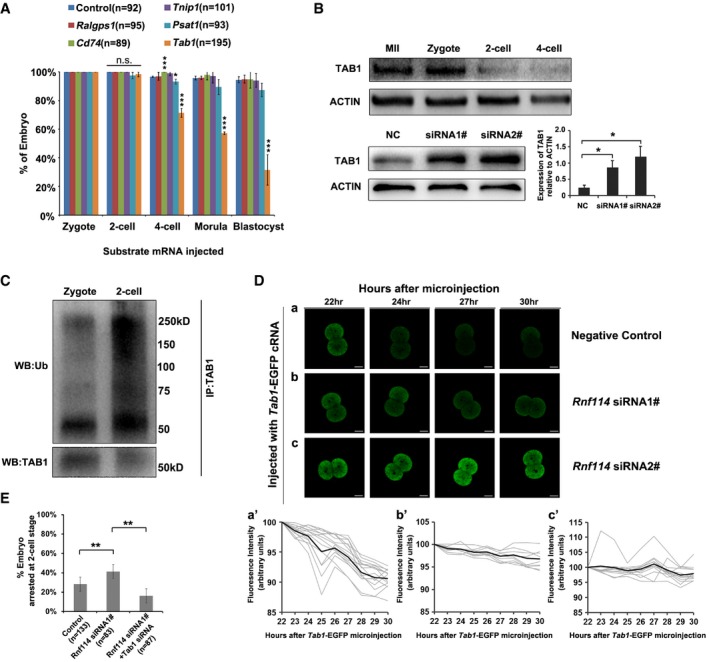
- Mouse zygotes were microinjected with sterile ddH2O (control group) or Ralgps1, Cd74, Tnip1, Psat1, or Tab1 mRNA. During 96–100 h of culturing, the percentages of embryos at various stages were counted under microscopy. The results show that overexpression of Tab1 impaired the early embryonic development. Three independent experiment replicates were performed, and error bars represent s.d.; *P < 0.05, ***P < 0.001 in unpaired two‐tailed t‐test.
- TAB1 reduced with mouse early embryonic development, while abnormally accumulated at two‐cell stage once treated with Rnf114 siRNAs. TAB1 was detected by Western blotting with anti‐TAB1 antibody. The relative level of TAB1 protein after Rnf114 siRNAs treatment were presented as mean ± s.e.m. (*P < 0.05 in unpaired two‐tailed t‐test, four independent experiment replicates).
- Detection of endogenous TAB1 ubiquitination at one‐cell or two‐cell stage. Endogenous TAB1 proteins were immunoprecipitated by anti‐TAB1 antibody at either one‐cell or two‐cell stage; the immunoprecipitates were detected by either anti‐TAB1 antibody or anti‐ubiquitin antibody. The results show that more Ub chains on TAB1 were detected in the two‐cell‐stage embryos than in the zygotes, when the TAB1 proteins were at comparable level.
- RNF114‐dependent TAB1 degradation during early embryo development. (a–c) Representative time‐lapse embryo images are shown for each treatment. Scale bar = 20 μm. (a'–c') Quantification of the GFP fluorescence signals for respective treatments in (a–c). n = 14, 9, or 12 for the NC, Rnf114 siRNA1#, or Rnf114 siRNA2# treatment group, respectively. Gray traces: Tab1‐GFP fluorescent signals were normalized to the first time point. Black traces: average signal of all embryos for each treatment.
- Tab1 knockdown partly rescued the early embryo development defect observed with Rnf114 knockdown. Both Rnf114 and Tab1 siRNAs were injected into the GV oocytes, then the oocytes were matured and were fertilized by IVF, and the fertilized eggs were transferred into KSOM media, cultured for 48–50 h. The percentages of embryos arrested at two‐cell stage were counted. Three independent experiment replicates were performed, and error bars represent s.d.; **P < 0.01 in unpaired two‐tailed t‐test.
Source data are available online for this figure.
Figure EV5. Endogenous ubiquitination of TAB1 in mouse two‐cell‐stage embryos.
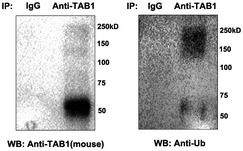
Cell lysates of mouse two‐cell‐stage embryos were immunoprecipitated by anti‐TAB1 antibody, followed by Western blot analysis utilizing specific antibodies against TAB1 and Ub. A parallel immunoprecipitation with IgG was performed as a control.
TAB1 clearance is required for the activation of NF‐κB pathway during MZT
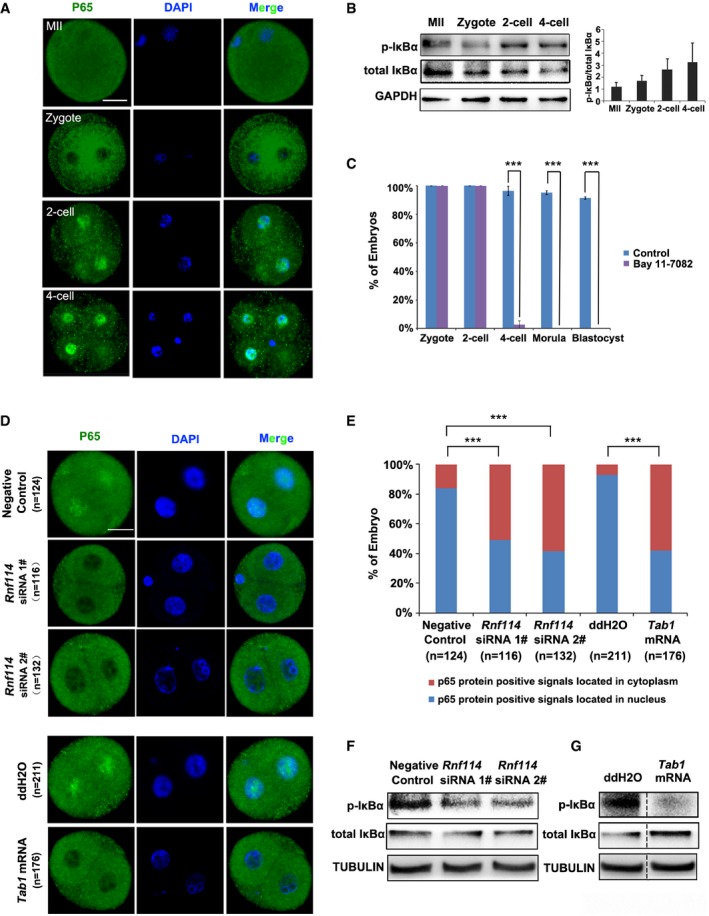
Next, we addressed the functional role of TAB1 clearance during MZT. TAB1 is a regulator of the MAP kinase kinase kinase MAP3K7/TAK1, which is an intracellular hub molecule that regulates both nuclear factor‐κB (NF‐κB) and mitogen‐activated protein kinase (MAPK) signaling pathways 24, 25. These pathways play key roles in development, cell survival, immune response, metabolism, and carcinogenesis 26. A previous study showed that TAB1 knockdown resulted in increased NF‐κB pathway activity, and excess amount of TAK1 blocks the spontaneous NF‐κB activation by a negative feedback manner in some cells 27. The NF‐κB/Rel transcription factors are present in the cytosol in an inactive state coexisted with the inhibitory IκB proteins, while the phosphorylation and proteolytic degradation of IκBα result in the release and nuclear translocation of active RelA (p65) 28, 29, 30. The activation and nuclear translocation of NF‐κB p65/p50 are key steps for the DNA binding of NF‐κB and transcription of downstream target genes 31. Interestingly, activation of the NF‐κB pathway is crucial to the early stage development of the mouse embryo 32. Similar to their results, we found that the NF‐κB pathway was quickly activated during early embryonic development; p65 was mainly located in the cytoplasm at MII and zygote stages, while at the two‐cell stage and thereafter it translocated into the nucleus (Fig 5A). With embryonic development, the protein level of inhibitory subunit IκBα decreased accompanied with upregulated phosphorylated form (p‐IκBα), and the ratio of p‐IκBα to total IκBα increased gradually form MII oocyte to four‐cell stage (Fig 5B). These results provide direct evidence for NF‐κB activation during MZT. Furthermore, zygotes treated with the p65 inhibitor Bay 11‐7082 33 were arrested at the two‐cell stage (Fig 5C). Together, these results suggest that the activation of the NF‐κB pathway might be essential to MZT.
Figure 5. Developmental defect caused by TAB1 accumulation might be due to failure of NF‐κB pathway activation.
-
ANF‐κB activation during MZT. Immunofluorescence staining of p65 shows nuclear translocation of p65 at the two‐cell stage and thereafter. Scale bar = 25 μm.
-
BThe protein levels of IκBα and its phosphorylated form (p‐IκBα) during mouse early embryonic development were detected by Western blotting, representative Western blotting of IκBα and p‐IκBα is shown on the left and the quantitation analysis (right panel) indicated that the ratio of p‐IκBα/total IκBα was gradually increased from MII oocytes to four‐cell‐stage embryos (three independent experiment replicates were performed, and error bars represent s.e.m.).
-
CNF‐κB pathway inhibition using the specific inhibitor Bay 11‐7082 resulted in two‐cell‐stage arrest. Three independent experiment replicates were performed, and error bars represent s.d.; ***P < 0.001 in unpaired two‐tailed t‐test.
-
D, ERepresentative immunofluorescence staining of p65 after knockdown of Rnf114 by Rnf114 siRNA #1 or siRNA #2 or overexpression of Tab1 by Tab1 mRNA injection (shown in D). Scale bar = 25 μm. The percentages of two‐cell‐stage embryos with p65 nuclear translocation (shown in E) were significantly decreased after Rnf114 knockdown or Tab1 overexpression (chi‐square test was performed for statistical analysis, ***P < 0.001).
-
FWestern blot analysis at two‐cell stage showed the levels of p‐IκBα were reduced in the Rnf114 siRNA #1‐ and siRNA #2‐injected groups.
-
GWestern blot analysis at two‐cell stage showed the levels of p‐IκBα were reduced in the exogenous Tab1 mRNA‐injected groups, and ddH2O was injected as control. Dotted line indicates a cropped lane.
Source data are available online for this figure.
We then analyzed the involvement of TAB1 and RNF114 in the regulation of NF‐κB activity during MZT. We found that in contrast to the predominant nuclear localization of p65 at the two‐cell stage in embryos derived from control zygotes, p65 nuclear translocation was clearly blocked when either RNF114 was knocked down or TAB1 was overexpressed by Tab1 mRNA microinjection (Fig 5D and E). Western blot of two‐cell‐stage embryos further revealed that the level of p‐IκBα was obviously reduced after RNF114 was knocked down (Fig 5F) or TAB1 was overexpressed (Fig 5G). Taken together, these results suggest that RNF114‐mediated ubiquitination and degradation of TAB1 might be essential to the activation of NF‐κB pathway during MZT, thus directly coupling maternal clearance to early embryo development.
A previous study reported that RNF114 overexpression could activate the NF‐κB pathway in HEK293T cells 34, while in T cells, it negatively regulates NF‐κB pathway by increasing the stability of A20 and IκBα 35. Our studies provide a novel mechanism for RNF114 function in NF‐κB pathway activation during MZT. The discrepancy for the effect of RNF114 on NF‐κB pathway activation among these studies might be due to the different status of these cells/tissues in response to different stimuli. The functional role of NF‐κB pathway during MZT has been unaddressed for some time as most of the null mutants in the NF‐κB pathway die before sexual maturity 36, thus preventing the study of their effect on MZT.
Materials and Methods
Animals and ethics statement
CD1 mice were maintained under a controlled environment of 20–25°C, 12/12‐h light/dark cycle, and 50–70% humidity, with free access to water and food. All animal procedures were approved by Nanjing Medical University, Institutional Animal Care and Use Committee, and conducted according to the Guide for the Care and Use of Laboratory Animals.
Embryo collection and culture
CD1 female mice (6–8 weeks old) were superovulated by injections of 10 IU HCG (human chorionic gonadotropin) followed by 10 IU PMSG (pregnant mare serum gonadotropin) 48 h later. MII oocytes were collected at 16 h post‐HCG from the ampullae of oviducts. Zygotes were collected at 16 h post‐HCG from the ampullae of oviducts of superovulated females that had been mated with CD1 males. The cumulus cells were removed by digestion with hyaluronidase (Sigma) for several minutes. Embryos were cultured in fresh KSOM medium (Millipore) at 37°C in a 5% CO2 in air atmosphere. Next, two‐cell‐, four‐cell‐, eight‐cell‐, morula‐, and blastocyst‐stage embryos were collected after 22–26, 48–50, 60–65, 70–75, and 96–100 h of culture, respectively.
To examine the effects of the NF‐κB inhibitors, zygotes were collected at 16 h post‐HCG and cultured in KSOM medium containing 10 μM Bay 11‐7082 (Beyotime Co., China). Embryos that were cultured in KSOM medium with only DMSO were used as controls.
siRNA microinjection
To knockdown RNF114, Rnf114 siRNAs (Ribobio Co., China) (siRNA1# 5′‐AUAGGAUGAAAUUCUUACGGCAGCC‐3′; siRNA2# 5′‐UUUGGAACAUGACGUCACAUGGGCC‐3′) were diluted in buffer to 20 mM. A quantity of 5–10 pl of siRNA was microinjected into the cytoplasm of mouse zygotes in M2 medium (Sigma), and zygotes were cultured in fresh KSOM medium. Non‐silencing siRNA (5′‐UUCUCCGAACGUGUCACGUTT‐3′) was used as the control. Zygotes were cultured for 24 h (two‐cell stage) and collected to assess knockdown efficiency by real‐time PCR and Western blotting. Rnf114‐specific primers were designed as 5′‐CGGCAGATCGAGAGCATAGAG‐3′ and 5′‐ TGGCCTTTACACCTTCCATGA‐3′. 18S rRNA was used as internal control. The relative changes in gene expression were calculated by method.
For rescue experiments, the mixture of Rnf114 siRNA 1# and the specific siRNA against Tab1 (5′‐GUGGAUGGGUUACAGGUUA‐3′) with proven effect was injected into the GV oocytes. To ensure the knockdown efficiency, the GV oocytes were cultured in M2 medium supplemented with 3.5 μM milrinone for 24 h to prevent germinal vesicle breakdown (GVBD) and then transferred to a milrinone‐free medium to mature. The mature oocytes were fertilized by IVF (in vitro fertilization) procedures, and the subsequent development was observed carefully.
Substrate protein microarrays
To identify the substrates of RNF114 E3 ligase, the ProtoArray® Human Protein Microarrays v5.0 (Invitrogen) was performed according to the manufacturer's protocol. The microarray contains over 9,000 unique human proteins on a nitrocellulose‐coated glass slide. Wild‐type Rnf114 was profiled in duplicate at 0.15 μg/μl, while mutant Rnf114 was profiled in one reaction. The reactions contained E1 (0.02 μg/μl), E2 (0.05 μg/μl), and biotin–ubiquitin (0.1 μg/μl). For the control array, only E1 and E2 were used in the reaction. Energy regeneration solution (Boston Biochem) was used at a final concentration of 1×. Slides were blocked in 4‐well QuadriPERM trays with 5 ml blocking buffer (50 mM HEPES pH 7.5, 200 mM NaCl, 0.08% Triton X‐100, 25% glycerol, 20 mM reduced glutathione, 1.0 mM DTT, 1% BSA) for 60 min at 4°C. Reactions were prepared in assay buffer (50 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgSO4, 1 mM DTT, 0.1% Tween 20, 1% BSA) and incubated at 30°C for 10 min; then, 120 μl of each reaction mixture was applied to the slides and overlaid with a LifterSlip™ (Erie Scientific), taking care not to produce bubbles. Slides were incubated at 30°C for 90 min. Arrays were then washed with three 5‐min washes using 5 ml 0.5% SDS and three 5‐min washes with 5 ml assay buffer. Arrays were then incubated with 1.0 ng/μl streptavidin Alexa Fluor®647 for 45 min at 4°C, followed by five 5‐min washes with 5 ml assay buffer per wash. Arrays were quickly rinsed in water to remove residual salts and dried by centrifugation at 200 g for 2 min in a tabletop centrifuge equipped with a plate rotor. The arrays were then scanned for data analysis.
Data acquisition/analysis
Arrays were scanned with an Axon GenePix 4000B fluorescent scanner (Molecular Devices). Data were acquired with GenePix 6.0 software (Molecular Devices) and processed in ProtoArray® Prospector. Significance call queries were performed by Prospector to identify hits on each protein array. The significance call determines which signals are greater than or equal to a value that is determined by calculating the median plus 3.0 standard deviations (using signal minus background values for all non‐control proteins) for all non‐control proteins on the array. Substrates are defined as proteins having positive significance calls that are not observed on the appropriate negative control. In some cases, hits not identified by the program are counted but only if manually verified by looking at the spots on the arrays. A protein was defined as exhibiting significant ubiquitination signal if it met the following set of threshold criteria: (i) the signal value on the array probed with E3 was least threefold higher than the signal value in the E1/E2 control assay; (ii) Z‐Factor > 0.5, which means the signal is at least twofold above the noise; (iii) the Z‐score or normalized fluorescent signal > 2.0 standard deviations above the mean human protein signal on the corresponding array; (iv) both the replicate spot CV and the inter‐assay CV < 50%; and (v) the CI P‐value < 0.05, correlating with a visually confirmable signal on the array.
Plasmids, protein expression, and purification
The expression plasmids of human or mouse Rnf114 and the eukaryotic expression plasmids of 13 substrates, including Flag‐Appl1, Flag‐Optn, Flag‐Tnip2, Flag‐Tom1 l2, Flag‐Nek10, Flag‐Ralgps1, Cd74, Dcn1, Epha1, Psat1, Rab3il1, Tab1, and Tnip1, were purchased from OriGene, GeneCopoeia, or FunGenome Company. We amplified human Rnf114 from EX‐W0860‐B04 T7 E. Coli N/A N‐GST EK pReceiver‐B04 cDNA and cloned it into both the pGEX‐4T‐2 vector with GST tag and the pRK vector with His tag, while mouse Rnf114 were also cloned into the pGEX‐4T‐2 and pRK vectors. In addition, Flag‐Cd74, Flag‐Dcn1, Flag‐Epha1, Flag‐Psat1, Flag‐Rab3il1, Flag‐Tab1, and Flag‐Tnip1 were, respectively, amplified and cloned into pcMV6 vector. pGEX‐GST‐Rnf114 or pRK‐His‐Rnf114 RING mutant was generated by site‐directed mutagenesis.
Prokaryotic expression plasmids of human WT or mutant Rnf114 were transformed into TOP10 competent cells using standard methods, and the transformed samples were plated onto agar plates containing appropriate antibiotics. A single colony was selected and incubated in LB medium with appropriate antibiotics until the culture reached the log phase, and isopropyl‐D‐thiogalactoside (IPTG) (1 mM final concentration) was added for 3 h to induce protein expression. Bacterial cell lysates were prepared by sonication and recombinant proteins were purified by previously described methods 37 and used for the protein microarray assay.
Cell transfection
HEK293T cells were grown in DMEM supplemented with 10% fetal bovine serum and maintained at 37°C in a humidified incubator with 5% CO2. Upon nearing confluence, cells were dissociated enzymatically with trypsin–EDTA and passaged. HEK293T cells were co‐transfected with plasmids expressing RGS‐His‐tagged WT or mutant Rnf114, EGFP, and each substrate including Flag‐Cd74, Flag‐Tab1, Flag‐Optn, Flag‐Tnip1, Flag‐Appl1, Flag‐Psat1, Flag‐Ralgps, Flag‐Dcn1, Flag‐Tom1 l2, or Flag‐Rab3il1 with Lipofectamine 2000 (Invitrogen). Cells were lysed in lysis buffer containing protease inhibitors and subjected to Western blotting with antibodies against FLAG or RGS‐His as reported previously 38 (detail information of antibodies is given in Table EV2).
In vitro ubiquitination assays
In vitro substrate‐independent ubiquitination assays were conducted as previously described 39. Briefly, ubiquitin‐activating enzyme E1 (0.02 μg/μl), Ubc4 (0.05 μg/μl), WT or mutant RNF114 protein (0.1 μg/μl), FLAG‐tagged ubiquitin (0.1 μg/μl), ATP (0.1 M), and substrate (0.02 μg/μl) were added into buffer containing 50 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2, 0.05% Nonidet P‐40, and 0.5 mM dithiothreitol. The reactions were incubated at 37°C for various times and then stopped by adding an equal volume of 2× SDS–PAGE loading buffer. The samples were then resolved by SDS–PAGE and blotted for ubiquitinated protein products using anti‐FLAG antibody.
In vitro substrate‐dependent ubiquitination assays were performed with anti‐FLAG immunoprecipitates as described previously 40. In brief, HEK293 cells were transfected with expression vectors of different potential substrate, and the cells extracted were precipitated with anti‐Flag M2 affinity agarose beads (Sigma‐Aldrich, USA). The resulting immunoprecipitates were added into the ubiquitination reaction system described above with or without pGEX‐GST‐Rnf114, His‐tagged ubiquitin was used instead to distinguish with the substrate protein, and the ubiquitinated protein was detected by both anti‐His and anti‐FLAG antibodies.
In vitro transcription
The ORFs of Tab1, Tnip1, Ralgps1, Psat1, and Cd74 genes were subcloned into pCS2+ vector, which has a myc tag, allowing in vitro transcription of polyadenylated mRNA 41. In addition, Tab1‐GFP was also cloned into pCS2+ vector. The constructs were linearized by SacII or KpnI and purified using a gel extract kit (Promega). SP6 message machine (Ambion) was used for producing capped mRNAs, and the mRNAs were purified by the RNeasy cleanup kit (Qiagen). Microinjection of mRNAs was performed as previously described 42. The same amount of RNase‐free ddH2O was injected as control. Western blotting was performed using anti‐myc tag antibody to confirm the overexpression of exogenous protein.
Immunofluorescence staining
Embryos were treated for 5 min at room temperature with 0.5% Triton X‐100 followed by 1% BSA in PBS for 1 h without washing, and then incubated with primary antibodies (details are given in Table EV2) overnight at 4°C. Embryos were then washed and incubated with secondary antibodies for 3 h at room temperature in the dark. After DAPI or PI staining, the embryos were washed, cover‐slipped with DABCO (diazabicyclooctane), and examined using a ZEISSLS M710 confocal laser‐scanning microscope (Carl ZEISS Micro Imaging GmbH, Jena, Germany).
Immunoprecipitation
Immunoprecipitation experiments were performed using Pierce Crosslink Immunoprecipitation kit (Pierce, #26147) according to the instruction of manufacturer. Briefly, mouse zygotes or two‐cell‐stage embryos were collected and lysed in buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP‐40, 5% glycerol; pH 7.4), and the lysates were cleared by centrifugation (10,000 g for 10 min) at 4°C. Supernatant was transferred into a new tube, and then, anti‐TAB1 antibody‐cross‐linked Protein A/G Plus Agarose was added to the samples and incubated at 4°C overnight. After elution, samples were analyzed with Western blotting. TAB1 IP‐WB antibody pair (Abnova, H00010454‐PW1) was used for immunoprecipitating and Western blotting, and protein ubiquitination was analyzed by Western blotting by mouse antibody against ubiquitin (Santa Cruz, 8017).
Live imaging and quantitative analysis of data
Zygotes were placed in three 15 μl drops of KSOM medium (Merck Millipore, Germany) on a glass‐bottom dish under mineral oil (Sigma‐Aldrich, USA). Time‐lapse images were acquired with an ECLIPSE TE2000‐PFS microscope (Nikon Corp., Japan), and a 37°C humidified chamber. Stacked frames of individual embryos were obtained every hour for 22–30 h after microinjection. Still images were generated with the NIS‐Elements Viewer (Nikon Corp., Japan).
Quantitative analysis of the fluorescence intensity was performed with ImageJ software, which is public‐domain software developed by the National Institute of Health (NIH) in the United States. To measure GFP signal, the area occupied by the embryo in each frame was defined manually, and the mean fluorescence intensity (MFI) of the signal within this area was measured. Values were corrected for background fluorescence by subtracting the MFI of the area surrounding the embryo at each time point. To control for the apparent differences in MFI levels between individual embryos caused by discrepancies in the amount of injected mRNA, background‐corrected MFIs were normalized to the intensity measured at the first time point.
Statistical analysis
Data were evaluated by unpaired two‐tailed t‐test or chi‐square test statistical analysis for comparisons between the experimental and control group. Statistical significance: n.s., not significant, P > 0.05; *P < 0.05; **P < 0.01: ***P < 0.001.
Author contributions
RH and WL conceived the project. YY, CZ, YW, WL, CL, LW, YL, YS, ML, SZ, YW, WZ, and JZ performed experiments and acquired data. All authors analyzed, interpreted, and concluded results. The manuscript was written by RH and WL with contributions from YY and CZ.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
This work was supported by National Nature Science Foundation of China (31671556, 31171113, 91519317) and China National Basic Research Program (2013CB911400, 2012CB944704).
EMBO Reports (2017) 18: 205–216
Contributor Information
Ran Huo, Email: huoran@njmu.edu.cn.
Wei Li, Email: leways@ioz.ac.cn.
References
- 1. Svoboda P, Franke V, Schultz RM (2015) Sculpting the transcriptome during the oocyte‐to‐embryo transition in mouse. Curr Top Dev Biol 113: 305–349 [DOI] [PubMed] [Google Scholar]
- 2. Walser CB, Lipshitz HD (2011) Transcript clearance during the maternal‐to‐zygotic transition. Curr Opin Genet Dev 21: 431–443 [DOI] [PubMed] [Google Scholar]
- 3. Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N (2008) Autophagy is essential for preimplantation development of mouse embryos. Science 321: 117–120 [DOI] [PubMed] [Google Scholar]
- 4. Huo LJ, Fan HY, Zhong ZS, Chen DY, Schatten H, Sun QY (2004) Ubiquitin‐proteasome pathway modulates mouse oocyte meiotic maturation and fertilization via regulation of MAPK cascade and cyclin B1 degradation. Mech Dev 121: 1275–1287 [DOI] [PubMed] [Google Scholar]
- 5. Shin SW, Tokoro M, Nishikawa S, Lee HH, Hatanaka Y, Nishihara T, Amano T, Anzai M, Kato H, Mitani T et al (2010) Inhibition of the ubiquitin‐proteasome system leads to delay of the onset of ZGA gene expression. J Reprod Dev 56: 655–663 [DOI] [PubMed] [Google Scholar]
- 6. Solter D, Hiiragi T, Evsikov AV, Moyer J, De Vries WN, Peaston AE, Knowles BB (2004) Epigenetic mechanisms in early mammalian development. Cold Spring Harb Symp Quant Biol 69: 11–17 [DOI] [PubMed] [Google Scholar]
- 7. Brooks WS, Helton ES, Banerjee S, Venable M, Johnson L, Schoeb TR, Kesterson RA, Crawford DF (2008) G2E3 is a dual function ubiquitin ligase required for early embryonic development. J Biol Chem 283: 22304–22315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dealy MJ, Nguyen KV, Lo J, Gstaiger M, Krek W, Elson D, Arbeit J, Kipreos ET, Johnson RS (1999) Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet 23: 245–248 [DOI] [PubMed] [Google Scholar]
- 9. Li B, Ruiz JC, Chun KT (2002) CUL‐4A is critical for early embryonic development. Mol Cell Biol 22: 4997–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S (2010) Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA 107: 17639–17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang P, Ni X, Guo Y, Guo X, Wang Y, Zhou Z, Huo R, Sha J (2009) Proteomic‐based identification of maternal proteins in mature mouse oocytes. BMC Genom 10: 348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giannini AL, Gao Y, Bijlmakers MJ (2008) T‐cell regulator RNF125/TRAC‐1 belongs to a novel family of ubiquitin ligases with zinc fingers and a ubiquitin‐binding domain. Biochem J 410: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao H, Li CC, Pardo J, Chu PC, Liao CX, Huang J, Dong JG, Zhou X, Huang Q, Huang B et al (2005) A novel E3 ubiquitin ligase TRAC‐1 positively regulates T cell activation. J Immunol 174: 5288–5297 [DOI] [PubMed] [Google Scholar]
- 14. Hatakeyama S, Nakayama KI (2003) Ubiquitylation as a quality control system for intracellular proteins. J Biochem 134: 1–8 [DOI] [PubMed] [Google Scholar]
- 15. Bergmann A (2010) The role of ubiquitylation for the control of cell death in Drosophila . Cell Death Differ 17: 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haglund K, Dikic I (2012) The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci 125: 265–275 [DOI] [PubMed] [Google Scholar]
- 17. Sheng K, Liang X, Huang S, Xu W (2014) The role of histone ubiquitination during spermatogenesis. Biomed Res Int 2014: 870695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han J, Kim YL, Lee KW, Her NG, Ha TK, Yoon S, Jeong SI, Lee JH, Kang MJ, Lee MG et al (2013) ZNF313 is a novel cell cycle activator with an E3 ligase activity inhibiting cellular senescence by destabilizing p21(WAF1.). Cell Death Differ 20: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schweitzer B, Meng L, Mattoon D, Rai AJ (2010) Immune response biomarker profiling application on ProtoArray protein microarrays. Methods Mol Biol 641: 243–252 [DOI] [PubMed] [Google Scholar]
- 20. Bauer M, Maj M, Wagner L, Cahill DJ, Linse S, O'Connell DJ (2011) Protein networks involved in vesicle fusion, transport, and storage revealed by array‐based proteomics. Methods Mol Biol 781: 47–58 [DOI] [PubMed] [Google Scholar]
- 21. Li W, Ye Y (2008) Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci 65: 2397–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer HJ, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157: 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- 24. Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K (1996) TAB1: an activator of the TAK1 MAPKKK in TGF‐beta signal transduction. Science 272: 1179–1182 [DOI] [PubMed] [Google Scholar]
- 25. Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin‐dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- 26. Roh YS, Song J, Seki E (2014) TAK1 regulates hepatic cell survival and carcinogenesis. J Gastroenterol 49: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreno‐Garcia ME, Sommer K, Rincon‐Arano H, Brault M, Ninomiya‐Tsuji J, Matesic LE, Rawlings DJ (2013) Kinase‐independent feedback of the TAK1/TAB1 complex on BCL10 turnover and NF‐kappaB activation. Mol Cell Biol 33: 1149–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghosh S, Baltimore D (1990) Activation in vitro of NF‐kappa B by phosphorylation of its inhibitor I kappa B. Nature 344: 678–682 [DOI] [PubMed] [Google Scholar]
- 29. Beg AA, Baldwin AS Jr (1993) The I kappa B proteins: multifunctional regulators of Rel/NF‐kappa B transcription factors. Genes Dev 7: 2064–2070 [DOI] [PubMed] [Google Scholar]
- 30. Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA (1995) Phosphorylation of human I kappa B‐alpha on serines 32 and 36 controls I kappa B‐alpha proteolysis and NF‐kappa B activation in response to diverse stimuli. EMBO J 14: 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oeckinghaus A, Ghosh S (2009) The NF‐kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1: a000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nishikimi A, Mukai J, Yamada M (1999) Nuclear translocation of nuclear factor kappa B in early 1‐cell mouse embryos. Biol Reprod 60: 1536–1541 [DOI] [PubMed] [Google Scholar]
- 33. Bokstrom H, Wiqvist N (1989) Preoperative dilatation of the cervix at legal abortion with a synthetic, fast‐swelling hygroscopic tent. Acta Obstet Gynecol Scand 68: 313–318 [DOI] [PubMed] [Google Scholar]
- 34. Bijlmakers MJ, Kanneganti SK, Barker JN, Trembath RC, Capon F (2011) Functional analysis of the RNF114 psoriasis susceptibility gene implicates innate immune responses to double‐stranded RNA in disease pathogenesis. Hum Mol Genet 20: 3129–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodriguez MS, Egana I, Lopitz‐Otsoa F, Aillet F, Lopez‐Mato MP, Dorronsoro A, Lobato‐Gil S, Sutherland JD, Barrio R, Trigueros C et al (2014) The RING ubiquitin E3 RNF114 interacts with A20 and modulates NF‐kappaB activity and T‐cell activation. Cell Death Dis 5: e1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gerondakis S, Grossmann M, Nakamura Y, Pohl T, Grumont R (1999) Genetic approaches in mice to understand Rel/NF‐kappaB and IkappaB function: transgenics and knockouts. Oncogene 18: 6888–6895 [DOI] [PubMed] [Google Scholar]
- 37. Ye J, Sui YF, Chen GS, Zhang XM (2003) Cloning and prokaryotic expression of heat shock protein 70 gene of Mycobacterium tuberculosis . Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 19: 443–445 [PubMed] [Google Scholar]
- 38. He H, Kong S, Liu F, Zhang S, Jiang Y, Liao Y, Li Q, Wang B, Zhou Z, Wang H et al (2015) Rbbp7 is required for uterine stromal decidualization in mice. Biol Reprod 93: 13 [DOI] [PubMed] [Google Scholar]
- 39. Liu W, Shang Y, Zeng Y, Liu C, Li Y, Zhai L, Wang P, Lou J, Xu P, Ye Y et al (2014) Dimeric Ube2 g2 simultaneously engages donor and acceptor ubiquitins to form Lys48‐linked ubiquitin chains. EMBO J 33: 46–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kohroki J, Nishiyama T, Nakamura T, Masuho Y (2005) ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett 579: 6796–6802 [DOI] [PubMed] [Google Scholar]
- 41. Wang Q, Huang S, Yang L, Zhao L, Yin Y, Liu Z, Chen Z, Zhang H (2008) Down‐regulation of Sonic hedgehog signaling pathway activity is involved in 5‐fluorouracil‐induced apoptosis and motility inhibition in Hep3B cells. Acta Biochim Biophys Sin (Shanghai) 40: 819–829 [PubMed] [Google Scholar]
- 42. Li S, Ou XH, Wei L, Wang ZB, Zhang QH, Ouyang YC, Hou Y, Schatten H, Sun QY (2012) Septin 7 is required for orderly meiosis in mouse oocytes. Cell Cycle 11: 3211–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 5


