SUMMARY
The monobactams, exemplified by the natural product sulfazecin, are the only class of β-lactam antibiotics not inactivated by metallo-β-lactamases, which confer bacteria with extended-spectrum β-lactam resistance. We screened a transposon mutagenesis library from Pseudomonas acidophila ATCC 31363 and isolated a sulfazecin-deficient mutant that revealed a gene cluster encoding two non-ribosomal peptide synthetases (NRPSs), a methyltransferase, a sulfotransferase, and a dioxygenase. Three modules and an aberrant C-terminal thioesterase (TE) domain are distributed across the two NRPSs. Biochemical examination of the adenylation (A) domains provided evidence that L-2,3-diaminopropionate, not L-serine as previously thought, is the direct source of the β-lactam ring of sulfazecin. ATP/PPi exchange assay also revealed an unusual substrate selectivity shift of one A domain when expressed with or without the immediately upstream condensation domain. Gene inactivation analysis defined a cluster of 13 open reading frames sufficient for sulfazecin production, precursor synthesis, self-resistance, and regulation. The identification of a key intermediate supported a proposed NRPS-mediated mechanism of sulfazecin biosynthesis and β-lactam ring formation distinct from the nocardicins, another NRPS-derived subclass of monocyclic β-lactam. These findings will serve as the basis for further biosynthetic research and potential engineering of these important antibiotics.
Graphical Abstract
INTRODUCTION
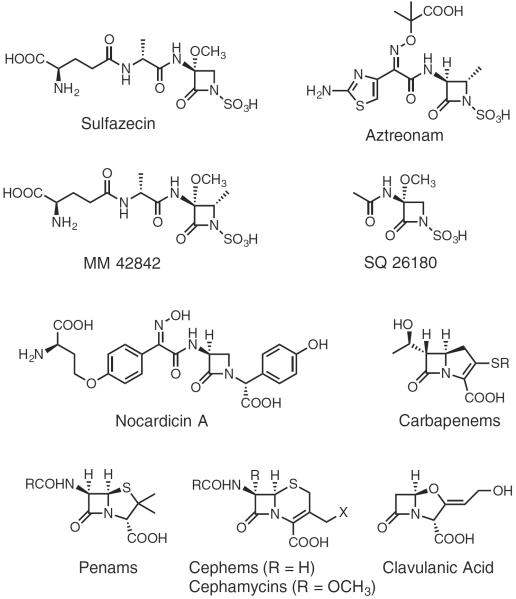
Antibiotics, particularly the β-lactam classes (Figure 1), are a cornerstone of modern medicine (Olsen, 2015). Their widespread use, however, for over half a century has created evolutionary pressure on bacteria to survive repeated exposure to these antimicrobial agents. Of the several manifestations of bacterial resistance currently known, the production of β-lactamases is the most important. These enzymes utilize mostly classical serine hydrolase activity directed against β-lactam drugs, cleaving the β-lactam ring and thus rendering them ineffective (King et al., 2016; McKenna, 2013). Recent reports of carbapenem-resistant Enterobacteriaceae (CREs) highlight the alarming rate at which bacteria are evolving resistance to currently used, last-resort antibiotics (Bebrone, 2007; King, 2013). CRE, among a growing number of other organisms, use the most effective class of β-lactamase: the metallo-β-lactamases (MBLs). MBLs invariably impart broad-spectrum antibiotic resistance to the producing host. There are currently no clinically approved MBL inhibitors, highlighting the challenge these resistance proteins present in treating bacterial infections.
Figure 1. β-Lactam Structures.
Examples of monobactams and representative structures of the other β-lactam antibiotic families.
The only β-lactam antibiotics that do not fall prey to MBLs are the monobactams (Bebrone, 2007; Docquier et al., 2003; Sykes and Bonner, 1985). The monobactams such as sulfazecin (Figure 1) belong to a subclass of monocyclic β-lactams that is distinguished by the presence of an N-sulfonated β-lactam ring and a variable C-3 side chain. The sulfamate functionality activates the β-lactam ring for reaction with, and successful binding to, the penicillin-binding proteins. Some monobactams such as the naturally occurring MM 42842 and synthetic aztreonam (Figure 1) harbor a C-4 methyl group, which provides enhanced chemical and β-lactamase stability (Sykes and Bonner, 1985). Aztreonam remains the only U.S. Food and Drug Administration-approved monobactam in clinical use since the isolation of naturally occurring monobactams. Apart from the 4α-methyl and 1-sulfonate groups found in natural monobactams, aztreonam is equipped with a third-generation cephalosporin side chain to improve activity against aerobic Gram-negative bacteria and Pseudomonas (Sykes and Bonner, 1985). It has proved efficacious in the clinical treatment of many multidrug-resistant infections, eradicating those caused by bacteria resistant to the most broadly active antibiotics, including meropenem, vancomycin, and imipenem (Araoka et al., 2012).
Sulfazecin is a hallmark of the naturally occurring monobactams. Since its discovery in the early 1980s in the producing strain Pseudomonas acidophila (Asai et al., 1981), however, very little research has been conducted on its molecular origin (Parker et al., 1986). The genomes of a few species related to monobactam producers have been fully sequenced (Brazilian National Genome Project Consortium, 2003). However, searching for isopenicillin synthetase (Leskiw et al., 1988) or β-lactam synthetase (Bachmann et al., 1998) paralogs resulted in no hits. Similarly, searching for homologs of NocB module 5 and the key non-ribosomal peptide synthetase (NRPS) components involved in the synthesis of the monocyclic β-lactam present in nocardicin A (Gaudelli et al., 2015) (Figure 1) in these genomes mostly resulted in enzymes involved in siderophore biosynthesis (Schwarzer et al., 2003).
Whole-cell radiochemical incorporation studies aimed at elucidating the biosynthetic origin of the β-lactam ring in SQ 26180 (Figure 1), a simple monobactam produced in Chromobacterium violaceum (Wells et al., 1982), indicated that 14C-serine was incorporated at this position without change in the oxidation state of the β-carbon (O'Sullivan et al., 1982), findings that parallel earlier experiments with nocardicin A (Townsend and Brown, 1981, 1983). 14C-Methionine yielded SQ 26180 with the radio-label in the 3α-methoxyl group. From these results it was proposed that the β-lactam ring in monobactams, such as the monocyclic nocardicins, is derived from serine and the methoxyl group from methionine (O'Sullivan et al., 1982). The source of sulfur in the sulfamate moiety was also investigated and it was found that inorganic sulfur could be utilized (O'Sullivan et al., 1983) and its addition to production media increased titers of sulfazecin in P. acidophila (Imada et al., 1980). A cell-free extract of Agrobacterium radiobacter, another monobactam producer, synthesized adenylyl-5′-phosphosulfate (APS) when incubated with sulfate, ATP, and appropriate co-factors, suggesting that the sulfamate group of monobactams is produced through APS or 3′-phosphoadenosine-5′-phosphosulfate (PAPS) (Imada et al., 1980; O'Sullivan et al., 1983).
As most CREs are Gram-negative bacteria and their resistances are mediated by MBLs, development of more potent monobactam derivatives could provide a viable tactic to combat these pathogens. To date, the biosynthesis of monobactams remains poorly understood and no biosynthetic gene clusters (BGCs) have been identified. In this paper, we describe the isolation and characterization of the sulfazecin BGC from Pseudomonas acidophila ATCC 31363. We demonstrate that, instead of serine, the β-lactam ring in sulfazecin is derived from L-2,3-diaminopropionate (L-2,3-Dap) and propose a mechanism of formation in the unusual thioesterase (TE) domain.
RESULTS AND DISCUSSION
Isolation of Sul−-Defective Mutants by Transposon Mutagenesis
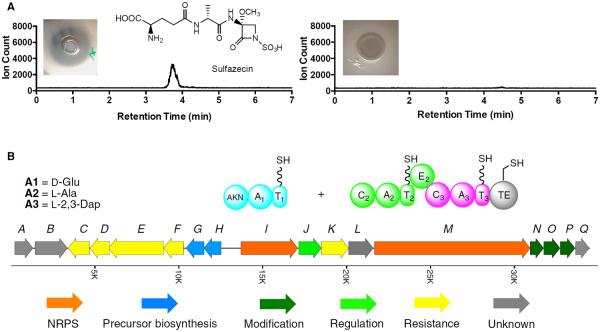
Owing to the limited biochemical data and the lack of genetic information about monobactam biosynthesis, a transposon mutagenesis approach was employed to generate mutants deficient in sulfazecin production. A total of 2,000 KanR/CarR transconjugants was analyzed for their ability to produce sulfazecin after mating with the Tn5 delivery vehicle pGS9 in E. coli PR47 (Willis et al., 1990). The 75 Sul− transconjugants identified by fermentation on solid medium were subjected to three more rounds of fermentation in liquid production medium (Figure S1). As shown in Figure 2A, one clone, P. acidophila Sul−-7, was eventually determined to be defective in sulfazecin synthesis by growth inhibition of an E. coli β-lactam extra-sensitive strain (ESS) and β-lactamase induction detected by a nitrocefin colorimetric assay (Aoki et al., 1976; Sykes and Bonner, 1985). The Sul− phenotype was further confirmed by high-resolution ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) after the fermentation broth was subjected to a reported sulfazecin-purification procedure (Figure 2A) (Asai et al., 1981).
Figure 2. Sulfazecin Production and Biosynthetic Gene Cluster.
(A) UPLC-HRMS analysis of wild-type P. acidophila (left) and P. acidophila Sul−-7 mutant (right). Insets are E. coli ESS growth-inhibition assays.
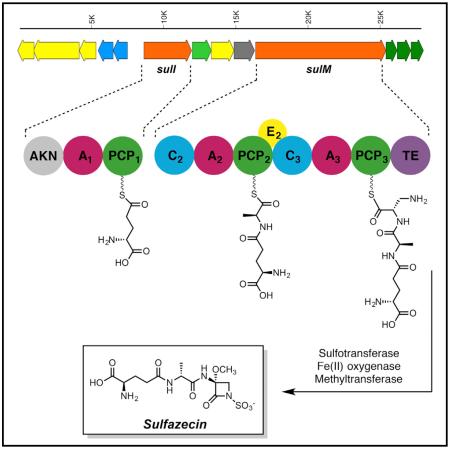
(B) Sulfazecin gene cluster from cosmid clone 1H4. The colored ORFs indicate the putative minimal gene cluster for sulfazecin biosynthesis, precursor synthesis, regulation, and resistance. The domain organization of the NRPSs is also shown, and the amino acid substrate selectively activated by each A domain. The AKN domain at the N terminus of SulI indicates the predicted adenylylsulfate kinase.
Molecular Characterization of a P. acidophila Sul−-7 Mutant
Total genomic DNA was isolated from P. acidophila Sul−-7. A 490-bp PCR product identical to the kanamycin-resistance cassette in Tn5 was obtained from the mutant as confirmed by DNA sequencing, but not from the wild-type strain. The genomic DNA was then digested with several restriction enzymes. Southern hybridizations using the kanamycin-resistance gene as a probe identified a positive fragment in NruI-digested genomic DNA (gDNA) containing 2.0-kb of P. acidophila genomic DNA flanking the Tn5 insertion site (Figure S2A). This DNA fragment was further subcloned as pBS/nruI4.5 and sequenced. FramePlot analysis showed one complete and one partial open reading frame (ORF), and a Blastp search revealed that the protein encoded by the complete ORF was homologous to hydroxyacylglutathione hydrolases (identity/positive: 80/86). The incomplete ORF located between the Tn5 and the complete ORF encoded an 80-AA peptide. Blastp analysis of this fragment showed homology to NRPSs from several secondary metabolic pathways (61%–78% identity), indicating that the Tn5 transposon had inserted into an NRPS responsible for the biosynthesis of sulfazecin (Figure S2B).
Cloning of the Sulfazecin Biosynthetic Gene Cluster
A gDNA cosmid library was constructed and screened using the 1.0-kb EcoNI-PshAI probe from the 2.0-kb Tn5 flanking region in pBS/nruI4.5. Sixteen positive clones were obtained from a 2,300-clone library. Sequence analysis revealed that the T3-ends of cosmids 1H4 and 12F10 encoded a hypothetical protein Osi_06252 and an MFS transporter, and the T7-ends encoded a long-chain fatty acid-CoA ligase and a membrane protein, respectively. These proteins are unlikely to be related to sulfazecin biosynthesis, suggesting that these cosmids contained the full-length sulfazecin gene cluster. Both cosmids were fully sequenced and the assembled data revealed that the insertional regions in cosmids 1H4 and 12F10 were 46,840 and 38,889 bp, respectively, with a G/C content of 63%. With a 24.45-kb overlap between the two inserts, the sequences spanned 60,368 bp of the P. acidophila genome (data not shown).
Sequence Analysis of the Sulfazecin, sul, Gene Cluster
FramePlot analysis of the 60-kb sequenced region revealed 40 complete ORFs, including two NRPS genes. The putative functions of deduced proteins from 17 genes flanking the two NRPS-encoding genes are summarized in Table 1 and Figure 2B: sulI and sulM encode proteins with over 80% similarity to NRPSs from various natural product biosynthetic pathways. The protein sequences were examined for conserved NRPS domains (Schwarzer et al., 2003). Closer examination using polyketide synthase (PKS)/NRPS analysis tools (nrps.igs.umaryland.edu) revealed one complete module in SulI (SulI M1) and two complete modules in SulM (SulM M2 and M3). Each module in SulM contains a condensation (C), adenylation (A), and thiolation (T) domain, with an additional epimerization (E) domain present in SulM M2. The overall sequence similarity of the TE domain is relatively low compared with TE domains from other NRPSs. The GXSXG active site signature, well conserved among known TE domains, is modified to AXCXG in the SulM TE, however, but still aligns with the GXSXG motif according to Blastp analysis. Similarly, the sulfazecin-like clusters identified by antiSMASH analysis all contain the same modification in their TE domains (Horsman et al., 2016). The Tn5 transposon was found inserted into the encoding region of the C2 domain of SulM in P. acidophila Sul−-7 (Figure S2A), indicating that sulM is essential for the biosynthesis of sulfazecin.
Table 1.
Deduced Functions and Homologs of the Gene Products of the Sulfazecin Cluster
| ORF | aa | Similarity | Proposed Function |
|---|---|---|---|
| sulA | 348 | acetyltransferase (71/82) | unknown |
|
| |||
| sulB | 615 | lipase (74/85) | resistance |
| outer membrane autotransporter (75/85) | |||
|
| |||
| sulC | 405 | membrane protein (80/85) | transporter, regulation |
| major facilitator superfamily (MFS)_1 (27/45) | |||
| MFS transporter (25/45) | |||
|
| |||
| sulD | 375 | β-lactamase (57/68) | resistance |
|
| |||
| sulE | 1,049 | acriflavine-resistance protein B (89/94) | resistance |
| multidrug-resistance protein MdtB (multidrug transporter mdtB) (88/93) | |||
|
| |||
| sulF | 382 | multidrug efflux RND membrane fusion protein MexE, partial (72/84) | resistance |
|
| |||
| sulG | 342 | ornithine cyclodeaminase (63/76) | L-2,3-diaminopropionate biosynthesis |
| 2,3-diaminopropionate biosynthesis protein SbnB | |||
| [2,3-diaminopropionate biosynthesis protein SbnB] | |||
|
| |||
| sulH | 329 | cysteine synthase (66/81) | L-2,3-diaminopropionate biosynthesis |
| 2,3-diaminopropionate biosynthesis protein SbnA (59/78) | |||
| [2,3-diaminopropionate biosynthesis protein SbnA] | |||
|
| |||
| sulI | 1,089 | NRPS (81/86) | NRPS, M1 |
|
| |||
| sulJ | 433 | hypothetical protein BTH_I1954 (81/88) | transporter |
| major facilitator superfamily protein (80/86) | |||
| MFS transporter (80/86) | |||
|
| |||
| sulK | 521 | RND efflux system outer membrane lipoprotein (56/71) | transporter |
|
| |||
| sulL | 470 | hydroxyacylglutathione hydrolase (81/86) | unknown |
|
| |||
| sulM | 2,984 | NRPS (72/80) | NRPS M2 and M3 |
|
| |||
| sulN | 256 | sulfotransferase family protein (77/83) | sulfotransferase |
|
| |||
| sulO | 311 | diguanylate cyclase (88/94) | dioxygenase |
| dioxygenase, TauD/TfdA (88/94) | |||
|
| |||
| sulP | 282 | methyltransferase (84/89) | methyltransferase |
|
| |||
| sulQ | 263 | hypothetical protein BPSL2225 (87/91) | unknown |
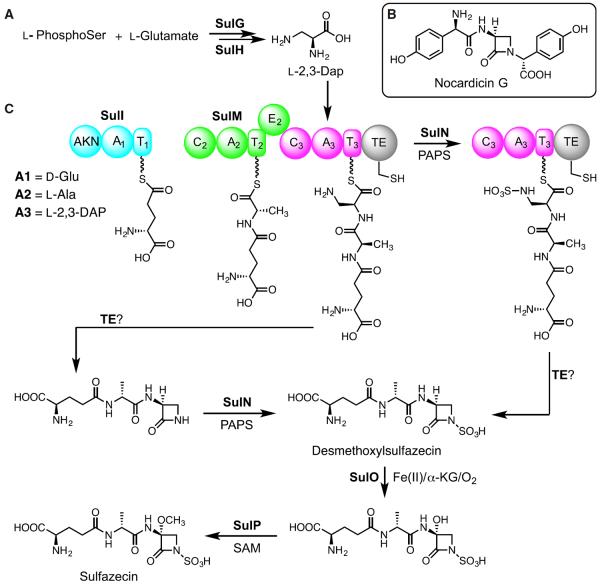
NRPS substrates can often be predicted by aligning eight conserved residues in the substrate-binding pockets of A domains contained within the NRPS (Challis et al., 2000). The structure of sulfazecin along with previous feeding studies suggest that alanine and serine are likely substrates of SulM (O'Sullivan et al., 1982). However, the substrate preference of SulM A2 (DV-P-N-F-I-M-V) and SulM A3 (D-V-W-E-I-N-A-D) could not be predicted by various NRPS analysis programs (Bachmann and Ravel, 2009; Challis et al., 2000; Rausch et al., 2005; Stachelhaus et al., 1999). Continued analysis revealed a single module consisting of an A and a T domain in SulI. The substrate selectivity of the lone SulI A1 domain was readily predicted as glutamate (D-A-M-H-V-G-G-T). Interestingly, a 652-AA domain located at the N terminus of SulI was not identified by NRPS domain prediction tools. This unusual domain is also present in at least one sulfazecin-like cluster identified by antiSMASH analysis (Weber et al., 2015) (Figure S6). Blastp search showed that this domain is homologous to adenylylsulfate kinases (55%–72% identity), enzymes responsible for the biosynthesis of PAPS (Geller et al., 1987). This unusual protein architecture has not been observed in other NRPSs before. Adenylylsulfate kinase in the gene cluster could be responsible for the simultaneous synthesis of additional PAPS to support sulfazecin biosynthesis.
Downstream of sulM lie three genes, sulN, sulO, and sulP. The sulN protein is highly similar to sulfotransferases, which contain two conserved sequence motifs, a 5′-phosphosulfate-binding motif located in a strand-loop-helix region, and a second motif similar to 3-phosphate-binding sites located in a strand-helix region (Negishi et al., 2001). SulN contains these conserved motifs (14PRTAGTTFG22 and 85LTLLREPVARLVSYYF100, residues that interact with PAPS are underlined) strongly suggesting that SulN acts as the sulfotransferase for N-sulfonation using PAPS as a co-substrate (Figure S3A). SulO shows high similarity to clavaminate synthase (CS)-like proteins and taurine catabolism dioxygenases. CS is a trifunctional Fe(II)/α-ketoglutarate (α-KG)-dependent oxygenase that carries out three non-sequential reactions in the biosynthesis of clavulanic acid (Baldwin et al., 1993; Busby and Townsend, 1996), the clinically important inhibitor of class A serine β-lactamases. Multiple alignments showed that the conserved residues for substrate binding, iron coordination, and catalysis (W99, T101, D126, H255, and R266) in these enzymes are all present in SulO (Figure S3B). Thus, we propose that SulO is responsible for C-3 hydroxylation during sulfazecin biosynthesis. Finally, Blastp analysis indicated that SulP is an S-adenosylmethionine-dependent methyltransferase. Installation of the 3α-methoxyl group into sulfazecin is, therefore, mechanistically analogous to the non-heme iron α-KG-dependent dioxygenase and methyltransferase that introduce the 7α-methoxyl group in cephamycin (O'Sullivan and Abraham, 1980). The 3α-methoxyl group is absent in some naturally occurring monobactams such as PB-5266A, B, and C (Kato et al., 1987), suggesting that methoxylation occurs after β-lactam ring formation, as known in cephamycin biosynthesis (O'Sullivan and Abraham, 1980).
It has been proposed that the β-lactam ring of sulfazecin is derived from L-serine (O'Sullivan et al., 1982). The source of the β-lactam nitrogen is not accounted for. However, NRPS A domain prediction failed to identify serine (or any amino acid) as the substrate of the SulM A3 domain. Two genes present in the sulfazecin cluster, sulG and sulH, encode proteins homologous to SbnA and SbnB. Kobylarz et al. (2014) recently demonstrated that SbnA uses PLP and the substrates L-phosphoserine and L-glutamate to produce a metabolite N-(1-amino-1-carboxyl-2-ethyl)-glutamic acid (ACEGA). SbnB uses NAD+ to oxidatively cleave ACEGA to yield α-KG and L-2,3-Dap. L-2,3-Dap is an unusual non-proteinogenic amino acid used as a building block by NRPSs during the biosynthesis of several antibiotics and siderophores, such as syringomycin, viomycin, capreomycin, and staphyloferrin B (Beasley et al., 2011; Gueznzi et al., 1998; Thomas et al., 2003; Wang and Gould, 1993). The structure of sulfazecin and the existence of L-2,3-Dap biosynthetic enzymes in its gene cluster strongly suggest that L-2,3-Dap, not L-serine, is the precursor of the sulfazecin and the substrate of the cryptic SulM A3.
A few genes (sulB, D, E, F, and K) encoding multidrug resistance, transporter, and efflux system member proteins are located in the gene cluster. Their functions appear to be self-resistance and product export. sulC and sulJ, encoding major facilitator superfamily proteins, could be involved in regulation. sulL encodes a protein highly similar to hydroxyacylglutathione hydrolases. Its function in the biosynthetic pathway cannot be assigned based on bioinformatic analysis and the structure of sulfazecin.
A Domain Substrate Selectivity of SulI and SulM
Heterologous expression of the three A domains housed in SulI and SulM as single or multidomain constructs was carried out in E. coli to experimentally determine their substrate selectivity using the standard ATP/PPi exchange assay (Lee and Lipmann, 1975). Based on the domain prediction (Figure 2B), each A-T di-domain of the three modules was cloned into pET29b or pET28b for expression of C- or N-terminal 6-His fusion proteins in E. coli Rosetta 2(DE3). Meanwhile, the SulM A2 monodomain and the SulM C2A2T2 tridomain were also expressed as N-terminal 6-His-tagged proteins (Figure S4D).
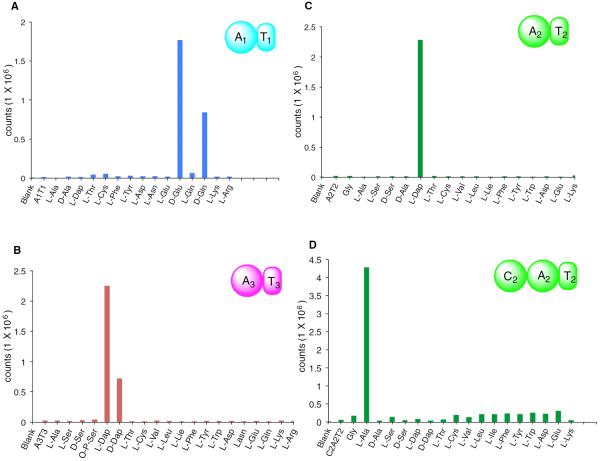
Bioinformatic analysis of the SulI A1 domain predicted glutamate to be its preferred substrate with high confidence. Very strong ATP/PPi exchange was obtained in the presence of D-Glu, while no activation of L-Glu was observed (Figure 3A). This high extent of stereoselection was unexpected but is in keeping with the observation that the glutamate moiety in sulfazecin has the D-configuration (Asai et al., 1981) and the absence of an E domain in module 1. Interestingly, the SulI A1T1 didomain in addition showed approximately 47% activity with D-Gln, suggesting that D-Gln could be an alternative substrate. To date, none of the known monobactam producers incorporate more than one side chain (Parker et al., 1986), but the incorporation of D-Gln in vitro suggests the biosynthetic pathway might be engineered to produce sulfazecin analogs in the future (Crusemann et al., 2013; Kries et al., 2015).
Figure 3. ATP/PPi Exchange Assays of A1T1, A2T2, A3T3, and C2A2T2.
(A) ATP/PPi exchange assay of A1T1.
(B) ATP/PPi exchange assay of A3T3.
(C) ATP/PPi exchange assay of A2T2.
(D) ATP/PPi exchange assay of C2A2T2. Blank, no enzyme; no amino acids, control.
As discussed above, the domain organization of SulM and the presence of enzymes responsible for the biosynthesis of L-2,3-Dap in the gene cluster suggested that L-2,3-Dap, instead of L-Ser, was likely the SulM A3 substrate. To test this proposition, SulM A3T3 was expressed and purified for the ATP/PPi exchange assay. As hoped, very strong ATP/PPi exchange activity was observed from L-2,3-Dap, but not from L-Ser. In addition, SulM A3T3 also activated D-2,3-Dap, but the activity was only ~30% that of its L-antipode (Figure 3B).
Finally, to probe the substrate selectivity of SulM A2, two constructs, SulM A2 and SulM A2T2, were expressed in E. coli as N-terminal 6-His proteins. Although not predicted, the structure of sulfazecin and the domain organization of SulM suggest that L-Ala is the substrate of SulM A2, which is epimerized to D-Ala by the immediately downstream E2 domain. The A domain residues involved in substrate binding in other alanine-activation domains are not well conserved (Figure S4A), often ambiguating or preventing substrate predictions by PKS/NRPS analysis programs (Xia et al., 2012). Surprisingly, the results of ATP/PPi exchange assays showed that, under standard conditions (Davidsen et al., 2013), L-2,3-Dap, not L-Ala, is the preferred substrate. This result was reproducible using either the SulM A2T2 or SulM A2 construct (Figures 3C and S4C).
The substrate selectivity was then examined using modified conditions for ATP/PPi exchange assay (Davidsen et al., 2013; Hou et al., 2011). The amino acid substrate and enzyme were incubated for 10 min at 30°C before reaction was initiated by addition of ATP, and the reaction was carried out at 30°C instead of the standard 23°C. Significant ATP/PPi exchange was observed for L-2,3-Dap, L-Asp, and L-Thr, but neither L-Ala nor D-Ala was activated by SulM A2 (Figure S4B). The overall high background and non-selective substrate activation under the altered conditions were reported previously as a result of pre-incubation of substrate and enzymes (Davidsen et al., 2013; Hou et al., 2011). ATP/PPi exchange assay using a cell-free extract from E. coli Rosetta 2(DE3) (pET28b) cells showed no amino acid activation activities, indicating that the observed activation of L-2,3-Dap by SulM A2 and SulM A2T2 was not due to contamination from the host cells (data not shown).
To further investigate this observation, the SulM C2A2T2 tridomain was over-produced and purified. Under standard ATP/PPi exchange conditions, the assay now clearly showed that L-Ala, not L-2,3-Dap, was selectively activated (Figure 3D). Related substrate selectivity shifts in other A domain constructs have been reported recently (Meyer et al., 2016). The gatekeeping and substrate activation roles of C domains are proposed to be due to C and A domain interactions (Meyer et al., 2016), likely the result of a conformational change upon amino acid binding in the SulM A2 domain.
Mutational Analysis of Sulfazecin Biosynthesis
More than half of the enzymes encoded in the sulfazecin gene cluster could be functionally assigned by bioinformatics analysis, including the NRPSs, sulfotransferase, dioxygenase, and methyltransferase. On the other hand, the gene(s) responsible for β-lactam formation and the boundaries of the BGC itself remained unknown. To define the boundaries of the sulfazecin BGC, sulC and sulQ, encoding a predicted membrane protein and a hypothetical protein, respectively, were inactivated by targeted gene disruption. The single-stranded disruption constructs (Figures S5A and S5C) were delivered into wild-type P. acidophila by electroporation. The GmR/KmR cells were grown for two rounds without antibiotic selection before screening for the GmR/KmS phenotype resulting from double crossover and loss of plasmids. The genotypes of the resulting knockout mutants, P. acidophila BGmD-2 and PGmR-1, were confirmed by PCR of the targeted regions followed by DNA sequence analysis (Figures S5A and S5C). Fermentation of these mutants showed that sulfazecin was still produced as indicated by nitrocefin assay (Figures S5A and S5C), suggesting that these genes are not essential for biosynthesis and genes located between sulD and sulP are sufficient for sulfazecin production in P. acidophila ATCC 31363.
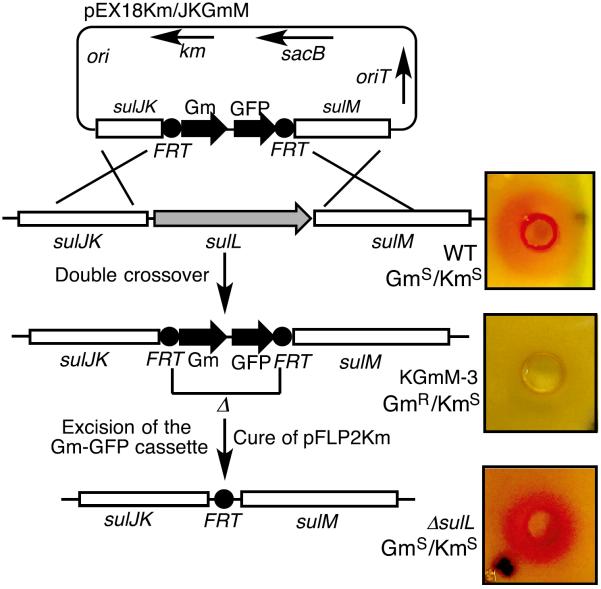
The encoding gene of SulL was targeted first by means of a gene-replacement approach with a GFP-Gm cassette. The disruption vector was transformed into the wild-type strain by conjugation. The exconjugants were selected by the GmR/KmS phenotype, and the targeted gene disruptions by double-crossover events were confirmed by PCR and DNA sequence analysis (Figure 4). Two of the resulting mutants, P. acidophila KGmM-3 and KGmM-4, were tested for sulfazecin production. Nitrocefin assay showed that antibiotic production was abolished in both mutants (Figure 4). Because sulL is located immediately upstream of sulM and appears to be co-transcribed with the latter, gene-replacement disruption of sulL could cause polar effects on downstream genes, such as sulM. Thus, an unmarked and plasmid-free mutant, P. acidophila ΔsulL was generated by Flp-catalyzed excision of the GFP-Gm markers in P. acidophila KGmM-3 (Figure 4). Expression of Flp recombinase from pFLP2Km acted at the FRT sites to catalyze excision of the GFP-Gm cassette leaving behind a short in-frame FRT-containing sequence (Hoang et al., 1998). The unmarked and plasmid-free mutant P. acidophila ΔsulL was confirmed by its GmS/KmS phenotype and by PCR amplification of the targeted region. DNA sequencing showed that sulL was completely deleted from the genome in the mutant. Fermentation of P. acidophila ΔsulL showed that sulfazecin was still produced in the mutant as indicated by the β-lactamase induction assay, suggesting that sulL is not essential for sulfazecin biosynthesis (Figure 4).
Figure 4. Construction of Gene Disruptants.
Construction of Gm-GFP integrant (P. acidophila KGmM-3)and unmarked plasmid-free(P.acidophila ΔsulL) mutants of sulL, and analysis for sulfazecin production by nitrocefin assays of each strain (Figure S5).
As a control, sulG, encoding one of the two enzymes responsible for the synthesis of L-2,3-Dap, was knocked out by a similar approach and confirmed by PCR and DNA sequence analysis. Bioassay on E. coli ESS, nitrocefin assay, and UPLC-HRMS confirmed that sulfazecin production was completely eliminated in the mutant P. acidophila FGmH-11 (Figures 5A and S5B). The production of sulfazecin could be restored, however, by chemical complementation in P. acidophila FGmH-11, as shown by bioassay and UPLC-HRMS when the fermentation broth was supplemented with L-2,3-Dap (Figure 5B). These results demonstrated that SulG is essential for sulfazecin biosynthesis and are consistent with the expectation that SulG (SbnB) and SulH (SbnA) are responsible for the synthesis of the direct β-lactam precursor L-2,3-Dap.
Figure 5. UPLC-HRMS Chromatograms and Antimicrobial Analyses of Products and Intermediates in Different P. acidophila Strains.
(A) P. acidophila FGmH-11, sulfazecin-deficient knockout.
(B) Sulfazecin produced in P. acidophila FGmH-11 supplemented with L-2,3-Dap.
(C) Desmethoxysulfazecin intermediate purified from the wild-type P. acidophila.
(D) Multiple sequence alignment of SulTE against TE domains in sulfazecin-like BGCs, and in PKS and NRPS TEs having high-resolution crystal structures. The active-site catalytic residue is labeled in red (*), and the GXSXG or AXCXG motifs are highlighted. SulTE, P. acidophila ATCC 31363; 31532, C. violesium 31532; DM48, B. gladioli ATCC 25417 DM48; E264, B. thailandensis E264; Y049, B. pseudomallei MSHR684 Y049; BP3921g, B. pseudomallei BP_3921g; 3QIT, curacin biosynthesis; 3ILS, aflatoxin PksA; 3QMV, prodiginine biosyn-thesis; 3LCR, tautomycetin biosynthesis; 1MNA, pikromycin biosynthesis.
Proposed Biosynthetic Pathway of Sulfazecin
The sulfazecin BGC has been mapped and characterized in the producer P. acidophila ATCC 31363. It is the first monobactam cluster to be described and whose core NRPS enzymes evince a mechanism of β-lactam ring biosynthesis different from the three other known pathways. In both clavam and carbapenem biosynthesis, the azetidinone ring arises in an ATP-dependent cyclization of a β-amino acid intermediate (Bachmann et al., 1998; McNaughton et al., 1998; Miller et al., 2003). Penicillin, cephalosporins, and cephamycins all devolve from the remarkable double oxidative cyclization of an NRPS-derived tripeptide to isopenicillin N accompanied by the release of two equivalents of water (White et al., 1982). Their common origins in serine (Townsend and Brown, 1981, 1983) superficially suggest that the monocyclic β-lactam of nocardicin G (Figure 6B), a precursor to nocardicin A (Townsend and Wilson, 1988), and the monobactams (O'Sullivan et al., 1982) are identical, or at least very similar. As the data demonstrate here, this long-held supposition is incorrect.
Figure 6. The Proposed Biogenetic Pathway to Sulfazecin.
(A) Biosynthesis of L-2,3-diaminopropionic acid (Dap). (B) Structure of nocardicin G, the first β-lactam containing intermediate in the pathway to nocardicin A. (C) Alternative possible biosynthetic routes to sulfazecin.
PAPS, 3′-phosphoadenosine-5′-phosphosulfate; SAM, S-adenosyl methionine; TE, thioesterase; α-KG, α-ketoglutaric acid; L-phosphoSer, L-phosphoserine.
The sulfazecin pathway is efficiently initiated by two NRPSs, SulI and SulM, comprising three canonical modules. A1 binds and activates D-Glu directly. This amino acid is abundantly available in bacteria to support peptidoglycan synthesis. Next, A2 incorporates L-Ala, which is epimerized in M2 to D-Ala as the dipeptide is transferred to M3, which selectively inserts L-2,3-Dap (Figure 6A) to form the D,D,L-tripeptide bound to T3. For the synthesis of sulfazecin, whether N-sulfonation occurs in trans on the M3-bound tripeptide, after β-lactam formation, or after hydrolytic release of the tripeptide but before β-lactam formation remains to be established (Figure 6C). Notwithstanding, the biosynthesis of the β-lactam ring in monobactams must differ from the C domain-catalyzed β-elimination/addition C-N bond formation proposed for nocardicin G and subsequent four-membered ring closure followed by C-terminal epimerization and hydrolytic release in the TE domain (Gaudelli et al., 2015). Introduction of the C-3 methoxyl group in sulfazecin likely occurs post-β-lactam in a notable parallel to cephamycin biosynthesis (O'Sullivan and Abraham, 1980). During the purification of sulfazecin from P. acidophila, we identified desmethoxylsulfazecin (Figure 5C), which lacks the 3α-methoxyl group. The observation of this cometabolite suggests that β-lactam ring formation and N-sulfonation precede C-3 oxidation and methylation.
Using the sulfazecin gene cluster as query, an antiSMASH analysis identified several new putative sulfazecin-like clusters, mostly in Burkholderia species (Figures 5D and S6). The core regions of these clusters are similar to sulfazecin, including NRPSs, sulfotransferase, dioxygenase, methyltransferase, and the resistance genes. Surprisingly, genes encoding SbnA and SbnB orthologs for L-2,3-Dap synthesis are missing in most clusters, suggesting that, because of absent crosstalk with another locus, they could be silent due to the lack of precursor biosynthetic pathways. Given the common AXCXG motif in place of the conserved GXSXG signature of conventional TEs in these clusters (Figure 5D), we speculate that transfer of the T3-bound tripeptide to the Cys, before N-sulfonation or after, maintains an active thioester in the TE poised for intramolecular nucleophilic addition to form the β-lactam ring (Figure 6C).
SIGNIFICANCE
The monobactams are the last class of β-lactam antibiotics for which a representative BGC has been found. Renewed interest in this category of drugs stems from their resistance to Zn2+ metallo-β-lactamases that confer extended-spectrum β-lactam resistance. The single monobactam in clinical use is aztreonam, which is manufactured entirely synthetically. With the sulfazecin gene cluster now in hand and allied biosynthetic systems identified, both fermentation and semi-synthesis methods can be contemplated for the efficient production of modified entities.
MbtH superfamily members can play essential roles in trans (and occasionally in cis) (Herbst et al., 2013) for A domain function (Baltz, 2011; Davidsen et al., 2013; Felnagle et al., 2010; Zhang et al., 2010). A second consideration was discovered in the course of this investigation that the domain context of a tested A domain can also affect the apparent substrate selectivity observed. When A2 and A2T2 were examined under conventional conditions, weak activity was observed favoring L-2,3-Dap and not L-Ala. Finally, the C2A2T2 tridomain, by more closely duplicating the in vivo context, proved selective and efficient for L-Ala. Related substrate selectivity experiments with A domains have been reported recently (Meyer et al., 2016).
The SulM TE domain contains an aberrant active site motif in which the expected Ser residue has been replaced by Cys. It is tempting to speculate that transfer of the intermediate tripeptide to the TE Cys would reside as the active thioester rather than an oxyester to facilitate both β-lactam formation and concomitant product release. While this second cyclization step resembles monocyclic β-lactam synthesis in nocardicin G, the prior step to form the immediate β-amino acid precursor is clearly different for the two pathways and, as a consequence, leads to terminal versus embedded azetidinone products. Mechanistic studies to investigate the former in monobactam biosynthesis and the timing of N-sulfonation will be reported in due course.
EXPERIMENTAL PROCEDURES
Production, Purification, and Characterization of Sulfazecin
Bacterial strains and plasmids used in this research are listed in Table S1. P. acidophila ATCC 31363 and its derived mutants were grown on King's B medium (Wilson et al., 1994). For liquid culture, the seed medium (Imada et al., 1980) was inoculated with a single colony, and grown at 28°C–30°C for 48 hr with rotation at 250 rpm. For production of sulfazecin, the fermentation medium (Imada et al., 1980) was inoculated with seed culture at a 20:1 ratio and the fermentation was carried out for 36–38 hr under the same conditions used for seed culturing. For chemical rescue of P. acidophila FGmH-11, 2 mM L-2,3-Dap was fed to 1 L of culture at 13 hr of fermentation.
Sulfazecin was purified with a modified method (Asai et al., 1981). Fermentation broth (4 L) was centrifuged at 9,000 rpm for 30 min at 4°C and the supernatant was pooled, adjusted to pH 4.0, and loaded on an active charcoal column (1,000 mL). The column was washed twice with 1 L of ddH2O and eluted with 2.0 L of acetone:ddH2O (50/50, v/v). Active fractions, determined by nitrocefin assay, were pooled and loaded on a column containing 200 mL of Dowex Cl−. The resin was washed twice with 200 mL of ddH2O and eluted with 500 mL of 5% NaCl. Active fractions were pooled and loaded on a 150-mL active charcoal column. The carbon was washed with 100 mL of ddH2O and eluted with 200 mL of acetone:ddH2O (50/50, v/v). The active fractions were either used directly for UPLC-HRMS chromatography (Waters ACQUITY/Xevo G-2 UPLC-MS) or after further purification by HPLC (Waters preparatory BEH Amide column and Agilent 1100 series HPLC).
Isolation of Sulfazecin-Deficient, sul−, Mutants
The CarR (carbenicillin)/KmR transconjugants were spotted on plates with sulfazecin-producing agar medium. After incubation at 30°C for 72 hr, plates were overlaid with nutrient agar seeded with E. coli ESS. Sulfazecin production in transconjugants was determined by the appearance of inhibition zones after growth of the overlaid plates at 37°C for 16 hr. The sul− transconjugants lacking inhibition zones were purified and inoculated into 3 mL of sulfazecin seed medium and grown at 30°C for 48 hr. Seed culture (100 μL) was transferred to 5 mL of sulfazecin-producing medium and fermented for 72 hr. The sulfazecin non-producers were determined by bioassay on E. coli ESS and by β-lactamase-induction assay with nitrocefin and further confirmed by UPLC-HRMS chromatography (Sykes and Wells, 1985).
ATP/PPi Exchange Assay
Each 150-μL reaction mixture consisted of 50 mM HEPES (pH 7.5), 1.5 mM DTT, 1.0 mM MgCl2, 40 mM KCl, 4 mM ATP, 5 mM NaPPi, 0.75 mM amino acid substrate, 1 μCi [32P] NaPPi, 5% glycerol, and 3.50–5.0 mM enzyme. The reaction was initiated by the addition of enzyme and followed by incubation at room temperature for 30 min. The reaction was quenched with 750 μL of a charcoal suspension (100 mM NaPPi, 350 mM HClO4, and 16 g/L of active charcoal [Norit]). The samples were vortexed and then centrifuged at 14,000 rpm for 5 min. The pellets were washed two to three times with 1 mL of washing solution (100 mM NaPPi, 350 mM HClO4). The pellets were resuspended with 500 μL ddH2O, and mixed with 5 mL of Opti-Fluor (NEN, PerkinElmer) in a 7-mL glass scintillation vial. Charcoal-bound radioactivity was measured on a Beckman LS 6500 scintillation counter.
A modified ATP/PPi exchange assay was used for the A1 domain. Each 150-μL reaction consisted of 5.0 μM A1, 2.5 mM amino acid test substrate, 1 mM DTT, 2.5 mM ATP, 10 mM MgCl2, 2.5 mM NaPPi, 2 μCi 32P-labeled NaPPi, 5% glycerol, and 50 mM Tris-HCl (pH 7.5). Prior to the addition of ATP, the reaction mixture was incubated at 30°C for 10 min. The reaction was initiated by adding ATP, and the reaction was incubated at 30°C for an additional 30 min. The ATP/PPi exchange reaction was quenched and prepared for scintillation counting as described in the standard procedure (Davidsen et al., 2013).
Targeted Gene Disruption and Characterization of Disruption Mutants
First, the pEX18Tc plasmid (Hoang et al., 1998) was modified by replacing the 800-bp tetracycline-resistant cassette with the 1,323-bp kanamycin-resistant cassette excised from SuperCos 1 cosmid. The resulting vector was named as pEX18Km.
The knockout vectors were constructed by Gibson assembling: two fragments sized between 1.5 and 3.0 kb upstream and downstream of the targeted gene and the Gm-GFP cassette were amplified by PCR designed by the NEBuilder server (http://nebuilder.neb.com/#) (Table S1). The disruption constructs were assembled into the BamHI or SmaI site in pEX18Km to form the final replacement vector. The Gm-GFP integrant vector was transformed into wild-type P. acidophila by electroporation or by conjugation. The transformed cells were grown in sulfazecin seed medium containing 15% sucrose for 12 hr at 28°C and plated on YM agar containing 200 μg/mL of gentamicin and 15% sucrose. The GmR clones were replicated on YM agar containing 50 μg/mL of kanamycin. The GmR/KmS clones were confirmed for double-crossover events by PCR and DNA sequencing analysis. To generate a plasmid-free and unmarked mutants, plasmid pFLP2Km, derived from pFLP2 (Hoang et al., 1998) by replacing the ampicillin-resistance gene with the kanamycin-resistance cassette, was transformed into the Gm-GFP integrant mutants. Flp-catalyzed excision of the Gm-GFP markers leaves a short FRT-containing sequence. Plasmid pFLP2Km was cured by generously streaking of GmS cells on YM agar supplemented with 5% sucrose followed by incubation at 30°C. Loss of pFLP2Km was tested by replicating clones on kanamycin-containing plates.
Supplementary Material
Highlights.
The first monobactam biosynthetic gene cluster is isolated and characterized
Two non-ribosomal peptide synthetases play essential biosynthetic roles
A new mechanism of β-lactam formation is catalyzed in an aberrant thioesterase domain
ACKNOWLEDGMENTS
We thank Professor D.K. Willis, University Wisconsin, for providing E. coli PR47 and Professor H.P. Schweizer, Colorado State University, for supplying plasmids pEX18Tc, pPS858, and pFLP2. We are grateful to Dr. K.A. Moshos for preparing nitrocefin, and to M. Lau for valuable comments and reference collection. This work was supported by the NIH (AI 014937).
Footnotes
ACCESSION NUMBERS The DNA sequences of the sulfazecin gene cluster have been deposited to the NCBI nucleotide sequence database under accession number GenBank: KX757706.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2016.11.010.
AUTHOR CONTRIBUTIONS R.F.L. and C.A.T. designed and directed the research; R.F.L. performed research and R.A.O. performed UPLC-HRMS analyses; R.F.L. and R.A.O. analyzed the data; and R.F.L., C.A.T., and R.A.O. wrote the paper.
REFERENCES
- Aoki H, Sakai H, Kohsaka M, Konomi T, Hosoda J. Nocardicin A, a new monocyclic β-lactam antibiotic, I. discovery, isolation and characterization. J. Antibiot. 1976;29:492–500. doi: 10.7164/antibiotics.29.492. [DOI] [PubMed] [Google Scholar]
- Araoka H, Baba M, Tateda K, Ishii Y, Oguri T, Okuzumi K, Oishi T, Mori S, Mitsuda T, Moriya K, et al. Monobactam and aminoglyco-side combination therapy against metallo-β-lactamase-producing multidrug-resistant Pseudomonas aeruginosa screened using a `break-point checkerboard plate'. Jpn. J. Infect. Dis. 2012;65:84–87. [Google Scholar]
- Asai M, Haibara K, Muroi M, Kintaka K, Kishi T. Sulfazecin, a novel β-lactam antibiotic of bacterial origin. Isolation and chemical characterization. J. Antibiot. 1981;34:621–627. doi: 10.7164/antibiotics.34.621. [DOI] [PubMed] [Google Scholar]
- Bachmann BO, Ravel J. In silico prediction of microbial secondary metabolic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- Bachmann BO, Li R-F, Townsend CA. β-Lactam synthetase: a new biosynthetic enzyme. Proc. Natl. Acad. Sci. USA. 1998;95:9082–9086. doi: 10.1073/pnas.95.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JE, Lloyd MD, Wha-Son B, Schofield CJ, Elson SW, Baggaley KH, Nicholson NH. A substrate analogue study on clavaminic acid synthase: possible clues to the biosynthetic origin of proclavamic acid. J. Chem. Soc. Chem. Commun. 1993:500–502. 1993. [Google Scholar]
- Baltz RH. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 2011;38:1747–1760. doi: 10.1007/s10295-011-1022-8. [DOI] [PubMed] [Google Scholar]
- Beasley F, Cheung J, Heinrichs D. Mutation of L-2,3-diaminopropionic acid synthasegenes blocks staphyloferrin B synthesis in Staphylococcus aureus. BMC Microbiol. 2011;11:199–211. doi: 10.1186/1471-2180-11-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebrone C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007;74:1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Brazilian National Genome Project Consortium The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA. 2003;100:11660–11665. doi: 10.1073/pnas.1832124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby RW, Townsend CA. A single monomeric iron center in clavaminate synthase catalyzes three nonsuccessive oxidative transformations. Bioorg. Med. Chem. 1996;4:1059–1064. doi: 10.1016/0968-0896(96)00088-0. [DOI] [PubMed] [Google Scholar]
- Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Crusemann M, Kohlhaas C, Piel J. Evolution-guided engineering of nonribosomal peptide synthetase adenylation domains. Chem. Sci. 2013;4:1041–1045. [Google Scholar]
- Davidsen JM, Bartley JM, Townsend CA. Non-ribosomal pro-peptide precursor in nocardicin A biosynthesis predicted from adenylation domain specificity dependent on the MbtH family protein NocI. J. Am. Chem. Soc. 2013;135:1749–1759. doi: 10.1021/ja307710d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier JD, Riccio ML, Mugnaioli C, Luzzaro F, Endimiani A, Toniolo A. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 2003:1522–1528. doi: 10.1128/AAC.47.5.1522-1528.2003. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felnagle EA, Barkei JJ, Park H, Podevels AM, McMahon MD, Drott DW, Thomas MG. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry. 2010;49:8815–8817. doi: 10.1021/bi1012854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Long DH, Townsend CA. β-Lactam formation by a non-ribosomal peptide synthetase during antibiotic biosynthesis. Nature. 2015;520:383–387. doi: 10.1038/nature14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller DH, Henry JG, Belch J, Schwartzj NB. Co-purification and characterization of ATP-sulfurylase and adenosine-5′-phosphosulfate kinase from rat chondrosarcoma. J. Biol. Chem. 1987;262:7374–7382. [PubMed] [Google Scholar]
- Gueznzi E, Galli G, Grgurina I, Gross DC, Grandi G. Characterization of the syringomycin synthetase gene cluster a link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem. 1998;273:32857–32863. doi: 10.1074/jbc.273.49.32857. [DOI] [PubMed] [Google Scholar]
- Herbst DA, Roll B, Zocher G, Stehle T, Heide L. Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J. Biol. Chem. 2013;288:1991–2003. doi: 10.1074/jbc.M112.420182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Horsman ME, Hari TPA, Boddy CN. Polyetide synthase and non-ribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate? Nat. Prod. Rep. 2016;33:183–202. doi: 10.1039/c4np00148f. [DOI] [PubMed] [Google Scholar]
- Hou J, Robbel L, Marahiel MA. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem. Biol. 2011;18:655–664. doi: 10.1016/j.chembiol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Imada A, Kazuaki K, Asai M. Antibiotic G-6302, U.S.P. 4229436 (Takeda Chemical Industries) 1980. pp. 1–11. [Google Scholar]
- Kato T, Hinoo H, Shoji J, Matsumoto K, Hattori T, Hirooka K, Kionda E. PB-5266 A, B and C, new monobactams. I. Taxonomy, fermentation and isolation. J. Antibiot. 1987;40:135–138. doi: 10.7164/antibiotics.40.135. [DOI] [PubMed] [Google Scholar]
- King DT. Targeting metallo-β-lactamase enzymes in antibiotic resistance. Future Med. Chem. 2013;5:1243–1263. doi: 10.4155/fmc.13.55. [DOI] [PubMed] [Google Scholar]
- King DT, Sobhanifar S, Strynadka NCJ. One ring to rule them all: current trends in combating bacterial resistance to the β-lactams. Protein Sci. 2016;25:787–803. doi: 10.1002/pro.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobylarz MJ, Grigg JC, Takayama SJ, Rai DK, Heinrichs DE, Murphy MEP. Synthesis of L-2,3-diaminopropionic acid, a siderophore and antibiotic precursor. Chem. Biol. 2014;21:379–388. doi: 10.1016/j.chembiol.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Kries H, Niquille DL, Hilvert D. A subdomain swap strategy for reengineering nonribosomal peptides. Chem. Biol. 2015;22:640–648. doi: 10.1016/j.chembiol.2015.04.015. [DOI] [PubMed] [Google Scholar]
- Lee SG, Lipmann F. Tyrocidine synthetase system. Methods Enzymol. 1975;43:585–602. doi: 10.1016/0076-6879(75)43121-4. [DOI] [PubMed] [Google Scholar]
- Leskiw BK, Aharonowitz Y, Mevarech M, Wolfe S, Vining LC, Westlake DWS, Jensen SE. Cloning and nucleotide sequence determination of the isopenicillin N synthetase gene from Streptomyces clavuligerus. Gene. 1988;62:187–196. doi: 10.1016/0378-1119(88)90557-4. [DOI] [PubMed] [Google Scholar]
- McKenna M. The last resort. Nature. 2013;499:394–396. doi: 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- McNaughton HJ, Thirkettle JE, Zhang Z, Schofield CJ, Jensen SE, Barton B, Greaves P. β-Lactam synthetase: implications for β-lactamase evolution. Chem. Commun. 1998:2325–2326. 1998. [Google Scholar]
- Meyer S, Kehr JC, Mainz A, Dehm D, Petras D, Sussmuth RD, Dittmann E. Biochemical dissection of the natural diversification of microcystin provides lessons for synthetic biology of NRPS. Cell Chem. Bio. 2016;23:462–471. doi: 10.1016/j.chembiol.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Miller MT, Gerratana B, Stapon A, Townsend CA, Rosenzweig AC. Crystal structure of carbapenam synthetase (CarA) J. Biol. Chem. 2003;278:40996–41002. doi: 10.1074/jbc.M307901200. [DOI] [PubMed] [Google Scholar]
- Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. Structure and function of sulfotransferases. Arch. Biochem. Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- O'Sullivan J, Abraham EP. The conversion of cephalosporins to 7 alpha-methoxycephalosporins by cell-free extracts of Streptomyces clavuligerus. Biochem. J. 1980;186:613–616. doi: 10.1042/bj1860613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan J, Gillum AM, Aklonis CA, Souser ML, Sykes RB. Biosynthesis of monobactam compounds: origin of the carbon atoms in the β-lactam ring. Antimicrob. Agents Chemother. 1982;21:558–564. doi: 10.1128/aac.21.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan J, Souser ML, Kao CC, Aklonis CA. Sulfur metabolism in the biosynthesis of monobactams. Antimicrob. Agents Chemother. 1983;23:598–602. doi: 10.1128/aac.23.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O. New promising β-lactamase inhibitors for clinical use. Eur. J. Clin. Microbiol. Infect Dis. 2015;34:1303–1308. doi: 10.1007/s10096-015-2375-0. [DOI] [PubMed] [Google Scholar]
- Parker WL, O'Sullivan J, Sykes RB. Naturally occurring monobactams. Adv. Appl. Microbiol. 1986;31:181–205. doi: 10.1016/s0065-2164(08)70442-8. [DOI] [PubMed] [Google Scholar]
- Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucl. Acid Res. 2005;33:5799–5807. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer D, Finking R, Marahiel MA. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 2003;20:275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- Sykes RB, Bonner DB. Discovery and development of the monobactams. Rev. Infect Dis. 1985;7:S579–S593. doi: 10.1093/clinids/7.supplement_4.s579. [DOI] [PubMed] [Google Scholar]
- Sykes RB, Wells JS. Screening for b-lactam antibiotics in nature. J. Antibiot. 1985;38:119–121. doi: 10.7164/antibiotics.38.119. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Chan YA, Ozanick SG. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob. Agents Chemother. 2003;47:2823–2830. doi: 10.1128/AAC.47.9.2823-2830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend CA, Brown AM. Biosynthetic studies of nocardicin A. J. Am. Chem. Soc. 1981;103:2873–2874. [Google Scholar]
- Townsend CA, Brown AM. Nocardicin A: biosynthetic experiments with amino acid precursors. J. Am. Chem. Soc. 1983;105:913–918. [Google Scholar]
- Townsend CA, Wilson BA. The role of nocardicin G in nocardicin A biosynthesis. J. Am. Chem. Soc. 1988;110:3320–3321. [Google Scholar]
- Wang M, Gould SJ. Biosynthesis of capreomycin. 2. Incorporation of L-serine, L-alanine, and L-2, 3-diaminopropionic acid. J. Org. Chem. 1993;58:5176–5180. [Google Scholar]
- Weber T, Blin K, Duddela S, Krug D, Kim H-U, Bruccoleri R, Lee S-Y, Fischbach MA, MÄller R, Wohlleben W, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucl. Acid Res. 2015;43:w237–w243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JS, Trejo WH, Principe PA, Georgopapadakou NH, Sykes RB. SQ 26,180, a novel monobactam. I taxonomy, fermentation and biological properties. J. Antibiot. 1982;35:184–188. doi: 10.7164/antibiotics.35.184. [DOI] [PubMed] [Google Scholar]
- White RL, John EMM, Baldwin JE, Abraham EP. Stoichiometry of oxygen consumption in the biosynthesis of isopenicillin from a tripeptide. Biochem. J. 1982;203:791–793. doi: 10.1042/bj2030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DK, Hrabak EM, Rich JJ, Barta TM, Lindow SE, Panopoulos NJ. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol. Plant Microbe Interact. 1990;3:149–156. [Google Scholar]
- Wilson KJ, Sessitsch A, Akkermans A. Molecular markers as tools to study the ecology of microorganisms. In: Ritz K, Dighton J, Giller KE, editors. Beyond the Biomass. British Society of Soil Science; 1994. pp. 149–156. [Google Scholar]
- Xia S, Ma Y, Zhang W, Yang Y, Wu S, Zhu M, Deng L, Li B, Liu Z, Qi C. Identification of Sare0718 as an alanine-activating adenylation domain in marine actinomycete Salinispora arenicola CNS-205. PLoS One. 2012;7:e37487. doi: 10.1371/journal.pone.0037487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Heemstra JRJ, Walsh CT, Imker HJ. Activation ofthe pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry. 2010;49:9946–9947. doi: 10.1021/bi101539b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.