Abstract
Physikalisch-Technische Bundesanstalt (PTB) database is electrocardiograms (ECGs) set from healthy volunteers and patients with different heart diseases. PTB is provided for research and teaching purposes by National Metrology Institute of Germany. The analysis method of complex QRS in ECG signals for diagnosis of heart disease is extremely important. In this article, a method on Shannon energy (SE) in order to detect QRS complex in 12 leads of ECG signal is provided. At first, this algorithm computes the Shannon energy (SE) and then makes an envelope of Shannon energy (SE) by using the defined threshold. Then, the signal peaks are determined. The efficiency of the algorithm is tested on 70 cases. Of all 12 standard leads, ECG signals include 840 leads of the PTB Diagnostic ECG Database (PTBDB). The algorithm shows that the Shannon energy (SE) sensitivity is equal to 99.924%, the detection error rate (DER) is equal to 0.155%, Positive Predictivity (+P) is equal to 99.922%, and Classification Accuracy (Acc) is equal to 99.846%.
1. Introduction
In recent years, cardiovascular disorders have been one of the major diseases threatening human life. Therefore, the detection of heart signal waves such as QRS complex is highly significant [1]. Electrocardiogram is used to detect most of heart disorders and shows the electrical activities of heart as a signal [2]. ECG signals contain a lot of information concerning heart diseases. The detection of special points and different parameters such as QRS complex are one of the basic topics and are of high importance, because they lead to the diagnosis of heart diseases. The QRS are used to diagnose many cardiac diseases and noncardiac pathologies such as autonomic malfunction vascular, respiratory (RR) assessment in cardiomyopathy and the normal ventricular myocardium, estimate the heart rate and heart rate variability analysis, and detect ST segment [3–5]. Heart problems usually involve leaking valves and blocked coronary arteries. This research is motivated by reasons expressed. Heart rate cycle consists of a P-wave, a QRS complex, T-wave, and sometimes U-wave [5]. Figure 1 shows schematic representation of normal ECG.
Figure 1.
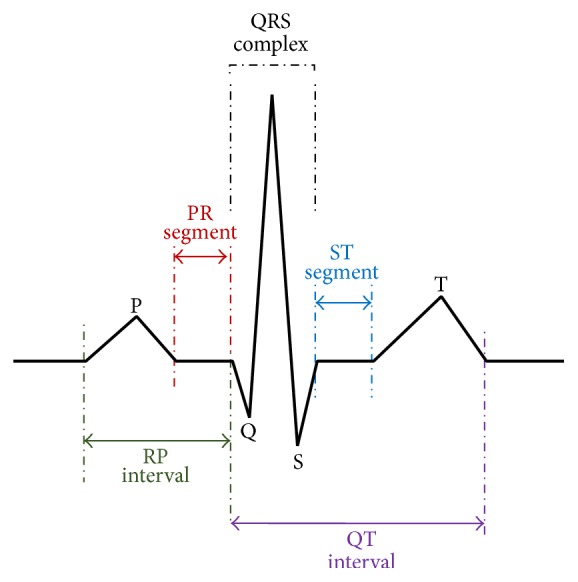
Schematic diagram of normal ECG.
Detecting any of heart signal waves may be difficult due to variable physiology, arrhythmia, disease, and noise. Therefore, in methods such as artificial neural networks and supportive vector machines, detection by the wave R is not always successful and true detection cannot be reached in different signals [6, 7].
The shape of the waves T, P, and QRS is well known; however, the time and frequency of these waves depend on the physiological and physical conditions. In addition, the signal may face polluted recordings with noises such as transmission lines [3].
In recent decades, various methods have been presented to improve the detection of heart signal waves, including Pan-Tompkins algorithm [7], Wavelet Transform, by usage of a constant scale in signal analysis, not considering the characteristics of the signal [8, 9], and artificial neural networks, containing of a series of interconnected simple processing units that each connection has a weight. Input layer, one or multiple hidden layers, and output layer constitute a neural network [10, 11]. Adaptive filter [12], called Hilbert-Huang Transform (HHT), is a new technique for extracting features that are nonlinear and nonstationary signals. This technique has a leakage in practical tasks [13]. Filter bank [14], a Hidden Markov Model (HMM), describes the process where direct observation is not possible, when sequence of symbols can observe HMM. It is used in many fields such as classification of heartbeat and apnea bradycardia detection in preterm infants [15]. Hermite Transform (HT) was recently used instead of Fourier Transform. HT shows better performance, when optimization is done properly [16]. Threshold method [17], Shannon energy envelope (SEE), is the average spectrum of energy and is better able to detect peaks in case of various QRS polarities and sudden changes in QRS amplitude. SEE detects R-peak with a better estimate [18]. S-Transform and Shannon energy (SSE) create a frequency-dependent regulation which is directly related with the Fourier spectrum. S-Transform includes short time Fourier Transform (STFT) and the Wavelet Transform (WT). SSE gives a smooth cover for P-waves and T-waves and completely decreases their influence [19]. Methods such as pattern matching are based on their comparing and contrasting. The calculations are complex and need manual classification [6].
In this paper, an algorithm based on Shannon energy has been proposed to improve the QRS complex detection and simplify the detection process. First, a band-pass filter is used for eliminating noise. Second, Shannon energy of ECG signal is calculated. Third, include moving averages and a differential for the envelope of step 2. Finally, with defining a threshold, peaks are detected. The proposed algorithm is tested on 115-second (to end) ECG signal of PTB Diagnostic ECG Database (PTBDB) [20, 21] and detection accuracy of 99.846% is obtained. The proposed technique results in good performance without being mathematically complex.
2. Method
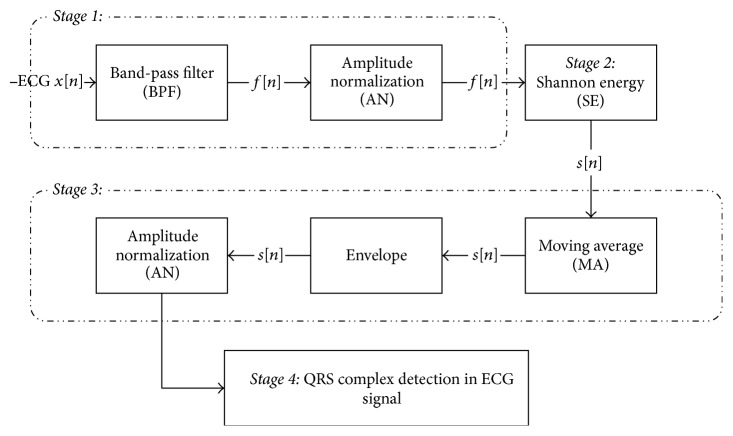
The block diagram of detecting QRS complex algorithm is shown in Figure 2. It includes four stages. Stage 1 includes band-pass digital filter and amplitude normalization. Stage 2 includes calculating Shannon energy of stage 1. In stage 3, with moving average and differencing, make a pack of Shannon energy, and in stage 4, with defining a threshold, QRS complex is detected.
Figure 2.
Block diagram to detect QRS complex.
2.1. Preparations Signal
Digital-analog conversion process is causing all kinds of noise interference and sometimes strongly affects the information. These interactions include frequency interference, muscle contraction, and wandering signals from the baseline or Gaussian white noise [5].
The ECG signal recorded from human beings is a poor signal and is often contaminated by noise. Frequency interference includes a narrow band from 48 to 60 Hz and harmonic interference, and the noise from muscle contraction occurs in 38 to 45 Hz. To eliminate this noise, notch filter is good [22]. Deep breathing, loosely connected electrodes, and sudden changes in voltage lead the baseline signal to be wondered (baseline drift) [5]. Random variable vector (mean) and chromatogram baseline estimation and denoising using sparsity (BEADS) algorithm [23] are good methods to eliminate baseline drift. The band-pass filter decreases efficacy of muscle contraction, frequency interference, baseline drift, and P-wave and T-wave interference [7, 24]. To repress these noises, Butterworth band-pass digital filter with stop-point set at 5 to 16 Hz is used. Butterworth has no ripple in band-pass. [25]. After band-pass filter, the signal is normalized with (1) in stage 1 [26].
| (1) |
where a[n] is a normalized amplitude; f[n] is an after processes band-pass filter (BPF). N denotes the number of samples.
2.2. Shannon Energy and Detection of QRS Complex
The proposed method is based on the use of signal energy. The signal square is very close to the signal energy. For discrete time signal energy is defined as follows:
| (2) |
Here, Ex expresses the signal energy, x(n) defined ECG data, and n is samples. ∑ represents sum from (−∞ ∞) [27]. To explain, we have the following:
| (3) |
Shannon energy calculates the average spectrum of the signal energy. In other words, discount the high components into the low components. So, input amplitude is not important. Shannon energy and Hilbert Transform (SEHT) provide a good accessory for detecting R-peak but this technique has a problem. SEHT needs high memory and has delays [28]. It is designed for solving our actual requirements. To find smooth Shannon energy, zero-phase filter and Shannon energy approximate are playing a basic role [24, 28].
Shannon energy (SE) calculates the energy of the local spectrum for each sample. Below is a calculation of Shannon energy:
| (4) |
where a[n] is after process normalization.
Energy that better approaches detection ranges in presence of noise or domains with more width results in fewer errors. Capacity to emphasize medium is the advantage of using Shannon energy rather than classic energy [18, 19]. The selected signal is normalized with (5) in stage 3 for decreasing the signal base and placing the signal below the baseline.
| (5) |
where μ is the random variable vector and σ defined standard deviation of the signal.
In stage 3, after computing Shannon energy, small spikes around the main peak of the energy are generated. These spikes make main peaks detection difficult. To eliminate this spike, Shannon energy is converted into energy package (Shannon energy envelope (SEE)). To overcome this problem, the Hilbert Transform is used. SEHT method is a simple and high accessory but the SEHT needs high memory and has delays, so it is unfit for real time detection [24, 28]. To smooth out the spikes, rectangular (h) with L length is used. Filtering operation is shown as follows:
| (6) |
where m[n] defines moving average, j is a constant, and S defines Shannon energy from previous steps. For spikes reducing and enveloping, the nonzero peaks obtained from differential get linked. In other words, diagnosed peaks are linked together.
Difference is defined below:
| (7) |
The sign is defined as follows:
| (8) |
where x is a real number.
In stage 4, positive peaks are QRS complex location. To detect QRS complex, a threshold (see (9)) is defined. In fact, samples with greater amplitude than the threshold are selected as output.
| (9) |
where κ is a constant.
3. Result
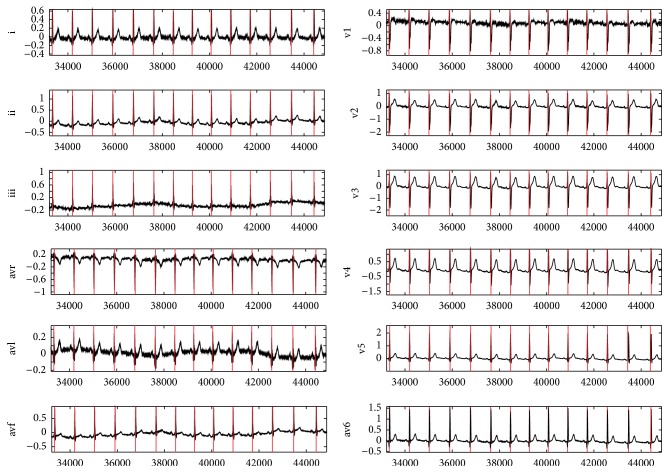
The experimental results are obtained after simulation on 70 healthy patients' signals for all 12 leads and using PTB Diagnostic ECG Database (PTBDB). The Physikalisch-Technische Bundesanstalt (PTB) is the National Metrology Institute of Germany. PTB database is provided for PhysioNet and has different morphologies. The ECGs in this database obtain 15 input channels including the conventional 12 leads (i, ii, iii, avr, avl, avf, v1, v2, v3, v4, v5, and v6) together with the 3 Frank lead ECGs (vx, vy, and vz). Input voltage is ±16 mV, input resistance is 100 Ω, ADC resolution is equal to 16 bits with 0.5 μ/LSB, and sampling frequency is equal to 1 KHz [20, 21]. The proposed algorithm was performed on a 2.4 GHz Intel core i3 CPU using GNU Octave version 4.0.2 [29]. A selected signal from patient 117 has a variety of physiology and baseline drift. Leads (i, ii, avl, avf, v3, v4, v5, and v6) of record s0291lrem and leads (i, ii, iii, avf, v1, v2, v4, v5, and v6) of record s0292lrem have high amplitude. Leads (i, avl, v2, v3, and v4) of record s0291lrem and leads (avr, avl, and avf) of record s0292lrem have a sharp and tall T-wave.
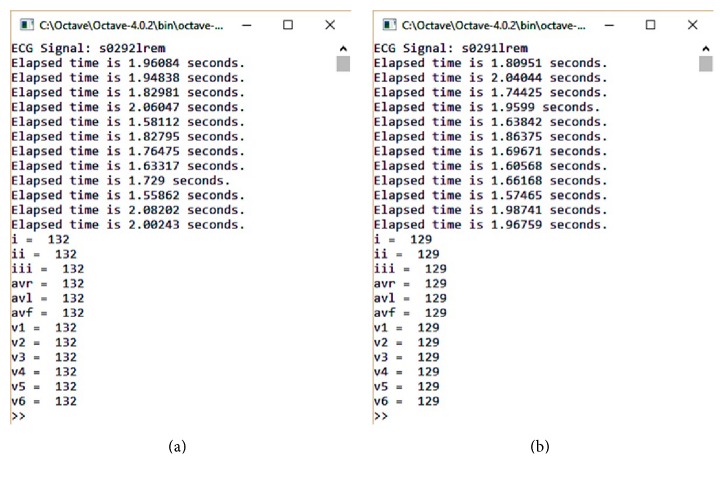
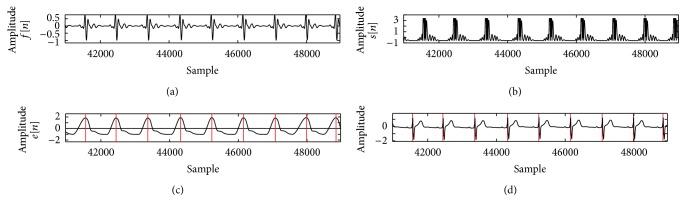
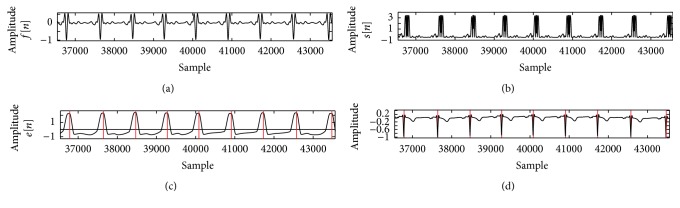
Figure 3 shows the result of simulation to detect each lead of patient 117 in Octave. Figures 4 and 5 show the process of ECG signal provision and peak detection. The QRS detection of the 12 channels of healthy ECG signal in patient 117 of the PTB database is reported in Table 1 and the Appendix. Detection of the 12 leads is shown in Figure 6. Figure 7 shows 3 leads of 3 cases.
Figure 3.
Simulation result. Time and number of peaks detection in each lead are shown. ((a) record s0292lrem; (b) record s0291lrem).
Figure 4.
Process of preparations of ECG signal (record s0291lrem, lead v3).
Figure 5.
Process of preparation of ECG signal (record s0292lrem, lead avr).
Table 1.
The QRS detection of ECG signal of the PTB database.
| Case | TP | FN | FP | DER% | Se% | +P | Acc |
|---|---|---|---|---|---|---|---|
| s0010_rem | 624 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0014lrem | 1987 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0015lrem | 1815 | 0 | 2 | 0.110 | 100.000 | 99.890 | 99.890 |
| s0017lrem | 1673 | 0 | 5 | 0.299 | 100.000 | 99.702 | 99.702 |
| s0020arem | 1906 | 4 | 21 | 1.312 | 99.791 | 98.910 | 98.705 |
| s0020brem | 1867 | 5 | 20 | 1.339 | 99.733 | 98.940 | 98.679 |
| s0021arem | 2207 | 1 | 0 | 0.045 | 99.955 | 100.000 | 99.955 |
| s0021brem | 2196 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0025lrem | 2382 | 6 | 0 | 0.252 | 99.749 | 100.000 | 99.749 |
| s0029lrem | 1638 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0031lrem | 2111 | 1 | 0 | 0.047 | 99.953 | 100.000 | 99.953 |
| s0035_rem | 552 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0036lrem | 2066 | 0 | 2 | 0.097 | 100.000 | 99.903 | 99.903 |
| s0037lrem | 1479 | 0 | 3 | 0.203 | 100.000 | 99.798 | 99.798 |
| s0038lrem | 1572 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0039lrem | 2088 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0042lrem | 1815 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0043lrem | 1212 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0044lrem | 1812 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0045lrem | 1968 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0046lrem | 1944 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0047lrem | 2651 | 1 | 0 | 0.038 | 99.962 | 100.000 | 99.962 |
| s0049lrem | 2040 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0050lrem | 1461 | 3 | 0 | 0.205 | 99.795 | 100.000 | 99.795 |
| s0051lrem | 1912 | 0 | 2 | 0.105 | 100.000 | 99.896 | 99.896 |
| s0052lrem | 1356 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0053lrem | 2148 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0054lrem | 1979 | 31 | 2 | 1.668 | 98.458 | 99.899 | 98.360 |
| s0055lrem | 1381 | 0 | 1 | 0.072 | 100.000 | 99.928 | 99.928 |
| s0056lrem | 1732 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0057lrem | 1896 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0058lrem | 2017 | 0 | 1 | 0.050 | 100.000 | 99.950 | 99.950 |
| s0059lrem | 1800 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0060lrem | 140 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0062lrem | 1488 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0063lrem | 1845 | 3 | 0 | 0.163 | 99.838 | 100.000 | 99.838 |
| s0064lrem | 1797 | 3 | 0 | 0.167 | 99.833 | 100.000 | 99.833 |
| s0065lrem | 1704 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0066lrem | 1513 | 0 | 1 | 0.066 | 100.000 | 99.934 | 99.934 |
| s0067lrem | 424 | 0 | 4 | 0.943 | 100.000 | 99.065 | 99.065 |
| s0068lrem | 1377 | 5 | 15 | 1.452 | 99.638 | 98.922 | 98.568 |
| s0069lrem | 1188 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0070lrem | 1983 | 0 | 1 | 0.050 | 100.000 | 99.950 | 99.950 |
| s0071lrem | 1848 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0072lrem | 2040 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0073lrem | 2125 | 5 | 0 | 0.235 | 99.765 | 100.000 | 99.765 |
| s0074lrem | 1140 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0075lrem | 1453 | 0 | 1 | 0.069 | 100.000 | 99.931 | 99.931 |
| s0076lrem | 1308 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0077lrem | 1692 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0078lrem | 1225 | 0 | 1 | 0.082 | 100.000 | 99.918 | 99.918 |
| s0079lrem | 1620 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0080lrem | 1556 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0082lrem | 1602 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0083lrem | 1465 | 1 | 0 | 0.068 | 99.932 | 100.000 | 99.932 |
| s0084lrem | 1464 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0085lrem | 1276 | 0 | 4 | 0.313 | 100.000 | 99.688 | 99.688 |
| s0097lrem | 2133 | 0 | 1 | 0.047 | 100.000 | 99.953 | 99.953 |
| s0101lrem | 1500 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0103lrem | 1273 | 0 | 2 | 0.157 | 100.000 | 99.843 | 99.843 |
| s0149lrem | 1572 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0152lrem | 1532 | 4 | 0 | 0.261 | 99.740 | 100.000 | 99.740 |
| s0087lrem | 1654 | 12 | 0 | 0.726 | 99.280 | 100.000 | 99.280 |
| s0088lrem | 1728 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0091lrem | 1380 | 1 | 1 | 0.145 | 99.928 | 99.928 | 99.855 |
| s0095lrem | 1797 | 3 | 0 | 0.167 | 99.833 | 100.000 | 99.833 |
| s0096lrem | 2603 | 1 | 0 | 0.038 | 99.962 | 100.000 | 99.962 |
| s0150lrem | 1583 | 1 | 0 | 0.063 | 99.937 | 100.000 | 99.937 |
| s0090lrem | 1358 | 0 | 2 | 0.147 | 100.000 | 99.853 | 99.853 |
| s0093lrem | 1249 | 0 | 1 | 0.080 | 100.000 | 99.920 | 99.920 |
| s0291lrem | 1548 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| s0292lrem | 1584 | 0 | 0 | 0.000 | 100.000 | 100.000 | 100.000 |
| Total | 119054 | 91 | 93 | 0.155 | 99.924 | 99.922 | 99.846 |
Figure 6.
Detected QRS complex of ECG data (record s0292lrem); red line defines QRS complex detection. y-axis represents the amplitude, and x-axis represents the sample.
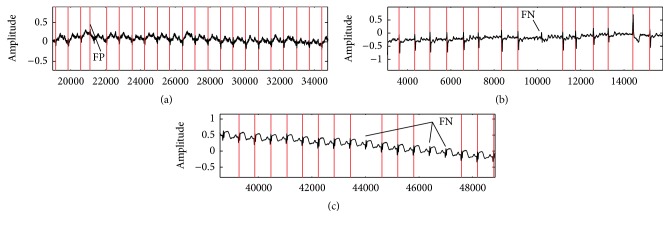
Figure 7.
(a) Detected QRS complex of ECG data (record s0020arem, lead avf). Records s0020arem and s0020brem include tall and sharp P-wave and T-wave. In this case, the QRS area has low energy. (b) Detected QRS complex of s0087lrem-lead 3. This case includes Irregular RR interval. (c) Lead v5 of s0089lrem. FN (false negative) is the number of not detected R peaks, and FP (false positive) is the number of noise spikes detected as R peaks. y-axis represents the amplitude, and x-axis represents the sample.
In order to define performance and efficiency of the algorithm, the Classification Accuracy (Acc), Positive Predictivity (+P), sensitivity (Se), and detection error rate were calculated by using the following equations:
| (10) |
Here, TP defines a true detected peak by the algorithm; FN (false negative) is the number of not detected R peaks, and FP (false positive) is the number of noise spikes detected as R peaks [3, 30].
Figures 4(a) and 5(a) show the output after the band-pass filter f[n] and normalized amplitude a[n]. Figures 4(b) and 5(b) show Shannon energy s[n] and normalized amplitude, and Figures 4(c) and 5(c) show after envelope e[n] signal. QRS complex of ECG signal is shown in Figures 4(d) and 5(d). Red line defines a detected peak. y-axis represents the amplitude, and x-axis represents the sample.
In this study, the proposed technique is tested on 840 leads of PTB Diagnostic ECG Database (PTBDB), and values achieved showed that sensitivity (Se) equals 99.924%, detection error rate (DER) equals 0.155%, Positive Predictivity (+P) equals 99.922%, and Classification Accuracy was 99.846%.
4. Conclusion
In the present study, the most common methods to remove noise in the ECG signal are evaluated. A Shannon energy-based approach to determine the QRS complex of the 12-lead ECG signal is provided. ECG signal is selected with a variety of physiology from the PTB Database and examined by Octave software. Accuracy and sensitivity achieved from Table 1 showed that the presented algorithm is fast and simple, without complex equations. This algorithm does not need a high memory and high hardware. Diagnosis time for each lead is approximately 2.5 seconds based on Octave. The results showed that algorithm detection has very little lag, less than 0.013 seconds, without error. This lag is generated from stage 3.
Appendix
See Table 2.
Table 2.
The QRS detection of the 12 channels of the PTB database.
(a).
| Leads | s0010_rem | s0014lrem | s0015lrem | s0017lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 52 | 0 | 0 | 166 | 0 | 0 | 151 | 0 | 0 | 139 | 0 | 0 |
| ii | 52 | 0 | 0 | 165 | 0 | 0 | 151 | 0 | 0 | 142 | 0 | 3 |
| iii | 52 | 0 | 0 | 165 | 0 | 0 | 152 | 0 | 1 | 139 | 0 | 0 |
| avr | 52 | 0 | 0 | 166 | 0 | 0 | 152 | 0 | 0 | 139 | 0 | 0 |
| avl | 52 | 0 | 0 | 166 | 0 | 0 | 152 | 0 | 1 | 139 | 0 | 0 |
| avf | 52 | 0 | 0 | 165 | 0 | 0 | 151 | 0 | 0 | 140 | 0 | 1 |
| v1 | 52 | 0 | 0 | 165 | 0 | 0 | 151 | 0 | 0 | 140 | 0 | 1 |
| v2 | 52 | 0 | 0 | 166 | 0 | 0 | 151 | 0 | 0 | 139 | 0 | 0 |
| v3 | 52 | 0 | 0 | 166 | 0 | 0 | 151 | 0 | 0 | 139 | 0 | 0 |
| v4 | 52 | 0 | 0 | 166 | 0 | 0 | 151 | 0 | 0 | 139 | 0 | 0 |
| v5 | 52 | 0 | 0 | 166 | 0 | 0 | 151 | 0 | 0 | 139 | 0 | 0 |
| v6 | 52 | 0 | 0 | 165 | 0 | 0 | 151 | 0 | 0 | 139 | 0 | 0 |
| Total | 624 | 0 | 0 | 1987 | 0 | 0 | 1815 | 0 | 2 | 1673 | 0 | 5 |
(b).
| Leads | s0020arem | s0020brem | s0021arem | s0021brem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| ii | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| iii | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| avr | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| avl | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| avf | 158 | 3 | 21 | 151 | 5 | 20 | 184 | 0 | 0 | 183 | 0 | 0 |
| v1 | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| v2 | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| v3 | 159 | 0 | 0 | 156 | 0 | 0 | 183 | 1 | 0 | 183 | 0 | 0 |
| v4 | 158 | 1 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| v5 | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| v6 | 159 | 0 | 0 | 156 | 0 | 0 | 184 | 0 | 0 | 183 | 0 | 0 |
| Total | 1906 | 4 | 21 | 1867 | 5 | 20 | 2207 | 1 | 0 | 2196 | 0 | 0 |
(c).
| Leads | s0025lrem | s0029lrem | s0031lrem | s0035_rem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 199 | 0 | 0 | 136 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| ii | 199 | 0 | 0 | 137 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| iii | 197 | 2 | 0 | 137 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| avr | 197 | 2 | 0 | 137 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| avl | 197 | 2 | 0 | 136 | 0 | 0 | 175 | 1 | 0 | 46 | 0 | 0 |
| avf | 199 | 0 | 0 | 137 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| v1 | 199 | 0 | 0 | 136 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| v2 | 199 | 0 | 0 | 136 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| v3 | 199 | 0 | 0 | 137 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| v4 | 199 | 0 | 0 | 136 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| v5 | 199 | 0 | 0 | 137 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| v6 | 199 | 0 | 0 | 136 | 0 | 0 | 176 | 0 | 0 | 46 | 0 | 0 |
| Total | 2382 | 6 | 0 | 1638 | 0 | 0 | 2111 | 1 | 0 | 552 | 0 | 0 |
(d).
| Leads | s0036lrem | s0037lrem | s0038lrem | s0039lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 173 | 0 | 1 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| ii | 172 | 0 | 0 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| iii | 172 | 0 | 0 | 124 | 0 | 1 | 131 | 0 | 0 | 174 | 0 | 0 |
| avr | 173 | 0 | 1 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| avl | 172 | 0 | 0 | 124 | 0 | 1 | 131 | 0 | 0 | 174 | 0 | 0 |
| avf | 172 | 0 | 0 | 124 | 0 | 1 | 131 | 0 | 0 | 174 | 0 | 0 |
| v1 | 172 | 0 | 0 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| v2 | 172 | 0 | 0 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| v3 | 172 | 0 | 0 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| v4 | 172 | 0 | 0 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| v5 | 172 | 0 | 0 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| v6 | 172 | 0 | 0 | 123 | 0 | 0 | 131 | 0 | 0 | 174 | 0 | 0 |
| Total | 2066 | 0 | 2 | 1479 | 0 | 3 | 1572 | 0 | 0 | 2088 | 0 | 0 |
(e).
| Leads | s0042lrem | s0043lrem | s0044lrem | s0045lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 152 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| ii | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| iii | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| avr | 152 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| avl | 152 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| avf | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| v1 | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| v2 | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| v3 | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| v4 | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| v5 | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| v6 | 151 | 0 | 0 | 101 | 0 | 0 | 151 | 0 | 0 | 164 | 0 | 0 |
| Total | 1815 | 0 | 0 | 1212 | 0 | 0 | 1812 | 0 | 0 | 1968 | 0 | 0 |
(f).
| Leads | s0046lrem | s0047lrem | s0049lrem | s0050lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 121 | 1 | 0 |
| ii | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 121 | 1 | 0 |
| iii | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| avr | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| avl | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| avf | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| v1 | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| v2 | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| v3 | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| v4 | 162 | 0 | 0 | 220 | 1 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| v5 | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 122 | 0 | 0 |
| v6 | 162 | 0 | 0 | 221 | 0 | 0 | 170 | 0 | 0 | 121 | 1 | 0 |
| Total | 1944 | 0 | 0 | 2651 | 1 | 0 | 2040 | 0 | 0 | 1461 | 3 | 0 |
(g).
| Leads | s0051lrem | s0052lrem | s0053lrem | s0054lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 166 | 2 | 0 |
| ii | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 154 | 10 | 2 |
| iii | 160 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 165 | 2 | 0 |
| avr | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 167 | 1 | 0 |
| avl | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 167 | 1 | 0 |
| avf | 161 | 0 | 2 | 113 | 0 | 0 | 179 | 0 | 0 | 164 | 4 | 0 |
| v1 | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 162 | 5 | 0 |
| v2 | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 164 | 4 | 0 |
| v3 | 160 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 166 | 2 | 0 |
| v4 | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 168 | 0 | 0 |
| v5 | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 168 | 0 | 0 |
| v6 | 159 | 0 | 0 | 113 | 0 | 0 | 179 | 0 | 0 | 168 | 0 | 0 |
| Total | 1912 | 0 | 2 | 1356 | 0 | 0 | 2148 | 0 | 0 | 1979 | 31 | 2 |
(h).
| Leads | s0055lrem | s0056lrem | s0057lrem | s0058lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 115 | 0 | 0 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| ii | 116 | 0 | 1 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| iii | 115 | 0 | 0 | 145 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| avr | 115 | 0 | 0 | 145 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| avl | 115 | 0 | 0 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| avf | 115 | 0 | 0 | 145 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| v1 | 115 | 0 | 0 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| v2 | 115 | 0 | 0 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| v3 | 115 | 0 | 0 | 145 | 0 | 0 | 158 | 0 | 0 | 169 | 0 | 1 |
| v4 | 115 | 0 | 0 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| v5 | 115 | 0 | 0 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| v6 | 115 | 0 | 0 | 144 | 0 | 0 | 158 | 0 | 0 | 168 | 0 | 0 |
| Total | 1381 | 0 | 1 | 1732 | 0 | 0 | 1896 | 0 | 0 | 2017 | 0 | 1 |
(i).
| Leads | s0059lrem | s0060lrem | s0090lrem | s0062lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| ii | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| iii | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| avr | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| avl | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| avf | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| v1 | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| v2 | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| v3 | 150 | 0 | 0 | 140 | 0 | 0 | 114 | 0 | 1 | 124 | 0 | 0 |
| v4 | 150 | 0 | 0 | 140 | 0 | 0 | 114 | 0 | 1 | 124 | 0 | 0 |
| v5 | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| v6 | 150 | 0 | 0 | 140 | 0 | 0 | 113 | 0 | 0 | 124 | 0 | 0 |
| Total | 1800 | 0 | 0 | 1680 | 0 | 0 | 1358 | 0 | 2 | 1488 | 0 | 0 |
(j).
| Leads | s0063lrem | s0064lrem | s0065lrem | s0066lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 154 | 0 | 0 | 149 | 1 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| ii | 151 | 3 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 127 | 0 | 1 |
| iii | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| avr | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| avl | 154 | 0 | 0 | 149 | 1 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| avf | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| v1 | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| v2 | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| v3 | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| v4 | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| v5 | 154 | 0 | 0 | 150 | 0 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| v6 | 154 | 0 | 0 | 149 | 1 | 0 | 142 | 0 | 0 | 126 | 0 | 0 |
| Total | 1845 | 3 | 0 | 1797 | 3 | 0 | 1704 | 0 | 0 | 1513 | 0 | 1 |
(k).
| Leads | s0067lrem | s0068lrem | s0069lrem | s0070lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 35 | 0 | 0 | 115 | 0 | 0 | 99 | 0 | 0 | 165 | 0 | 0 |
| ii | 35 | 0 | 0 | 115 | 0 | 0 | 99 | 0 | 0 | 165 | 0 | 0 |
| iii | 36 | 0 | 1 | 113 | 2 | 11 | 99 | 0 | 0 | 165 | 0 | 0 |
| avr | 35 | 0 | 0 | 115 | 0 | 0 | 99 | 0 | 0 | 165 | 0 | 0 |
| avl | 36 | 0 | 1 | 116 | 0 | 2 | 99 | 0 | 0 | 165 | 0 | 0 |
| avf | 36 | 0 | 1 | 115 | 0 | 0 | 99 | 0 | 0 | 165 | 0 | 0 |
| v1 | 35 | 0 | 0 | 116 | 0 | 2 | 99 | 0 | 0 | 165 | 0 | 0 |
| v2 | 35 | 0 | 0 | 114 | 1 | 0 | 99 | 0 | 0 | 167 | 0 | 1 |
| v3 | 36 | 0 | 1 | 115 | 0 | 0 | 99 | 0 | 0 | 166 | 0 | 0 |
| v4 | 35 | 0 | 0 | 114 | 1 | 0 | 99 | 0 | 0 | 165 | 0 | 0 |
| v5 | 35 | 0 | 0 | 114 | 1 | 0 | 99 | 0 | 0 | 165 | 0 | 0 |
| v6 | 35 | 0 | 0 | 115 | 0 | 0 | 99 | 0 | 0 | 165 | 0 | 0 |
| Total | 424 | 0 | 4 | 1377 | 5 | 15 | 1188 | 0 | 0 | 1983 | 0 | 1 |
(l).
| Leads | s0071lrem | s0072lrem | s0073lrem | s0074lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 154 | 0 | 0 | 170 | 0 | 0 | 177 | 0 | 0 | 95 | 0 | 0 |
| ii | 154 | 0 | 0 | 170 | 0 | 0 | 177 | 0 | 0 | 95 | 0 | 0 |
| iii | 154 | 0 | 0 | 170 | 0 | 0 | 173 | 5 | 0 | 95 | 0 | 0 |
| avr | 154 | 0 | 0 | 170 | 0 | 0 | 178 | 0 | 0 | 95 | 0 | 0 |
| avl | 154 | 0 | 0 | 170 | 0 | 0 | 177 | 0 | 0 | 95 | 0 | 0 |
| avf | 154 | 0 | 0 | 170 | 0 | 0 | 177 | 0 | 0 | 95 | 0 | 0 |
| v1 | 154 | 0 | 0 | 170 | 0 | 0 | 178 | 0 | 0 | 95 | 0 | 0 |
| v2 | 154 | 0 | 0 | 170 | 0 | 0 | 178 | 0 | 0 | 95 | 0 | 0 |
| v3 | 154 | 0 | 0 | 170 | 0 | 0 | 178 | 0 | 0 | 95 | 0 | 0 |
| v4 | 154 | 0 | 0 | 170 | 0 | 0 | 177 | 0 | 0 | 95 | 0 | 0 |
| v5 | 154 | 0 | 0 | 170 | 0 | 0 | 177 | 0 | 0 | 95 | 0 | 0 |
| v6 | 154 | 0 | 0 | 170 | 0 | 0 | 178 | 0 | 0 | 95 | 0 | 0 |
| Total | 1848 | 0 | 0 | 2040 | 0 | 0 | 2125 | 5 | 0 | 1140 | 0 | 0 |
(m).
| Leads | s0075lrem | s0076lrem | s0077lrem | s0078lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| ii | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 103 | 0 | 1 |
| iii | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| avr | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| avl | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| avf | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| v1 | 122 | 0 | 1 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| v2 | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| v3 | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| v4 | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| v5 | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| v6 | 121 | 0 | 0 | 109 | 0 | 0 | 141 | 0 | 0 | 102 | 0 | 0 |
| Total | 1453 | 0 | 1 | 1308 | 0 | 0 | 1692 | 0 | 0 | 1225 | 0 | 1 |
(n).
| Leads | s0079lrem | s0080lrem | s0093lrem | s0082lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 133 | 0 | 0 |
| ii | 135 | 0 | 0 | 129 | 0 | 0 | 104 | 0 | 0 | 133 | 0 | 0 |
| iii | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 134 | 0 | 0 |
| avr | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 134 | 0 | 0 |
| avl | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 133 | 0 | 0 |
| avf | 135 | 0 | 0 | 129 | 0 | 0 | 105 | 0 | 1 | 133 | 0 | 0 |
| v1 | 135 | 0 | 0 | 129 | 0 | 0 | 104 | 0 | 0 | 134 | 0 | 0 |
| v2 | 135 | 0 | 0 | 129 | 0 | 0 | 104 | 0 | 0 | 133 | 0 | 0 |
| v3 | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 134 | 0 | 0 |
| v4 | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 134 | 0 | 0 |
| v5 | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 134 | 0 | 0 |
| v6 | 135 | 0 | 0 | 130 | 0 | 0 | 104 | 0 | 0 | 133 | 0 | 0 |
| Total | 1620 | 0 | 0 | 1556 | 0 | 0 | 1249 | 0 | 1 | 1602 | 0 | 0 |
(o).
| Leads | s0083lrem | s0084lrem | s0085lrem | s0097lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 178 | 0 | 1 |
| ii | 123 | 1 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 177 | 0 | 0 |
| iii | 122 | 0 | 0 | 122 | 0 | 0 | 108 | 0 | 2 | 178 | 0 | 0 |
| avr | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 178 | 0 | 0 |
| avl | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 178 | 0 | 0 |
| avf | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 178 | 0 | 0 |
| v1 | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 177 | 0 | 0 |
| v2 | 122 | 0 | 0 | 122 | 0 | 0 | 108 | 0 | 2 | 178 | 0 | 0 |
| v3 | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 178 | 0 | 0 |
| v4 | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 178 | 0 | 0 |
| v5 | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 178 | 0 | 0 |
| v6 | 122 | 0 | 0 | 122 | 0 | 0 | 106 | 0 | 0 | 177 | 0 | 0 |
| Total | 1465 | 1 | 0 | 1464 | 0 | 0 | 1276 | 0 | 4 | 2133 | 0 | 1 |
(p).
| Leads | s0101lrem | s0103lrem | s0149lrem | s0152lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 125 | 0 | 0 | 106 | 0 | 1 | 131 | 0 | 0 | 128 | 0 | 0 |
| ii | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| iii | 125 | 0 | 0 | 107 | 0 | 1 | 131 | 0 | 0 | 128 | 0 | 0 |
| avr | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| avl | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| avf | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 124 | 4 | 0 |
| v1 | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| v2 | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| v3 | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| v4 | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| v5 | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| v6 | 125 | 0 | 0 | 106 | 0 | 0 | 131 | 0 | 0 | 128 | 0 | 0 |
| Total | 1500 | 0 | 0 | 1273 | 0 | 2 | 1572 | 0 | 0 | 1532 | 4 | 0 |
(q).
| Leads | s0087lrem | s0088lrem | s0089lrem | s0091lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 142 | 0 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 114 | 1 | 0 |
| ii | 138 | 4 | 0 | 144 | 0 | 0 | 194 | 2 | 0 | 115 | 0 | 0 |
| iii | 136 | 6 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| avr | 138 | 0 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| avl | 138 | 0 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| avf | 137 | 1 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 116 | 0 | 1 |
| v1 | 136 | 0 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| v2 | 138 | 0 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| v3 | 137 | 1 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| v4 | 138 | 0 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| v5 | 138 | 0 | 0 | 144 | 0 | 0 | 156 | 40 | 0 | 115 | 0 | 0 |
| v6 | 138 | 0 | 0 | 144 | 0 | 0 | 196 | 0 | 0 | 115 | 0 | 0 |
| Total | 1654 | 12 | 0 | 1728 | 0 | 0 | 2310 | 42 | 0 | 1380 | 1 | 1 |
(r).
| Leads | s0095lrem | s0096lrem | s0150lrem | s0150lrem | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | |
| i | 149 | 1 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| ii | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| iii | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| avr | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| avl | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| avf | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| v1 | 149 | 1 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| v2 | 149 | 1 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 131 | 1 | 0 |
| v3 | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| v4 | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| v5 | 150 | 0 | 0 | 216 | 1 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| v6 | 150 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 132 | 0 | 0 |
| Total | 1797 | 3 | 0 | 2603 | 1 | 0 | 0 | 0 | 0 | 1583 | 1 | 0 |
(s).
| Leads | s0291lrem | s0292lrem | ||||
|---|---|---|---|---|---|---|
| TP | FN | FP | TP | FN | FP | |
| i | 129 | 0 | 0 | 132 | 0 | 0 |
| ii | 129 | 0 | 0 | 132 | 0 | 0 |
| iii | 129 | 0 | 0 | 132 | 0 | 0 |
| avr | 129 | 0 | 0 | 132 | 0 | 0 |
| avl | 129 | 0 | 0 | 132 | 0 | 0 |
| avf | 129 | 0 | 0 | 132 | 0 | 0 |
| v1 | 129 | 0 | 0 | 132 | 0 | 0 |
| v2 | 129 | 0 | 0 | 132 | 0 | 0 |
| v3 | 129 | 0 | 0 | 132 | 0 | 0 |
| v4 | 129 | 0 | 0 | 132 | 0 | 0 |
| v5 | 129 | 0 | 0 | 132 | 0 | 0 |
| v6 | 129 | 0 | 0 | 132 | 0 | 0 |
| Total | 1548 | 0 | 0 | 1584 | 0 | 0 |
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Wang K., Ma S., Feng J., Zhang W., Fan M., Zhao D. Design of ECG signal acquisition system based on DSP. Proceedings of the International Workshop on Information and Electronics Engineering (IWIEE '12); March 2012; Harbin, China. pp. 3763–3767. [DOI] [Google Scholar]
- 2.Yeh Y.-C., Wang W.-J. QRS complexes detection for ECG signal: the difference operation method. Computer Methods and Programs in Biomedicine. 2008;91(3):245–254. doi: 10.1016/j.cmpb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Yochum M., Renaud C., Jacquir S. Automatic detection of P, QRS and T patterns in 12 leads ECG signal based on CWT. Biomedical Signal Processing and Control. 2016;25:46–53. doi: 10.1016/j.bspc.2015.10.011. [DOI] [Google Scholar]
- 4.Jin F., Sugavaneswaran L., Krishnan S., Chauhan V. S. Quantification of fragmented QRS complex using intrinsic time-scale decomposition. Biomedical Signal Processing and Control. 2017;31:513–523. doi: 10.1016/j.bspc.2016.09.015. [DOI] [Google Scholar]
- 5.Zhang H. An improved QRS wave group detection algorithm and matlab implementation. Physics Procedia. 2012;25:1010–1016. doi: 10.1016/j.phpro.2012.03.192. [DOI] [Google Scholar]
- 6.Sadhukhan D., Mitra M. R-peak detection algorithm for Ecg using double difference and RR interval processing. Procedia Technology. 2012;4:873–877. doi: 10.1016/j.protcy.2012.05.143. [DOI] [Google Scholar]
- 7.Pan J., Tompkins W. J. A real-time QRS detection algorithm. IEEE Transactions on Biomedical Engineering. 1985;32(3):230–236. doi: 10.1109/tbme.1985.325532. [DOI] [PubMed] [Google Scholar]
- 8.Zou Y., Han J., Xuan S., et al. An energy-efficient design for ECG recording and R-peak detection based on wavelet transform. IEEE Transactions on Circuits and Systems II: Express Briefs. 2015;62(2):119–123. doi: 10.1109/TCSII.2014.2368619. [DOI] [Google Scholar]
- 9.Yan J., Lu L. Improved Hilbert-Huang transform based weak signal detection methodology and its application on incipient fault diagnosis and ECG signal analysis. Signal Processing. 2014;98:74–87. doi: 10.1016/j.sigpro.2013.11.012. [DOI] [Google Scholar]
- 10.Ribas Ripoll V. J., Wojdel A., Romero E., Ramos P., Brugada J. ECG assessment based on neural networks with pretraining. Applied Soft Computing. 2016;49:399–406. doi: 10.1016/j.asoc.2016.08.013. [DOI] [Google Scholar]
- 11.Mateo J., Torres A. M., García M. A., Santos J. L. Noise removal in electroencephalogram signals using an artificial neural network based on the simultaneous perturbation method. Neural Computing and Applications. 2016;27(7):1941–1957. doi: 10.1007/s00521-015-1988-7. [DOI] [Google Scholar]
- 12.Köhler B.-U., Hennig C., Orglmeister R. The principles of software QRS detection. IEEE Engineering in Medicine and Biology Magazine. 2002;21(1):42–57. doi: 10.1109/51.993193. [DOI] [PubMed] [Google Scholar]
- 13.Bo J., Cao X., Wan Y., et al. Investigation performance on electrocardiogram signal processing based on an advanced algorithm combining wavelet packet transform (WPT) and Hilbert-Huang Transform (HHT) Lecture Notes in Electrical Engineering. 2014;269:959–968. doi: 10.1007/978-94-007-7618-0_94. [DOI] [Google Scholar]
- 14.Lagerholm M., Peterson G. Clustering ECG complexes using hermite functions and self-organizing maps. IEEE Transactions on Biomedical Engineering. 2000;47(7):838–848. doi: 10.1109/10.846677. [DOI] [PubMed] [Google Scholar]
- 15.Akhbari M., Shamsollahi M. B., Sayadi O., Armoundas A. A., Jutten C. ECG segmentation and fiducial point extraction using multi hidden Markov model. Computers in Biology and Medicine. 2016;79:21–29. doi: 10.1016/j.compbiomed.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Brajović M., Orović I., Daković M., Stanković S. On the parameterization of Hermite transform with application to the compression of QRS complexes. Signal Processing. 2017;131:113–119. doi: 10.1016/j.sigpro.2016.08.007. [DOI] [Google Scholar]
- 17.Jane R., Blasi A., Garcia J., Laguna P. Computers in Cardiology 1997. Piscataway, NJ, USA: IEEE Computer Society Press; 1997. Evaluation of an automatic threshold based detector of waveform limits in Holter ECG with the QT database; pp. 295–298. [Google Scholar]
- 18.Manikandan M. S., Soman K. P. A novel method for detecting R-peaks in electrocardiogram (ECG) signal. Biomedical Signal Processing and Control. 2012;7(2):118–128. doi: 10.1016/j.bspc.2011.03.004. [DOI] [Google Scholar]
- 19.Zidelmal Z., Amirou A., Ould-Abdeslam D., Moukadem A., Dieterlen A. QRS detection using S-Transform and Shannon energy. Computer Methods and Programs in Biomedicine. 2014;116(1):1–9. doi: 10.1016/j.cmpb.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Goldberger A. L., Amaral L. A., Glass L., et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101(23):E215–E220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 21.PhysioNet. The PTB Diagnostic ECG Database (ptbdb), http://physionet.org/physiobank/database/ptbdb/
- 22.Verma A. R., Singh Y. Adaptive tunable notch filter for ECG signal enhancement. Procedia Computer Science. 57:332–337. doi: 10.1016/j.procs.2015.07.347. 3rd International Conference on Recent Trends in Computing 2015 (ICRTC-2015), 2015. [DOI] [Google Scholar]
- 23.Ning X., Selesnick I. W., Duval L. Chromatogram baseline estimation and denoising using sparsity (BEADS) Chemometrics and Intelligent Laboratory Systems. 2014;139:156–167. doi: 10.1016/j.chemolab.2014.09.014. [DOI] [Google Scholar]
- 24.Zhu H., Dong J. An R-peak detection method based on peaks of Shannon energy envelope. Biomedical Signal Processing and Control. 2013;8(5):466–474. doi: 10.1016/j.bspc.2013.01.001. [DOI] [Google Scholar]
- 25.Schlichthärle D. Digital Filters: Basics and Design. Berlin, Germany: Springer; 2011. Analog filters; pp. 19–83. [Google Scholar]
- 26.Ricardo Ferro B. T., Ramírez Aguilera A., Fernández De La Vara Prieto R. R. Automated detection of the onset and systolic peak in the pulse wave using Hilbert transform. Biomedical Signal Processing and Control. 2015;20, article no. 672:78–84. doi: 10.1016/j.bspc.2015.04.009. [DOI] [Google Scholar]
- 27.Ingle V. K., Proakis J. G. Digital Signal Processing Using MATLAB. Boston, Mass, USA: Brooks/Cole; 2000. [Google Scholar]
- 28.Rakshit M., Panigrahy D., Sahu P. K. An improved method for R-peak detection by using Shannon energy envelope. Sādhanā. Academy Proceedings in Engineering Sciences. 2016;41(5):469–477. [Google Scholar]
- 29.GNU-Octave. https://www.gnu.org/software/octave/
- 30.Merah M., Abdelmalik T. A., Larbi B. H. R-peaks detection based on stationary wavelet transform. Computer Methods and Programs in Biomedicine. 2015;121(3):149–160. doi: 10.1016/j.cmpb.2015.06.003. [DOI] [PubMed] [Google Scholar]