Abstract
Pancreatic ductal adenocarcinoma (PDAC) has generally a poor prognosis, but recent data suggest that there are molecular subtypes differing in clinical outcome. This study examines the association between histopathologic heterogeneity, genetic profile, and survival. Tumor histology from 177 resected PDAC patients with follow-up data was subclassified according to predominant growth pattern, and four key genes were analyzed. PDACs were classified as conventional (51%), combined with a predominant component (41%), variants and special carcinomas (8%). Patients with combined PDACs and a dominant cribriform component survived longer than patients with conventional or other combined PDACs. Genetic alterations in at least two out of four genes were found in 95% of the patients (KRAS 93%, TP53 79%, CDKN2A/p16 75%, SMAD4 37%). Patients with less than four mutations survived significantly longer (p = 0.04) than those with alterations in all four genes. Patients with either wildtype KRAS or CDKN2A/p16 lived significantly longer than those with alterations in these genes (p = 0.018 and p = 0.006, respectively). Our data suggest that the number of altered genes, the mutational status of KRAS and certain morphological subtypes correlate with the outcome of patients with PDAC. Future pathology reporting of PDAC should therefore include the KRAS status and a detailed morphological description.
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive tumor with dismal prognosis. The overall 5-year survival rate is only 6% and after curative surgery less than 25%1, making PDAC one of the most lethal tumors among solid malignancies2. This poor outcome is related to multiple factors, including resistance to chemotherapy and the relatively late stage of diagnosis due to unspecific symptoms and aggressive tumor biology1.
Over the last decade major improvements have been made in understanding the mechanisms of molecular carcinogenesis in PDACs3,4,5,6,7. The first milestone was the discovery of the molecular fingerprint of PDAC that included the common mutations in KRAS, SMAD4, TP53 and CDKN2A/p168. Recent advances in gene sequencing9 by introduction of high-throughput molecular methods allowed to further address the genetic complexity of PDAC10. The first global analysis of 24 advanced PDACs using comprehensive exome sequencing revealed a high mutation rate with an average of 63 mutations per case connected to 12 core signaling pathways10. In 2011, Collisson et al. showed that PDACs and murine PDAC cell lines may be stratified by their transcriptional profiles into subtypes with different clinical outcome and drug response11. The stratification of these subtypes was associated with three patterns that were termed classical, quasi-mesenchymal and exocrine-like. The terminology of this subdivision, however, is hardly understandable regarding its morphological meaning. In other recent studies, it was reported that intact KRAS and the number of mutations in key genes may have prognostic value in PDAC patients12,13,14,15,16. However, it is unclear so far, whether there are genotype-phenotype correlations that are based on the identification of PDACs with special growth patterns or morphological variants with better survival than classical PDACs, beyond the established pathological parameters of staging and grading17.
This study that only involved PDAC patients in whom surgical treatment could be performed, focuses on detailed histological investigation and molecular examination of the mutational status of KRAS, CDKN2A/p16, TP53 and SMAD4 in correlation with survival and accurate morphological subtyping. The presented findings show that, according to the analyzed parameters, prognostic relevant subtypes of PDAC can be identified.
Results
The patients’ clinicopathologic features are summarized in Table 1. Female (45.8%) and male (54.2%) patients were equally represented with a median age at diagnosis of 68 years. Most patients presented with advanced stage of the disease (≥UICC 2b: 73.4%, 130/177). A minority of patients (9.4%) received neoadjuvant treatment (details see Supplementary Table 1). All patients were judged resectable and underwent major pancreatic surgery: pylorus-preserving partial pancreaticoduodenectomy (ppWhipple) 55.4% (98/177), partial pancreaticoduodenectomy (Whipple, classic) 9% (16/177), distal pancreatectomy 18.6% (33/177) and total pancreatectomy 17% (30/177).
Table 1. Clinicopathological features of 177 patients with resected pancreatic adenocarcinomas.
| N, 177 | % | |
|---|---|---|
| Sex | ||
| Male | 96 | 54.2% |
| Female | 81 | 45.8% |
| Age median in years | 68 (31–88) | |
| Tumor characteristics | ||
| pT | ||
| 1 | 4 | 2.3% |
| 2 | 11 | 6.2% |
| 3 | 142 | 80.2% |
| 4 | 20 | 11.3% |
| pN | ||
| 0 | 52 | 29.4% |
| 1 | 125 | 70.6% |
| Grading | ||
| 1 | 13 | 7.3% |
| 2 | 72 | 40.7% |
| 3 | 92 | 52.0% |
| Resection margin | ||
| 0 | 90 | 50.8% |
| 1 | 61 | 34.5% |
| 2 | 2 | 1.1% |
| X | 24 | 13.6% |
Histological features
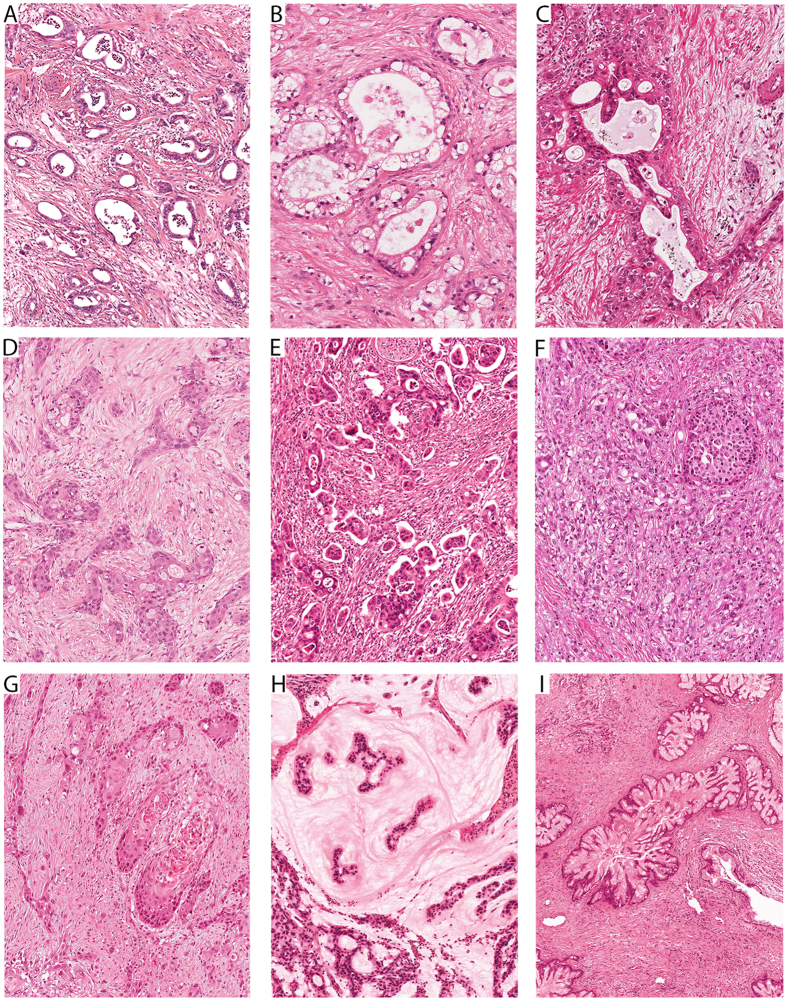
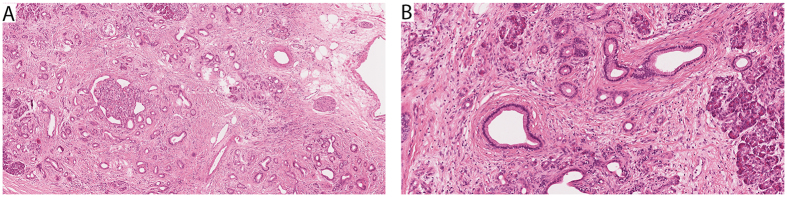
The results are summarized in Table 2 and examples for each pattern are shown in Figs 1 and 2. The majority of the cases (92.1%, 163/177) were classified as either conventional PDACs (91/177, 51%), predominantly graded G2 (44%) or G3 (48%), or combined PDACs (72/177, 41%), predominantly graded G2 (37%) or G3 (61%). Two of the conventional PDACs contained at the periphery concomitant small (<1.5 cm) gastric type IPMNs and four had retention cysts. The combined PDACs showed as dominant histologic feature a conspicuous cribriform (17/177), clear-cell (16/177), papillary (17/177), gyriform (8/177), micropapillary (2/177) or complex (12/177) component (Fig. 1A–F). While the cribriform and clear cell combined PDACs were all G2 tumors, the gyriform, papillary, micropapillary and complex combined PDACs accounted for most (90%) of the G3 tumors. One PDAC with a clear cell component was associated with a pancreato-biliary type IPMN, and another PDAC with a complex component with a gastric type IPMN. A minority of tumors (7.9%, 14/177) fulfilled the criteria of PDAC variants and special pancreatic carcinomas. There were two G3-adenosquamous carcinomas (Fig. 1G), two G1-colloid carcinomas (Fig. 1H), six G2-papillary carcinomas (Fig. 1I), one G2-medullary carcinoma (not shown), and three G1-tubular carcinomas (Fig. 2). One of the colloid carcinomas was associated with an intestinal type IPMN. Two thirds of the papillary carcinomas (4/6) were associated with IPMNs, either of gastric (1/6), intestinal (1/6) or pancreato-biliary type (2/6). The three tubular adenocarcinomas were composed of well-differentiated open tubules that infiltrated the parenchyma diffusely and were accompanied by abundant cellular desmoplastic stroma (Fig. 2). The tubules had oval, rounded or angular shapes and were lined by a single layer of mostly cuboidal cells, with little nuclear pleomorphism, inconspicuous nucleoli and scanty mitotic figures.
Table 2. Histological types of pancreatic ductal adenocarcinoma: frequency and survival.
| Tumor type | Frequency | % | Type of associated IPMN | Median survival (months) |
|---|---|---|---|---|
| Conventional ductal adenocarcinoma | 91 | 51.4 | 2 gastric | 22.7 |
| Combined ductal adenocarcinoma | ||||
| with cribriform component | 17 | 9.6 | 28.7 | |
| with papillary component | 17 | 9.6 | 13.9 | |
| with clear-cell component | 16 | 9.0 | 1 pancreato-biliary | 17.6 |
| with complex component | 12 | 6.7 | 1 gastric | 10.7 |
| with gyriform component | 8 | 4.5 | 12.5 | |
| with micropapillary component | 2 | 1.1 | 16.1 | |
| Variants and special carcinomas | ||||
| Adenosquamous carcinoma* | 2 | 1.1 | 4.1 | |
| Colloidal/mucinous carcinoma* | 2 | 1.1 | 1 intestinal | >64.3** |
| Medullary carcinoma* | 1 | 0.5 | >75.1** | |
| Tubular carcinoma | 3 | 1.7 | >55.3** | |
| Papillary carcinoma | 6 | 3.4 | 2 pancreato-biliary, 1 intestinal, 1 gastric | 20.6 |
| All tumors | 177 | 100 | ||
Figure 1. Spectrum of histologic patterns in pancreatic ductal adenocarcinomas (PDACs).
(A) Conventional PDAC. (B–F) Combined PDACs with a dominant histological component: (B) Clear-cell component. (C) Cribriform component. (D) Gyriform component. (E) Micropapillary component. (F) Complex component. (G) Adenosquamous carcinoma. (H) Colloid carcinoma. (I) Papillary carcinoma.
Figure 2. Tubular adenocarcinoma of the pancreas.
(A) Low power view showing groups of well-differentiated infiltrating tubules surrounded by small cuffs of desmoplastic stroma. (B) Infiltrating well differentiated neoplastic glands closely imitating normal ducts.
No morphological differences were observed between patients who received neoadjuvant therapy and those without neoadjuvant treatment (data not shown).
Molecular features
KRAS, TP53, CDKN2A/p16 and SMAD4 mutations were found in various frequencies and combinations in all but one tumor (case # 62; confirmed by NGS). Most PDACs carried multiple mutations (two: 47/177, 25.6%; three: 85/177, 48%; four: 36/177, 20.3%), while tumors with one mutation (8/177, 4.5%) were rare.
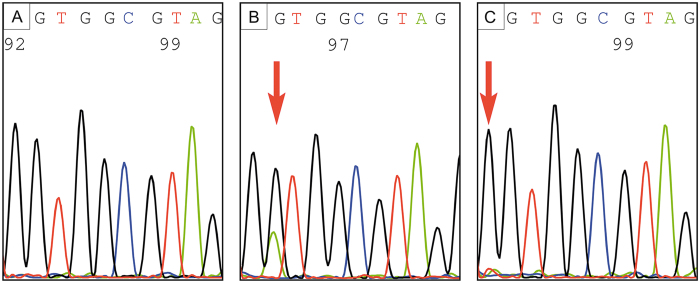
KRAS was most frequently mutated (Table 3, Fig. 3), mainly in exon 2 (91%), and rarely in exon 3 (8.5%) and only once in both exons. No mutations were detected in exon 4. Almost all (98.8%, 162/164) KRAS mutations were detected by HRMA and Sanger sequencing (with expected detection limits of 10% and 15–20%, respectively (29)). Two low-level mutations (mutations with a low frequency in sequence reads) were identified by NGS (see also Fig. 3). All tumors with wildtype KRAS showed intact BRAF (codon 600).
Table 3. Molecular characteristics of pancreatic adenocarcinomas.
| N, 177 | % | ||
|---|---|---|---|
| KRAS | Wildtype | 13 | 7.3 |
| Mutated | 164 | 93 | |
| TP53 | Intact | 38 | 21.5 |
| Altered | 139 | 78.5 | |
| CDKN2A/p16 | Intact | 43 | 24 |
| Altered | 133 | 75 | |
| n.a. | 1 | 0.6 | |
| SMAD4 | Intact | 112 | 63 |
| Altered | 65 | 37 |
Figure 3. KRAS mutation analysis (direct sequencing) of codon 12 of exon 2.
(A) Hotspot of exon 2 shows KRAS wildtype, (B) KRAS mutation (p.G12D, arrow), or (C) low-level mutation (p.G12C, arrow, faint red signal), as confirmed by next generation sequencing (NGS) (case #190).
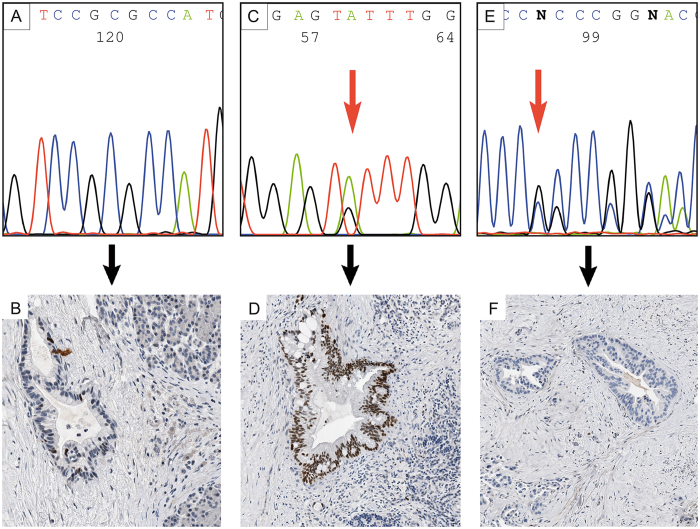
TP53 wildtype and TP53 mutations were associated with distinct p53 immunolabelling patterns depicted in Fig. 4. TP53 Mutations were found in 139/177 tumors and detected in the known hotspots (exon 5–8), but not in exon 9. Mutation type 1 (defined as nuclear expression in ≥25% of tumor cells and missense mutation) was more common (52%, 92/177) than mutation type 2 (26.6%, 47/177; defined as absence of expression and presence of an intragenic deletion, a nonsense, a frameshift or splice site mutation) (Fig. 4).
Figure 4. TP53 analyses.
(A,B) TP53 wildtype is associated with nuclear TP53 expression in up to 24% of neoplastic cells. (C,D) TP53 missense mutation (p.Y205C; exon 6) is associated with nuclear TP53 overexpression in ≥25% of neoplastic cells (mutation type 1). (E,F) TP53 intragenic deletion or nonsense, frameshift or splicesite mutations (mutation type 2; here represented by an insertion-frameshift mutation p.P153*fs28; exon 5) is associated with loss of nuclear TP53 expression in neoplastic cells.
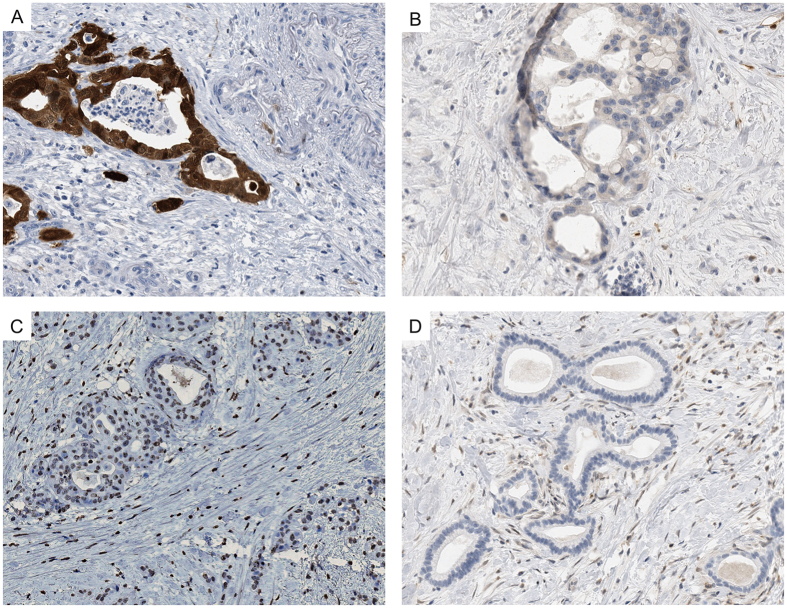
CDKN2A/p16 alterations, recorded in 75% of the tumors, were detected by loss of protein expression (130 tumors), which was confirmed by loss of heterozygosity (LOH) in 58 tumors. Three additional tumors showed intact expression despite presence of LOH. Loss of SMAD4 expression was recorded in one third of the cases (Table 3, Fig. 5).
Figure 5. Immunohistochemical analysis of CDKN2A/p16 and SMAD4.
(A) Strong nuclear and cytoplasmatic staining of CDKN2A/p16 in neoplastic cells indicating the presence of an intact gene. (B) No labeling of CDKN2A/p16 in neoplastic cells indicating either a deletion, inactivating mutation, or promoter hypermethylation. (C) Nuclear SMAD4 immunolabeling of neoplastic cells indicating the presence of an intact protein. (D) Loss of SMAD4 expression in >90% of neoplastic cells indicating a deletion or inactivating mutation of the gene.
Interestingly, altered CDKN2A/p16 was significantly more common in patients with positive lymph nodes (p = 0.02). No significant differences between lymph node negative and positive patients were observed for number of mutations, KRAS, TP53, SMAD4 and tumor morphology.
Four investigated metastases (3 from the liver and one from the peritoneum) had the same KRAS mutations as the primary. In case # 5 a low-level mutation in the primary tumor (p.G12D, 3%) was associated with a high-level mutation (p.G12D) in the liver metastasis. In case # 9 intact TP53 in the primary tumor was associated with a strong (>90%) nuclear labeling in the peritoneal metastasis.
No differences were observed in the mutational status of patients who received neoadjuvant therapy compared to the large group of patients without neoadjuvant therapy (data not shown).
Correlation of molecular with morphologic features
KRAS wildtype was significantly more commonly detected in variants than classical PDACs (conventional or combined) (p = 0.035) (Table 4). In detail, the group of KRAS wildtype tumors (7.3%, 13/177) included conventional PDACs (7/13), combined PDACs (3/13), and one colloid, one medullary and one tubular carcinoma. The KRAS wildtype tubular carcinoma and one KRAS-mutated tubular carcinoma also lacked alterations in the three other genes (Table 5), a result that was confirmed by additional extended gene analysis. One of the two KRAS-mutated tubular carcinomas harbored CDKN2A/p16 mutations (Table 5). All papillary carcinomas were associated with mutated KRAS and lacked mutated GNAS18, irrespective of the presence of an associated IPMN. CDKN2A/p16 alterations were more strongly associated (p = 0.016) with combined PDACs (60/71) than with conventional PDACs (29/91) (Table 4).
Table 4. Comparison between morphological phenotype, genotype, grading and survival.
| Morphological phenotype | Conventional ductal adenocarcinoma (without components) (A1) | Combined ductal adenocarcinoma (with additional components) (A2) | Variants (B) |
|---|---|---|---|
| N | 91 | 72 | 14 |
| KRAS | |||
| Wildtype | 7 | 3 | 3 |
| Mutated | 84 | 69 | 11 |
| p-value A1 vs A21 | 0.35 | ||
| p-value A vs B1 | 0.035 | ||
| CDKN2A/p16 | |||
| Intact | 29 | 11 | 3 |
| Altered | 62 | 60 | 11 |
| p-value A1 vs A21 | 0.016 | ||
| p-value A vs B1 | 0.78 | ||
| TP53 | |||
| Intact | 22 | 11 | 5 |
| Altered | 69 | 61 | 9 |
| p-value A1 vs A21 | 0.16 | ||
| p-value A vs B1 | 0.17 | ||
| SMAD4 | |||
| Intact | 57 | 45 | 10 |
| Altered | 34 | 27 | 4 |
| p-value A1 vs A21 | 0.98 | ||
| p-value A vs B1 | 0.51 | ||
| Number of mutations | |||
| 0 | 0 | 0 | 1 |
| 1 | 5 | 1 | 2 |
| 2 | 30 | 14 | 3 |
| 3 | 40 | 40 | 5 |
| 4 | 16 | 17 | 3 |
| p-value A1 vs A21 | 0.09 | ||
| p-value A vs B1 | 0.004 | ||
| Pathologic stage | |||
| Primary tumour | |||
| T1 | 2 | 2 | 0 |
| T2 | 3 | 8 | 0 |
| T3 | 74 | 55 | 14 |
| T4 | 12 | 7 | 0 |
| p-value A1 vs A21 | 0.23 | ||
| p-value A vs B1 | 0.55 | ||
| Nodal status | |||
| N0 | 24 | 22 | 7 |
| N1 | 67 | 50 | 7 |
| p-value A1 vs A21 | 0.60 | ||
| p-value A vs B1 | 0.12 | ||
| Grading | |||
| G1 | 7 | 1 | 5 |
| G2 | 4 | 27 | 6 |
| G3 | 44 | 44 | 3 |
| p-value A1 vs A21 | 0.08 | ||
| p-value A vs B1 | <0.001 | ||
| Survival | |||
| (months) | 22.7 | 15.5 | 34.1 |
| p-value A1 vs A22 | 0.07 | ||
| p-value A vs B2 | 0.06 | ||
1 Chi2-test/Fisher’s exact test, 2Log-rank-test.
Table 5. Clinicopathologic and molecular features of pancreatic tubular adenocarcinoma.
| ID | Sex | Age | Survival (months) | Site/Size (cm) | Grading | pT | pN | cM | KRAS | TP53 | CDKN2A/p16 | SMAD4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # 44 | F | 82.6 | 19.3 | head/4.2 | G1 | T3 | N1 | M0 | mut (p.G12V) | intact | altered | intact |
| # 62 | M | 74.9 | >68.6, still alive | head/3.8 | G1 | T3 | N0 | M0 | wt | intact | intact | intact |
| # 190 | M | 50.0 | >55.3, still alive | head/3.5 | G1 | T3 | N1 | M0 | mut (p.G12C low-level mutation, 6%) | intact | intact | intact |
Clinical outcome correlated to morphologic and molecular features
At the time of survival analysis, 33 of 146 patients (22.6%) were alive. Median follow-up for all patients was 19.8 months (2.5–75.1 months) and median follow-up for patients alive was 49.4 months (25.9–75.1 months). Overall survival rates were 72% for one year, 29% for three years and 21% for five years.
Patients with colloid, medullary, tubular or papillary carcinoma survived significantly longer than patients with conventional and combined PDACs (p = 0.04) (Table 2). Longest survival was seen in two of the patients with a tubular carcinoma. They were still alive at the completion of the study, with a survival of >68.8 and >55.3 months, respectively. On the contrary, patients with adenosquamous carcinomas had an extremely poor outcome (4.1 and 10.0 months) (Table 2).
Detailed analysis of the large group of patients with conventional and combined PDACs revealed that patients with a conventional PDAC and those with a cribriform type combined PDAC showed the most favorable overall median survival (22.7 and 28.7 months), followed by combined PDACs with clear-cell and papillary components (17.6 and 13.9 months) (Table 2). Combined PDACs with gyriform and complex components were associated with poor survival (12.5 and 10 months).
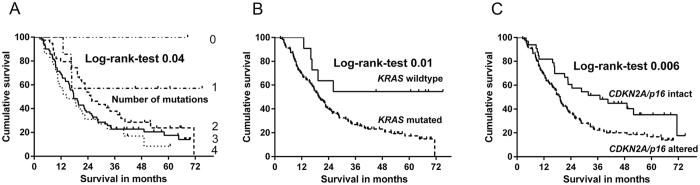
The median patient survival correlated significantly with the number of mutations (p = 0.04, Fig. 6A). Patients with no or low number of mutations (no mutation >68.6; one mutation >45; two mutations >25.3 months) survived longer than patients with tumors that harbored three (17.6 months) or four mutations (14.5 months).
Figure 6. Kaplan-Meier survival curves in pancreatic adenocarcinomas correlated with molecular status.
(A) Significant correlation of median patient survival with number of mutations. (B) Patients with KRAS wildtype have a significantly better overall survival than patients with mutated KRAS. (C) Patients with intact CDKNA2/p16 have a significantly better overall survival than patients with altered CDKNA2/p16.
Correlation of patient outcome and molecular features revealed a significant survival benefit in patients with wildtype KRAS (7.3%, 11/146) compared with that of KRAS mutated patients (92.7%, 135/146) (p = 0.018, median survival >45 vs. 19.7 months, Fig. 6B). Likewise, patients with intact CDKN2A/p16 (22.8%, 33/145) lived significantly longer than those with altered CDKN2A/p16 (77.2%, 112/145) (p = 0.006; 36.9 vs. 18.8 months, Fig. 6C). Significant prognostic results were further obtained by the combination of CDKN2A/p16 and KRAS. All patients from the cohort followed for survival with an intact status of both genes (n = 4) were still alive at the end of the study. A median overall survival could therefore not be calculated. No prognostic significance was observed for mutated TP53 (with no prognostic difference between TP53 type 1 and type 2 mutations) and SMAD4.
Univariate survival analysis using log-rank test showed significant p-values for KRAS, CDKN2A/p16, number of mutations, pN and grading. In a backward selected multivariate Cox proportional Hazard model altered KRAS (HR 2.77, 95%-CI: 1.11–6.90; p = 0.019), pN1 (HR 2.66; 95%-CI: 1.67–4.26; p < 0.001) and grading G3 (HR 3.66, 95%-CI: 1.57–8.54; p = 0.001) were independent predictive variables for survival.
Discussion
Reliable prognostic markers for pancreatic ductal adenocarcinoma (PDAC) patients are so far rare. By analyzing the expression profile of primary PDACs, and human and mouse PDAC cell lines, Collisson recently identified molecular subtypes of PDAC that differed in clinical outcome and drug response11. Other studies revealed an association between expression as well as mutational status of key tumor suppressor and oncogenes and patient survival16,19. In all these studies, the molecular profile of PDACs is not or only vaguely correlated to the individual morphology of the tumors.
Here we present data of a correlative study on histopathology, molecular profile, and survival in PDACs and related carcinomas of 177 resected patients, with the aim to find prognostic relevant features. As expected, the overall outcome of our patient cohort was bad. Nearly two thirds of patients survived less than 24 months and the 5-year survival did not exceed 21%. However, within this cohort there were patients who survived for up to four years and longer, and whose tumors had a special histopathology and/or molecular status.
Histopathological heterogeneity in PDACs has long been recognized, but it has not been defined in detail, -with the exception of the definition of histological grade and histological variants. Here we classified the pancreatic carcinomas according to a defined growth pattern into three groups. The first group included the conventional type PDACs, which showed an equal mixture of various histological elements (for details see Material and Methods). The second group encompassed the combined PDACs, which were characterized by a dominant histological component (defined as involving more than 30% of the tumor area). The third group contained PDAC variants (i.e. adenosquamous carcinoma, colloid carcinoma, papillary carcinoma) and special adenocarcinomas such as medullary carcinoma and tubular carcinoma. By analyzing the survival of patients who were ascribed to the various morphological types, we found and confirmed that the colloid carcinoma, the medullary carcinoma and the tubular carcinoma showed a better outcome than conventional PDACs and particularly adenosquamous carcinoma20,21,22,23. Patient with a conventional PDAC or a PDAC with dominant cribriform component survived longer than patients with combined PDAC and other histological components, such as the clear cell, the papillary, the gyriform and, in particular, the complex component.
PDACs typically harbor KRAS mutations, followed by mutations of CDKN2A/p16, SMAD4 and TP5310. These genes are considered the driver genes of PDAC. In addition, there is a multitude of other, but much less frequent, gene alterations, as revealed by whole genome sequencing analysis6,7. In our patients, in whom the molecular results were obtained by Sanger methodology and completed in selected cases by NGS of a large PDAC gene panel including the 40 most commonly mutated genes, KRAS was found to be mutated in 92.7%, TP53 in 78.5%, CDKN2A/p16 in 72.9% and SMAD4 in 37.3%. Most of these alterations coexisted in individual tumors and two third of the PDAC patients (68%) harbored alterations in three or all four genes.
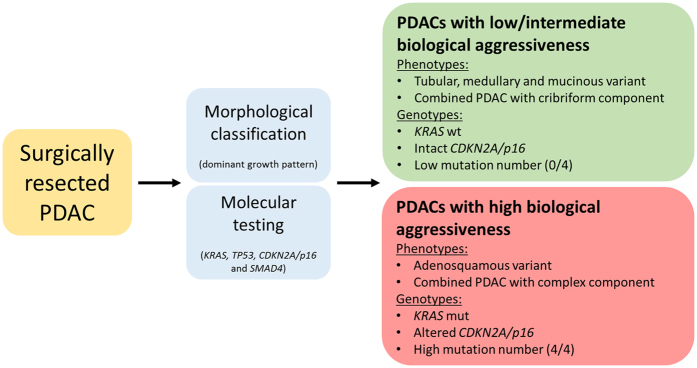
Correlation of the individual mutational status with the respective histopathology and survival data revealed a number of findings with prognostic significance. First, the number of mutations per tumor was of prognostic relevance. Patients with one, two or three mutations survived longer than those with alterations in all four genes (>45 vs. 25.3 vs. 17.6 vs. 14.5 months). Although these results confirm previous reports16, so far the correlation between gene status and phenotype has not been analysed in detail. Among the tumors with low number of mutations and prolonged survival, there are particularly colloid carcinomas, medullary carcinomas and tubular type carcinomas, while adenosquamous carcinomas, papillary carcinomas, and the combined PDAC with a complex pattern belong to the carcinomas with a high mutational frequency and poor survival (Fig. 7). Medullary and colloid carcinomas are known for their low prevalence of somatic mutations and good prognosis20,21,22,24. Conversely, adenosquamous carcinoma is well known for its many somatic mutations and aggressive behavior25,26. The papillary carcinoma variant seems in many cases to be the invasive component of an IPMN27, since IPMNs either of the pancreato-biliary (2/6), intestinal (1/6) or gastric type (1/6) were found in four of our six cases. However, it may also occur without an associated IPMN, since in two cases an associated IPMN was not found, and this was also true in another recently reported series of 10 papillary cystic PDACs28.
Figure 7. Risk stratification after surgical resection of PDAC.
Proposed scheme to identify PDACs with low/intermediate and high biological aggressiveness based on morphological classification and molecular testing of key genes.
Second, patients with either wild type KRAS or CDKN2A/p16 had a better outcome than those with mutations in these genes, while loss of SMAD4 and intact or altered TP53 was of no prognostic relevance. The statistical significance of wildtype KRAS proved to be so strong that multivariate analysis identified the mutational status of KRAS as an independent prognostic marker. This result confirmed the recently reported survival benefit for PDAC patients with intact KRAS12,13,14,15. The largest of these studies by Sinn, included 153 PDAC patients and reported a significantly decreased median survival of 12.7 months for patients with mutated KRAS versus 20.7 months for patients with wild type KRAS15. In our study, the corresponding data were 19.7 months for mutated KRAS versus >45 months with wild type KRAS. The longer survival in our patients may be due to a better selection on the basis of a more extensive KRAS analysis including exon 2 to 4, in contrast to Sinn’s study that only focused on KRAS exon 2. This methodological difference probably also explains, why in Sinn’s study the KRAS mutation rate of 68% is much lower than in our study with a rate of 93%.
Among the tumors with a wildtype KRAS status were a medullary carcinoma, whose particular genetic and biologic features have been described20,21,22,23, and a tubular adenocarcinoma. This latter type of carcinoma, which corresponds by grade to a G1 tumor (WHO 2010), may be considered a conventional PDAC with a strictly well differentiated tubular differentiation. It shows great similarities regarding its morphology, low frequency, rare mutations and long survival to the tubular type carcinoma of the breast29,30,31,32. One of the three tubular adenocarcinomas that we identified, not only had an intact KRAS gene, but also harbored no alterations in the 40 most commonly altered PDAC genes, including CDKN2A/p16, TP53 and SMAD4. Moreover, the patient with this tumor is still alive (see Table 4, case # 62), with a follow-up for >68.6 months. The other two patients with tubular carcinomas had KRAS and/or CDKN2A/p16 alterations, but no loss of SMAD4 or TP53 mutations (details see Table 4), and survived for >55.3 and 19.3 months.
The relevance of mutated KRAS as a prognosticator in PDAC, a feature shared with bile duct cancer33, is biologically most likely related to its driver function. Evidence that KRAS, in interaction with TP53, CDKN2A/p16 and SMAD4, is a driver gene comes from genetically engineered KRAS mouse models of pancreatic cancer10,34,35,36,37,38,39,40,41. Moreover, recently developed mouse models in which mutated KRAS can be switched on and off, have impressively demonstrated that continuous oncogenic KRAS signaling is essential for both progression and maintenance of PDAC42,43 and its metastases44.
Patients with wildtype CDKN2A/p16, like patients with intact KRAS, lived significantly longer than those with altered genes (p = 0.006; 36.9 months vs. 18.8 months). Though multivariate analysis failed to identify this gene constellation as an independent prognostic factor, CDKN2A/p16 seems to play a role in patient outcome. All our patients without alterations in CDKN2A/p16 and KRAS were still alive at the completion of the study. On the other hand, loss of CDKN2A/p16, as was recently reported, seems to be associated with lymphatic invasion and widespread metastasis19 and was significantly associated with lymphatic spread in our study. Interestingly, subgroup analysis revealed a significant higher CDKN2A/p16 mutation rate in combined PDACs with a dominant histological component compared to conventional PDACs. This suggests that a driver gene might be linked to the presence of dominant histological components in PDACs.
The prognostic relevance of SMAD4 has been the subject of controversial discussions over the last years. While several studies associated the loss of SMAD4 with poor prognosis or early metastatic disease14,19,45,46, others were unable to confirm these results16,47,48. Likewise, no differences in the survival data in patients with intact and loss of SMAD4 were observed in our study (p = 0.15, median overall survival 22.2 vs. 17.6 months).
In summary, our findings specify the prognostic relationship between the histopathology and molecular profile, based on the morphologic stratification of PDACs in subtypes and variants, and the mutational status of the four driver genes, KRAS, CDKN2A/p16, SMAD4 and TP53. Because our data suggest that PDAC subgroups can be identified (low to intermediate aggressive PDACs versus highly aggressive PDACs, see Fig. 7), tailored therapy options may be discussed. Patients with altered CDKN2A/p16 might benefit from more aggressive preoperative therapies, whereas patients with wildtype KRAS might best be treated by upfront surgery followed by adjuvant therapies. In view of these potential therapies it should be considered to include the status of the four driver genes into the pathological reporting of PDACs in the future12,13,14,15,49. Given the potential clinical implications of our results, validation in independent PDAC cohorts is of utmost importance.
Material and Methods
The study was approved by the ethics committee of the TU München, Germany (documents no. 1926/2007 and 126/2016S). Written informed consent was obtained from all patients. All methods were performed in accordance with the relevant guidelines and regulations.
Patients
From 07/2007 to 07/2011 200 patients underwent an elective pancreatic resection at the Department of Surgery, Klinikum rechts der Isar, TU München, Germany, with a final histopathologic diagnosis of PDAC. Associated and concomitant intraductal papillary mucinous neoplasms (IPMN)50 were reported. IPMNs with only minimally invasive component were excluded. TNM and grading followed the WHO recommendations17. Patients with a family history of PDAC were excluded from the study.
Clinical data and follow up
Clinical, demographic and macroscopic information was obtained from a patient database and by reviewing the medical charts and pathology reports. Follow up on the patients’ conditions was obtained by clinical record, directly contacting the patients and/or their physicians. Patients with neoadjuvant therapy (n = 14, details see Supplementary Table 1), distant metastasis/UICC stage IV disease (n = 11, thereof six liver metastases, four peritoneal metastases and one pulmonary metastasis, details see Supplementary Table 2) and/or arterial resection (n = 4), perioperative death/death due to complications (n = 1) and recurrence surgery (n = 1) were excluded from survival analysis. Three patients were lost to follow-up.
Histologic analysis
All PDACs were histologically classified into conventional PDACs, combined PDACs in which, in addition to the classical tubular growth pattern, a special histologic component was present in more than 30% of the tumor area, and variants with a special pattern in at least 50% of the tumor area. PDACs with a conventional morphology were largely composed of well- to moderately developed tubular and duct-like structures and showed only few other structures, such as glands with clear cell morphology, cribriform architecture, papillary epithelial lining and individual pleomorphic cells17. Combined PDACs with dominant histological features showed either a clear-cell, cribriform, gyriform, papillary, micropapillary or complex component against a background of tubular architecture (see Fig. 1). The complex component was characterized by small irregular glands mixed with solid or cribriform cell sheets and individual pleomorphic cells. PDAC variants included colloid, adenosquamous, and papillary carcinoma (see Fig. 1A–F). Among the pancreatic carcinomas that have not yet been regarded as PDAC variants are medullary and tubular carcinoma. The tubular adenocarcinoma is separated from conventional PDAC by its entirely well differentiated tubular architecture that is characterized by small tubular glands diffusely infiltrating the pancreatic parenchyma and difficult to distinguish from equally sized normal ducts (see Fig. 2).
Immunohistochemical analysis
All stainings were run on an automated immunostainer with an iVIEW DAB detection kit (Ventana Medical Systems, Roche, Mannheim, Germany) according to the company’s protocols for open procedures with slight modifications, for details see Supplementary materials and methods.
Molecular analysis
Molecular analysis was performed on extracted DNA from manually microdissected FFPE tumor tissue, for details see Supplementary materials and methods and Supplementary Table 1.
Statistical analyses
The statistical analyses performed are described in Supplementary materials and methods.
Additional Information
How to cite this article: Schlitter, A. M. et al. Molecular, morphological and survival analysis of 177 resected pancreatic ductal adenocarcinomas (PDACs): Identification of prognostic subtypes. Sci. Rep. 7, 41064; doi: 10.1038/srep41064 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was partly supported by a grant from Novartis Pharma GmbH, Nurnberg (to A.M.S., G.K. and I.E.). We are grateful to Professor Gisela Keller for advice and helpful discussion, to Petra Meyer, Daniela Angermeier and Birgit Geist for excellent technical assistance and Henrike Jahns for her contribution in figure preparation.
Footnotes
The authors declare no competing financial interests.
Author Contributions A.S., N.P., V.E., I.R. and M.B. carried out experiments and analyzed data. C.J., B.K., BO.K. and K.S. collected and analyzed data. A.M.S., G.K. and I.E. designed the study, conceived out experiments and analyzed data. C.W.M. and J.K. designed the study. A.M.S., A.S. and G.K. wrote the manuscript. All authors had final approval of the submitted version.
References
- Spath C. et al. Strategies to improve the outcome in locally advanced pancreatic cancer. Minerva Chir 70, 97–106 (2015). [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D. & Jemal A. Cancer statistics, 2015. CA Cancer J Clin 65, 5–29, doi: 10.3322/caac.21254 (2015). [DOI] [PubMed] [Google Scholar]
- Esposito I., Konukiewitz B., Schlitter A. M. & Kloppel G. Pathology of pancreatic ductal adenocarcinoma: facts, challenges and future developments. World J Gastroenterol 20, 13833–13841, doi: 10.3748/wjg.v20.i38.13833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito I., Segler A., Steiger K. & Kloppel G. Pathology, genetics and precursors of human and experimental pancreatic neoplasms: An update. Pancreatology, doi: 10.1016/j.pan.2015.08.007 (2015). [DOI] [PubMed] [Google Scholar]
- Wood L. D. & Hruban R. H. Pathology and molecular genetics of pancreatic neoplasms. Cancer J 18, 492–501, doi: 10.1097/PPO.0b013e31827459b6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz A. K. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 6, 6744, doi: 10.1038/ncomms7744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501, doi: 10.1038/nature14169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblum E. et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res 57, 1731–1734 (1997). [PubMed] [Google Scholar]
- Venter J. C. et al. The sequence of the human genome. Science 291, 1304–1351, doi: 10.1126/science.1058040 (2001). [DOI] [PubMed] [Google Scholar]
- Jones S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806, doi: 10.1126/science.1164368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 17, 500–503, doi: 10.1038/nm.2344 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. et al. Impact of epidermal growth factor receptor (EGFR) kinase mutations, EGFR gene amplifications, and KRAS mutations on survival of pancreatic adenocarcinoma. Cancer 109, 1561–1569, doi: 10.1002/cncr.22559 (2007). [DOI] [PubMed] [Google Scholar]
- Ogura T. et al. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J Gastroenterol 48, 640–646, doi: 10.1007/s00535-012-0664-2 (2013). [DOI] [PubMed] [Google Scholar]
- Shin S. H. et al. Genetic alterations of K-ras, p53, c-erbB-2, and DPC4 in pancreatic ductal adenocarcinoma and their correlation with patient survival. Pancreas 42, 216–222, doi: 10.1097/MPA.0b013e31825b6ab0 (2013). [DOI] [PubMed] [Google Scholar]
- Sinn B. V. et al. KRAS mutations in codon 12 or 13 are associated with worse prognosis in pancreatic ductal adenocarcinoma. Pancreas 43, 578–583, doi: 10.1097/MPA.0000000000000077 (2014). [DOI] [PubMed] [Google Scholar]
- Yachida S. et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res 18, 6339–6347, doi: 10.1158/1078-0432.CCR-12-1215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruban R. H. et al. In WHO Classification of Tumors of the Digestive System (eds F.T. Bosman, F. Carneiro, R.H. Hruban & N.D. Theise) 292–299 (IARC, 2010). [Google Scholar]
- Dal Molin M. et al. Clinicopathological correlates of activating GNAS mutations in intraductal papillary mucinous neoplasm (IPMN) of the pancreas. Ann Surg Oncol 20, 3802–3808, doi: 10.1245/s10434-013-3096-1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M. et al. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann Surg 258, 336–346, doi: 10.1097/SLA.0b013e3182827a65 (2013). [DOI] [PubMed] [Google Scholar]
- Calhoun E. S. et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol 163, 1255–1260, doi: 10.1016/S0002-9440(10)63485-2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggins M. et al. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+. Am J Pathol 152, 1501–1507 (1998). [PMC free article] [PubMed] [Google Scholar]
- Wilentz R. E. et al. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am J Pathol 156, 1641–1651, doi: 10.1016/S0002-9440(10)65035-3 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA 108, 21188–21193, doi: 10.1073/pnas.1118046108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adsay N. V. et al. Colloid (mucinous noncystic) carcinoma of the pancreas. Am J Surg Pathol 25, 26–42 (2001). [DOI] [PubMed] [Google Scholar]
- Borazanci E. et al. Adenosquamous carcinoma of the pancreas: Molecular characterization of 23 patients along with a literature review. World J Gastrointest Oncol 7, 132–140, doi: 10.4251/wjgo.v7.i9.132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trikudanathan G. & Dasanu C. A. Adenosquamous carcinoma of the pancreas: a distinct clinicopathologic entity. South Med J 103, 903–910, doi: 10.1097/SMJ.0b013e3181ebadbd (2010). [DOI] [PubMed] [Google Scholar]
- Kloppel G., Basturk O., Schlitter A. M., Konukiewitz B. & Esposito I. Intraductal neoplasms of the pancreas. Semin Diagn Pathol 31, 452–466, doi: 10.1053/j.semdp.2014.08.005 (2014). [DOI] [PubMed] [Google Scholar]
- Kelly P. J. et al. Cystic papillary pattern in pancreatic ductal adenocarcinoma: a heretofore undescribed morphologic pattern that mimics intraductal papillary mucinous carcinoma. Am J Surg Pathol 36, 696–701, doi: 10.1097/PAS.0b013e318249ce1c (2012). [DOI] [PubMed] [Google Scholar]
- Rakha E. A., Pinder S. E., Shi S. J. & Tsuda H. In WHO Classiciation of Tumours of the Breast Vol. 4 (eds S.R. Lankhani et al.) (IARC, 2012). [Google Scholar]
- Rakha E. A. et al. Tubular carcinoma of the breast: further evidence to support its excellent prognosis. J Clin Oncol 28, 99–104, doi: 10.1200/JCO.2009.23.5051 (2010). [DOI] [PubMed] [Google Scholar]
- Riener M. O. et al. Microarray comparative genomic hybridization analysis of tubular breast carcinoma shows recurrent loss of the CDH13 locus on 16q. Hum Pathol 39, 1621–1629, doi: 10.1016/j.humpath.2008.02.021 (2008). [DOI] [PubMed] [Google Scholar]
- Diab S. G. et al. Tumor characteristics and clinical outcome of tubular and mucinous breast carcinomas. J Clin Oncol 17, 1442–1448 (1999). [DOI] [PubMed] [Google Scholar]
- Churi C. R. et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 9, e115383, doi: 10.1371/journal.pone.0115383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405, doi: 10.1038/nature11547 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642, doi: 10.1038/nature05327 (2006). [DOI] [PubMed] [Google Scholar]
- Guerra C. et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302, doi: 10.1016/j.ccr.2007.01.012 (2007). [DOI] [PubMed] [Google Scholar]
- Hingorani S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- Mazur P. K. & Siveke J. T. Genetically engineered mouse models of pancreatic cancer: unravelling tumour biology and progressing translational oncology. Gut 61, 1488–1500, doi: 10.1136/gutjnl-2011-300756 (2012). [DOI] [PubMed] [Google Scholar]
- Morris J. P. t., Wang S. C. & Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer 10, 683–695, doi: 10.1038/nrc2899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y., Grabocka E. & Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 11, 761–774, doi: 10.1038/nrc3106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler B. et al. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc Natl Acad Sci USA 105, 10137–10142, doi: 10.1073/pnas.0800487105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. A. et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 122, 639–653, doi: 10.1172/JCI59227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H. et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670, doi: 10.1016/j.cell.2012.01.058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. A. et al. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One 7, e49707, doi: 10.1371/journal.pone.0049707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone B. A. et al. Loss of SMAD4 staining in pre-operative cell blocks is associated with distant metastases following pancreaticoduodenectomy with venous resection for pancreatic cancer. J Surg Oncol 110, 171–175, doi: 10.1002/jso.23606 (2014). [DOI] [PubMed] [Google Scholar]
- Singh P., Srinivasan R. & Wig J. D. SMAD4 genetic alterations predict a worse prognosis in patients with pancreatic ductal adenocarcinoma. Pancreas 41, 541–546, doi: 10.1097/MPA.0b013e318247d6af (2012). [DOI] [PubMed] [Google Scholar]
- Biankin A. V. et al. DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J Clin Oncol 20, 4531–4542 (2002). [DOI] [PubMed] [Google Scholar]
- Winter J. M. et al. Failure patterns in resected pancreas adenocarcinoma: lack of predicted benefit to SMAD4 expression. Ann Surg 258, 331–335, doi: 10.1097/SLA.0b013e31827fe9ce (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500, doi: 10.1016/j.ccr.2009.03.022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basturk O. et al. A Revised Classification System and Recommendations From the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am J Surg Pathol 39, 1730–1741, doi: 10.1097/PAS.0000000000000533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.