Abstract
Background
Hepatocellular carcinoma (HCC) is a common malignant tumor in the world, especially in China. As a member of the inhibitor of differentiation (Id) family, Id4 has been reported to function in many cancer types, but relatively little is known about its role in HCC. The purpose of this study was to investigate the potential relationship between Id4 and HCC development and the underlying mechanism involving the function of Id4 in HCC.
Methods
We used quantitative real-time polymerase chain reaction and Western blotting to examine the RNA and protein expression of Id4. In addition, we used Cell Counting Kit-8 assay and colony formation assay to identify the function of Id4 in the regulation of cell proliferation in human HCC.
Results
We found that the expression of Id4 protein was up-regulated in tumor tissues from HCC patients. Overexpression of Id4 promoted HCC cell proliferation, clonogenicity in vitro, and tumorigenicity in vivo. Id4 knockdown experiments showed that silencing Id4 blocked the proliferation and colony formation ability of HCC cells in vitro. Furthermore, overexpression of CCAAT/enhancer-binding protein β inhibited Id4 expression in HCC cells.
Conclusion
Id4 may be developed as a potent therapeutic agent for the treatment of HCC, but more details about the underlying mechanisms of action are needed.
Keywords: Hepatocellular carcinoma, Id4, Proliferation
Background
Since the inhibitor of differentiation (Id) genes were first identified in 1990, more than 25 years of research has established that Id proteins are critical regulators in normal development and in cancer [1]. The Id family, a class of the helix-loop-helix (HLH) family, has four members; they lack a basic DNA-binding domain and function by forming heterodimers with other HLH family members to inhibit transcriptional activity [2]. Deregulation of Ids has been reported in many types of human cancers, such as prostate cancer, breast cancer, and ovarian cancer, and it may be strongly associated with poor prognosis and disease grade [3, 4].
Based on the sequence and structural property analyses, Id4 is a remote homologue of other Ids despite sharing the conserved HLH domain, which suggests a potential novel role for Id4 in the development of cancer [5]. Some data have shown that Id1, Id2, and Id3 enhance proliferation and block differentiation in certain cell types, whereas Id4 has an opposite function [6].
Worldwide and in China, hepatocellular carcinoma (HCC), which accounts for most (70%–90%) primary liver cancers, is a leading cause of cancer death [7]. As reported previously, Id proteins are involved in the development of HCC. Lee et al. [8] first reported the overexpression of Id1 in HCC, which results in the inactivation of the p16INK4a/retinoblastoma pathway and leads to aberrant proliferation of HCC cells. Analysis of clinical samples showed a complex expression profile of Id1, Id2, and Id3 and their relevance to the de-differentiation in HCC, which was different from the expression profile in breast, prostate, and colon carcinogenesis [9].
Recent studies have also demonstrated the function of Id4 in breast and prostate cancers. Beger et al. [10] found that, in breast cancer, Id4 played a role in the down-regulation of breast cancer 1 (BRCA1); its overexpression enhanced the tumorigenic potential of cells [11]. Moreover, Junankar et al. [12] found that Id4 could be a vital regulator of mammary stem cells, because it could suppress the factors that contributed to luminal differentiation, maintaining the mammary stem cell pool; these results are consistent with the earlier finding demonstrating the positive role of Ids in cell growth but negative role in cell differentiation [10]. These findings show that Id4 is a tumor promoter in breast cancer but that its function varies depending on the subtype and the developmental stage of cancer. On the contrary, other studies showed that Id4 has a tumor-suppressive effect. In prostate cancer, Id4 was down-regulated because of promoter hypermethylation, which provided evidence that Id4 may be a tumor suppressor [13, 14]. However, the expression pattern and function of Id4 protein in HCC have not yet been determined.
In this study, we explored the potential relationship between Id4 and HCC development as well as the underlying mechanism involving the function of Id4 in HCC.
Methods
Cell lines and cell culture
The human HCC cell line SMMC-7721 was obtained from the Cell Bank of the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The PLC/PRF/5, HepG2, SK-HEP-1, and Hep3B2.1-7 cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). The Huh7 cell line was obtained from the Riken Cell Bank (Tsukuba, Japan). The MHCC-97L and MHCC-LM3 cell lines were kindly provided by the Liver Cancer Institute, Zhongshan Hospital of Fudan University (Shanghai, China). All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) at 37 °C in a 5% CO2 incubator. Standard transient transfections for all cell lines were conducted using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from tissues and cells using TRIzol reagents (Invitrogen, Carlsbad, CA, USA). One microgram of total RNA was reversely transcribed with the Prime Script RT Reagent Kit (Perfect Real Time) (TaKaRa Biotechnology, Dalian, China). Polymerase chain reaction (PCR) analysis was performed using specific primers for the Id4 gene: forward, 5′-GTGCGATATGAACGACTGCT-3′, and reverse, 5′-CAGGATCTCCACTTTGCTGA-3′. The expression levels were normalized using human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as an internal control: forward, 5′-AGAAGGCTGGGGCTCATTTG-3′, and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′.
Protein isolation and Western blotting
After specific treatments, proteins (20 µg) were separated using 12% SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) and transferred to nitrocellulose membrane by an electroblotting Bradford assay, according to the manufacturer’s instructions (Sigma-Aldrich). The anti-Id4 monoclonal antibody (sc-365656, 1:100) and anti-C/EBPβ (CCAAT/enhancer-binding protein β) polyclonal antibody (sc-150, 1:400) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and the β-actin antibody (A3854, 1:10,000) was purchased from Sigma-Aldrich.
Patient samples
Twenty-seven human HCC tissue samples were obtained from the Qidong Liver Cancer Institute (Qidong, Jiangsu, China). Tumor tissues and adjacent non-tumor tissues were used to detect the Id4 mRNA and protein levels by real-time PCR and Western blotting. All procedures were performed under consensus agreements and in accordance with the China Ethical Review Committee.
Immunohistochemical analysis
Fifty-seven human HCC tissue specimens were collected from patients who underwent surgical treatment at Qidong Liver Cancer Institute or at the First Affiliated Hospital of Zhejiang University (Hangzhou, Zhejiang, China). The 57 HCC patients, including 52 males and 5 females (mean age 45.0 years, ranging from 21.0 to 70.0 years), were followed up from November 21, 2001 to November 3, 2010. No patient received preoperative chemotherapy or radiotherapy. Informed consent was obtained from all patients, and the study was approved by the Ethics Committee of Fudan University.
Anti-Id4 polyclonal antibody (sc-491) was purchased from Santa Cruz Biotechnology. Immunohistochemical (IHC) analysis and signal evaluation were performed according to our previously described procedures [15]. All the HCC tissue slides were observed and photographed using an Axioskop 2 microscope (Carl Zeiss, Oberkochen, Germany). The IHC results were determined according to both staining intensity and the percentage of positive cells as described previously [15].
Plasmid constructs for overexpression of Id4 and C/EBPβ and RNA interference of Id4
The full-length human Id4 and C/EBPβ open reading frame (ORF) were respectively generated and cloned into the lentiviral vector pWPLX (Addgene, Cambridge, MA, USA) at the BamHI and EcoRI sites. The primers of Id4 ORF used for cloning and testing were as follows. Forward: 5′-GGATCCATGAAGGCGGTGAGCCCG-3′; reverse: 5′- GAATTCTCAGCGGCACAGAATGCT-3′. The primers of C/EBPβ (LAP1) ORF used for cloning and testing were as follows. Forward: 5′-CGCGGATCCATGCAACGCCTGGTGGCCT-3′; reverse: 5′-CCGGAATTCCTAGCAGTGGCCGGAGGAG-3′. We ordered two small-interfering RNAs (siRNAs) and two short-hairpin RNAs (shRNAs) targeting Id4, which were synthesized and constructed, respectively, by the GenePharma (Shanghai, China). The shId4 and shNC sequence (siId4-1, 5′-GCACGUUCAUAAACAUUCUTT-3′; siId4-2, 5′-CCCAACAAGAAAGUCAGCATT-3′; and siNC, 5′-TTCTCCGAACGTGTCACGT-3′) were cloned into the lentiviral vector pLVTHM (Addgene, Cambridge, MA, USA) to construct pLVTHM-shId4 and pLVTHM-shNC. To verify the effect of overexpression or gene silencing, real-time PCR and Western blotting were performed.
Colony formation assays
For colony formation assays, 500 SMMC-7721, 2000 MHCC-97L, and 3000 Huh7 cells per well were seeded in 6-well plates and cultured at 37 °C for 10–14 days. Then, the cells were fixed in 10% formaldehyde for 20 min and stained for 30 min with Giemsa solution (Sigma-Aldrich). Each measurement was performed in triplicate, and the experiments were each conducted three times.
Cell growth assay
Cell proliferation analyses were performed using a WST-8 Cell Counting Kit-8 (CCK-8, Ruian Biotech, Shanghai, China). Cells (800 for SMMC-7721, 2000 for MHCC-97L, and 1500 for Huh7) suspended in DMEM (100 μL) with 10% fetal bovine serum were seeded in 96-well plates and incubated. After 24 h, 10 μL of CCK-8 solution was added to each well, and the cultures were incubated at 37 °C for 2 h. Absorbance was measured at 450 nm for 7 days. The relative absorbance value was calculated, and the absorbance value measured on the first day was used as a control. Each measurement was performed in triplicate, and the experiments were each conducted three times.
Tumor xenograft assay
Six- to eight-week-old male BALB/c (nu/nu) mice were housed and treated under specific pathogen-free conditions at the Experimental Animal Center of Shanghai Jiaotong University School of Medicine (Shanghai, China). They were randomly divided into groups (eight mice per group) and maintained under standard conditions according to institutional animal guidelines. SMMC-7721-Id4 cells and their pWPXL vector control (SMMC-7721-pWPXL) cells (2 × 106 cells per mouse) were separately injected subcutaneously into the right flank of nude mice. After 5 weeks, the mice were euthanized, and the xenograft tumors were weighted.
Statistical analysis
Data were analyzed using SPSS 13.0 software (IBM Corporation, New York, NY, USA). Results are presented as mean ± standard deviation and compared using Student’s t test. The overall survival was calculated from the 4th month after hepatectomy to the date of death or the last follow-up. Univariate survival analysis was performed according to the Kaplan–Meier method, and differences in survival curves were assessed with the log-rank test. P values less than 0.05 were considered statistically significant.
Results
Id4 expression in HCC samples and cell lines
In 27 pairs of human HCC specimens, we detected the expression of Id4 by real-time PCR and Western blotting. Although Id4 mRNA expression was up-regulated in adjacent non-cancer tissues as compared with cancer tissues from 27 cases (Fig. 1a), no significant difference was found between the two groups of tissues (Fig. 1b). However, 63.0% (17/27) of tumor samples showed up-regulated expression of Id4 protein; 18.5% (5/27) showed no difference; and the remaining 18.5% (5/27) showed a decrease of Id4 expression, compared with the corresponding non-cancerous liver samples (Fig. 1c, d). This may be partially due to the genetic heterogeneity in patients or contamination of tumor cells in some analyzed adjunctive liver tissues. Additionally, the Pearson correlation analysis results showed that protein levels of Id4 did not correlate with mRNA levels in tumor (r = −0.108, P = 0.592) or non-tumor (r = −0.010, P = 0.960) tissues.
Fig. 1.
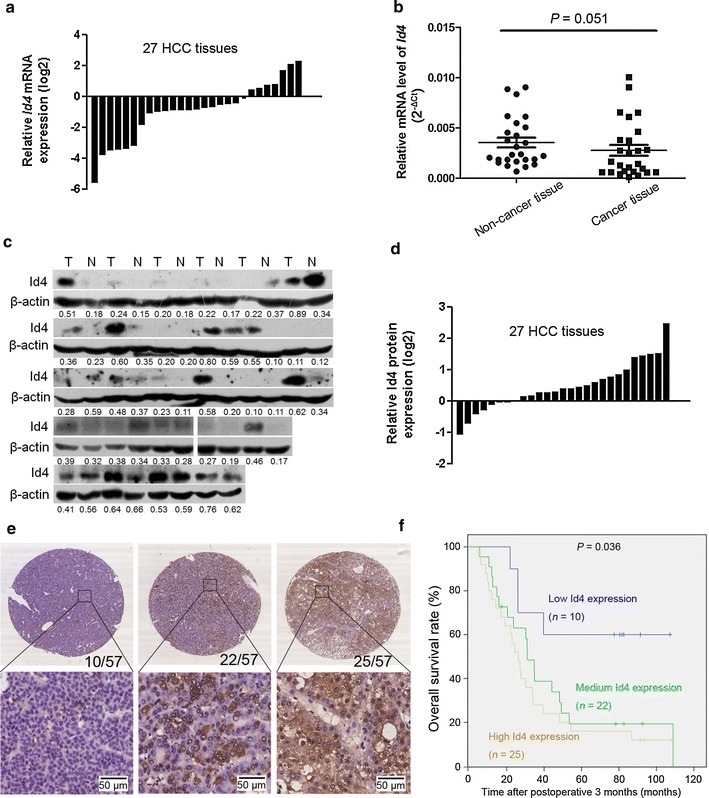
Inhibitor of differentiation 4 (Id4) was frequently up-regulated in hepatocellular carcinoma (HCC) cancer tissues at the protein level but not at the mRNA level. a The relative Id4 mRNA expression in HCC tumor tissues is compared with the adjacent non-cancer tissues. b Compared with the adjacent non-cancer tissues, the mRNA expression of Id4 is not significantly up-regulated in HCC tumor tissues. Paired Student’s t test, P = 0.051. ns not significant. c The up-regulated protein expression of Id4 is observed in 63.0% (17/27) of tumor tissues. Densitometry of Western blotting (measured by Image J) showed the relative protein expression (the numbers, IntDenId4/IntDenActin). d The relative Id4 protein expression in HCC tissues is compared with that in the adjacent non-cancer tissues. Densitometry of Western blotting (measured by Image J) showed the relative protein expression (IntDenId4/IntDenActin). e Representative Id4 immunostaining in a HCC tissue array containing 57 samples: no positive immunostaining is detected in 10 cases; Id4-positive cells are scattered or focally clustered in 22 cases; and diffuse staining pattern is present in 25 cases. The number of cases in each subset is shown in the right corner of the upper panel. Original magnifications: up ×4 and down ×40. T: tumor tissues; N adjacent non-cancer tissues. f Kaplan–Meier survival curve showing the overall survival of 57 HCC patients according to the Id4 expression level. *P < 0.05
To characterize the Id4 expression patterns, immunohistologic analysis was performed in a tissue array. Of the 57 HCC samples, Id4 expression was detected in 47 (82.5%) cases but not detected in 10 (17.5%) cases (Fig. 1e). In the 47 Id4-positive HCC samples, Id4-positive cells were scattered or focally clustered in 22 (38.6%) cases, and a diffuse staining pattern was present in 25 (43.9%) cases, which indicated that Id4 protein was overexpressed in most HCC tissues. To evaluate the association between the expression levels of Id4 and patient survival, a univariate analysis of overall survival was performed using the Kaplan–Meier method, and a log-rank test was performed. The result showed that patients with low Id4 expression had a higher survival rate when compared with medium or high Id4 expression groups (P = 0.036) (Fig. 1f). As an important transcriptional regulator [16], the increased level of Id4 expression may provide a clue regarding the role of Id4 in the development of liver cancer.
We also analyzed the expression of Id4 in HCC cell lines by real-time PCR and Western blotting (Fig. 2a, b). Id4 protein expression was markedly high in three cell lines (especially Huh7), and the mRNA level of Id4 was in line with the protein expression. A positive correlation was found between Id4 mRNA and protein expression in HCC cell lines (r = 0.799, P = 0.017), which was different from the expression determined in patient tissue samples.
Fig. 2.
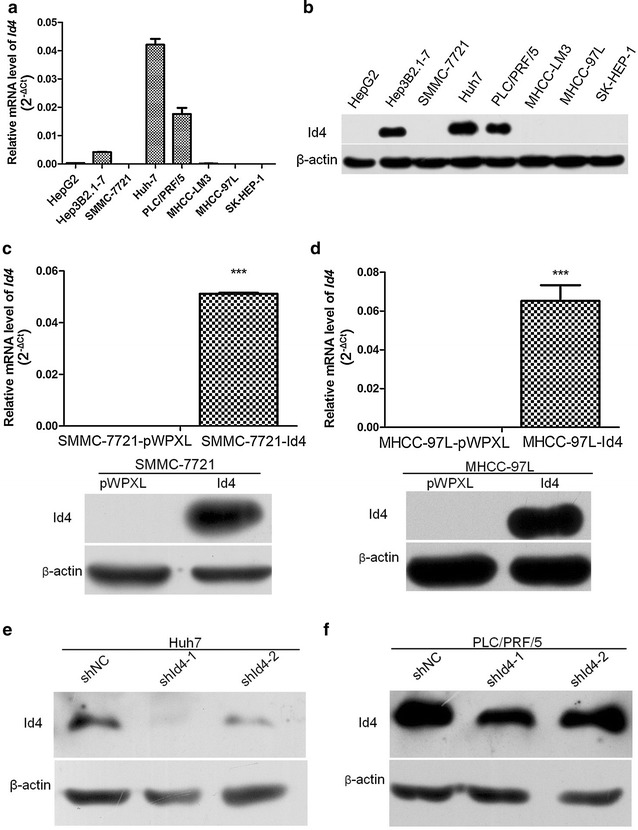
Expression of Id4 in HCC cell lines. a, b The mRNA and protein levels of Id4 in eight HCC cell lines. c, d Compared with the empty vector group, transfection with pMPXL-Id4 markedly up-regulates the expression of Id4 in SMMC-7721 and MHCC-97L. e, f In Huh7 and PLC/PRF/5 cells, stable transfection with shId4 markedly down-regulates the expression of Id4. ***P < 0.001
Id4 stimulated proliferation in vitro
Two cell lines with low Id4 expression, SMMC-7721 and MHCC-97L, were chosen for the construction of cell lines (SMMC-7721-Id4 and MHCC-97L-Id4) with overexpression of the Id4 gene. Then, total RNA and proteins were collected, and the steady state of Id4 was verified by real-time PCR and Western blotting (Fig. 2c, d). Two siRNA sequences were designed and synthesized, and endogenous Id4 expression was silenced in Huh7 and PLC/PRF/5 cells. Using Western blotting, we confirmed the transfection and Id4 knockdown efficiency in Huh7 and PLC/PRF/5 cells (Fig. 2e, f).
To assess the effect of Id4 overexpression on HCC cell growth, we performed CCK-8 assays and colony formation assays. From the fourth day to the seventh day after culturing, significant differences (P = 0.046, 0.04, 0.041, and 0.009) were observed in proliferation between the SMMC-7721-pWPXL and the SMMC-7721-Id4 groups (Fig. 3a). A similar result (P = 0.040, 0.025, 0.015, and < 0.001) was obtained in MHCC-97L cells (Fig. 3b). A significant decrease in proliferation was observed in the Huh7-shId4-1 (P = 0.015, 0.022, 0.018, and 0.019), Huh7-shId4-2 (P = 0.019 and 0.024), and PLC/PRF/5-shId4-1 (P = 0.011 and 0.024) cells when compared with the control cells transfected with shNC (Fig. 3c, d). Furthermore, HCC cells with Id4 overexpression formed more clones than the control cells transfected with empty vector pWPXL (SMMC-7721, P < 0.001; MHCC-97L, P = 0.028) (Fig. 3e). Moreover, when compared with the control cells transfected with shNC, the cells with down-regulated expression of Id4 (transfected with shId4-1 and shId4-2) formed significantly less number of cell clones in Huh7 (P = 0.023 and 0.029) and PLC/PRF/5 (P = 0.012 and 0.002) cells (Fig. 3f), which suggested that Id4 knockdown inhibited the cell growth ability of HCC cells. These results showed that increased accumulation of Id4 contributed to proliferation and clonogenicity in SMMC-7721 and MHCC-97L cells.
Fig. 3.
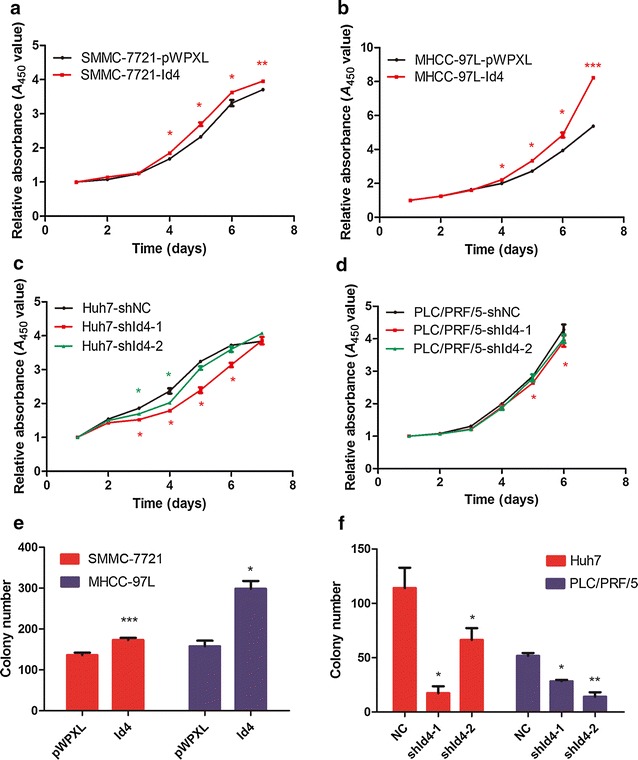
Id4 promoted proliferation and colony formation of HCC cells in vitro. The Cell Counting Kit-8 (CCK-8) assay was performed to examine cell proliferation. a, b Up-regulation of Id4 significantly promotes proliferation of SMMC-7721 and MHCC-97L cells. c, d Id4 knockdown significantly suppresses cell proliferation in Huh7 and PLC/PRF/5. e The number of colonies increased significantly in SMMC-7721-Id4 and MHCC-97L-Id4 groups. f The numbers of colonies decreased remarkably after Id4 silence in Huh7 and PLC/PRF/5groups. *P < 0.05, **P < 0.01, ***P < 0.001
C/EBPβ overexpression inhibited Id4 expression in Huh7 cells
To investigate the mechanism that regulates Id4 expression in HCC cells, we analyzed the Id4 promoter region using bioinformatics methods. Bioinformatics analysis showed that there were C/EBPβ-binding sites in the Id4 gene promoter region (Table 1). In Huh7 cells transfected with pWPXL-C/EBPβ, real-time PCR and Western blotting analysis showed that C/EBPβ mRNA and protein expression were significantly induced (Fig. 4a, b). Furthermore, Id4 mRNA and protein expression were inhibited in the C/EBPβ-overexpressing Huh7 cells, which indicated that C/EBPβ may regulate Id4 expression in HCC cells in a direct or indirect manner (Fig. 4c, d).
Table 1.
Predicted C/EBPß-binding sites in the Id4 promoter sequence

Predicted binding sites of C/EBPß protein in the Id4 promoter sequence are highlighted in yellow. The JASPAR database (http://jaspar.genereg.net/) was used in the bioinformatics analysis of DNA sequence
Fig. 4.
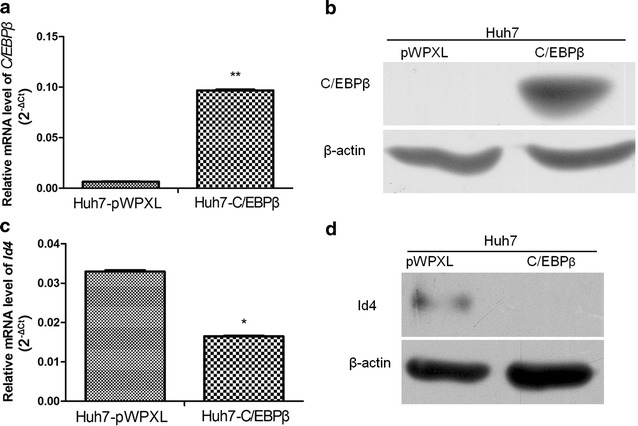
CCAAT/enhancer-binding protein β (C/EBPβ) overexpression inhibited Id4 expression in Huh7 cells. a, b Real-time PCR and Western blotting analysis show that C/EBPβ mRNA and protein expression are significantly induced in Huh7 cells. c, d Id4 mRNA and protein expression are inhibited in the C/EBPβ-overexpressing Huh7 cells. *P < 0.05, **P < 0.01
Id4 enhanced tumorigenicity potential in vivo
To determine whether overexpression of Id4 has an effect on tumor growth in vivo, animal experiments was carried out. The result showed that Xenograft tumors in the SMMC-7721-pWPXL group were smaller than that in the SMMC-7721-Id4 group (Fig. 5). A significant difference in xenograft tumor weight was observed between the two groups (0.23 ± 0.24 g in the SMMC-7721-pWPXL group and 0.42 ± 0.26 g in the SMMC-7721-Id4 group, P = 0.003). Our results indicate that Id4 overexpression significantly promoted the tumorigenicity of SMMC-7721 cells in this model system.
Fig. 5.
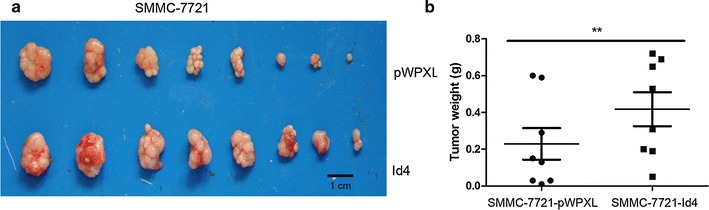
Overexpression of Id4 enhanced the tumorigenic potential of HCC cells in vivo. SMMC-7721 cells that stably expressed Id4 were injected subcutaneously into nude mice; the cells transfected with pWPXL vector were used as a control. After 5 weeks, the mice were euthanized, and the tumors were removed. a The xenograft tumors formed in nude mice. b The wet weight of the tumors. **P < 0.01
Discussion
In this study, we found high expression of Id4 in three cell lines (Huh7, PLC/PRF/5, and Hep3B2.1-7), which was similar to our previous findings of the expression trend of cluster of differentiation 133 (CD133) protein in HCC [17]. We also found that the expression of Id4 protein was up-regulated in HCC patient tumor samples. Additionally, immunohistochemical analysis showed that Id4 protein was overexpressed in most HCC patient samples, which provides new insights into the role of Id proteins in human HCC [9]. Although a positive correlation was observed between the Id4 mRNA and protein expression level in HCC cell lines, Id4 mRNA and protein expression showed no significant correlation in the HCC patient samples. We speculate that this may be partially due to the complex cell types comprising the tumor tissues. Alternatively, the discrepancy between the Id4 mRNA levels and protein levels may be a result of mediating activity by miRNAs or the different half-lives of proteins [18, 19]; either way, the detailed mechanism needs to be verified in further investigations. These findings indicate that Id4 might be a potent therapeutic agent in HCC.
Previous findings show that, in cancer, there are many modes of activation of Id genes, such as transcriptional induction by oncoproteins or growth factor-directed signals, which provide convincing support for the theory that, in some contexts, Id genes function as oncogenes [20]. Moreover, many studies have suggested that Id proteins are involved in the cell cycle by interacting with cyclin D1, p21, or other molecules to promote progression through the S phase [21, 22]. An increased level of Id4 has been observed in basal-like breast cancer, triple-negative breast cancer [23], glioblastoma [24], primary serous ovarian cancer [25], and melanoma [26], leading to the qualification of Id4 as a proto-oncogene. The positive function of Id4 in tumor aggressiveness is best understood in breast cancer [21], but very little evidence is available regarding the role of Id4 protein in HCC.
To explore the role of the Id4 gene in HCC, we used CCK-8 assays, colony formation assays, and the animal model, and found that sustained Id4 overexpression was sufficient to increase HCC cell growth, enhance colony formation ability, and contribute to tumorigenesis. On the other hand, the Id4 knockdown experiment showed the opposite results that silencing Id4 blocked the proliferation and colony formation ability of HCC cells. Therefore, it is reasonable to conclude that Id4 might function as a proto-oncogene in HCC.
C/EBPβ is an important transcriptional factor that has been implicated in many biological activities, including cancer progression. Loss of C/EBPβ regulation in breast cancer promoted the disease’s malignant progression by inducing epithelial-mesenchymal transition [27]. Our previous study showed that C/EBPβ could repress HCC cell migration and invasion by directly binding to the orosomucoid 2 (ORM2) promoter and inducing ORM2 overexpression [28]. Many other studies have shown that C/EBPβ regulates transcriptional repression [29, 30]. For example, in the presence of a pro-inflammatory stimulus, C/EBPβ overexpression led to a decrease in CD200R1 expression in microglial cells [30]. In our study, we found that Id4 expression was inhibited in the C/EBPβ-overexpressing Huh7 cells. Therefore, we speculate that C/EBPβ may regulate Id4 expression directly or indirectly in HCC cells; however, this needs to be verified by further investigations.
Overwhelming evidence suggests that Id proteins can be master regulators of cancer stem cells. The previous study showed that Id1/Id3 regulates colon cancer-initiating cells (CC-ICs) by p21, and that, in colon cancer, Id1/Id3 protects the tumor-initiating ability of CC-ICs from oxaliplatin [31]. Furthermore, in glioma, Jeon et al. [32] found a new Id4-miR-9-SOX2 regulatory pathway that could affect the self-renewal activity of glioma cells, the chemoresistance of glioma stem cell (GSCs), and the maintenance of the stemness of induced GSCs. In our previous studies, we showed that Ikaros suppresses CD133 expression and that Id4 is one of the differentially expressed genes between the control and the Ikaros-overexpressing groups [17]. With reference to the expression of Id4 and CD133 and the function of CD133 in HCC [33], we speculate that the Id4 gene has a similar effect of CD133 in HCC, but a great deal of work is needed to address this hypothesis.
In conclusion, we showed an increase in Id4 protein expression in HCC tissues and a promoting effect of Id4 on proliferation in HCC cell lines in vitro and in vivo. Further investigation of the mechanisms and clinical value is greatly needed.
Authors’ contributions
YZ performed real-time PCR, Western blotting, CCK-8 assay, colony formation assay, and statistical analysis and drafted the manuscript. FZ and MY performed the tumor formation assay. LZ helped draft the manuscript. CG prepared the mRNA and protein from HCC patients and performed immunohistochemical staining. XL analyzed the effect of C/EBPβ on Id4 expression. TC collected the HCC tissues for tissue microarray. JL designed the study. All authors read and approved the final manuscript.
Acknowledgements
This work was supported in part by grants from the National Key Program for Basic Research of China (973) (No. 2015CB553905), the National Natural Science Foundation of China (Nos. 81272438, 81472726, 81301859), the Key Discipline and Specialty Foundation of Shanghai Municipal Commission of Health and Family Planning, the National KeySci-Tech Special Project of China (No. 2013ZX10002–011), and the SKLORG Research Foundation (Nos. 91-12-04, 91-13-02, 91-14-09).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Yang Zhang and Li-Xing Zhang contributed equally to this work
Contributor Information
Yang Zhang, Email: zhangyang3182@163.com.
Li-Xing Zhang, Email: zhanglixing601@163.com.
Xiao-Qin Liu, Email: liuxiaoqin106484@163.com.
Fang-Yu Zhao, Email: 531397429@qq.com.
Chao Ge, Email: chaoge1127@163.com.
Tao-Yang Chen, Email: taoyangchen@yeah.net.
Ming Yao, Email: myao@shsci.org.
Jin-Jun Li, Phone: +86-21-64432140, Email: jjli@shsci.org.
References
- 1.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61(1):49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- 2.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5(8):603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 3.Sharma P, Patel D, Chaudhary J. Id1 and Id3 expression is associated with increasing grade of prostate cancer: Id3 preferentially regulates CDKN1B. Cancer Med. 2012;1(2):187–197. doi: 10.1002/cam4.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasskarl J, Munger K. Id proteins–tumor markers or oncogenes? Cancer Biol Ther. 2002;1(2):91–96. doi: 10.4161/cbt.50. [DOI] [PubMed] [Google Scholar]
- 5.Patel D, Morton DJ, Carey J, Havrda MC, Chaudhary J. Inhibitor of differentiation 4 (ID4): from development to cancer. Biochim Biophys Acta. 2015;1855(1):92–103. doi: 10.1016/j.bbcan.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma P, Chinaranagari S, Chaudhary J. Inhibitor of differentiation 4 (ID4) acts as an inhibitor of ID-1, -2 and -3 and promotes basic helix loop helix (bHLH) E47 DNA binding and transcriptional activity. Biochimie. 2015;112:139–150. doi: 10.1016/j.biochi.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo TT, Zheng RS, Zhang SW, Zeng HM, Chen WQ. Incidence and mortality of liver cancer in China in 2011. Chin J Cancer. 2015;34(11):508–513. doi: 10.1186/s40880-015-0056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee TK, Man K, Ling MT, Wang XH, Wong YC, Lo CM, et al. Over-expression of Id-1 induces cell proliferation in hepatocellular carcinoma through inactivation of p16INK4a/RB pathway. Carcinogenesis. 2003;24(11):1729–1736. doi: 10.1093/carcin/bgg145. [DOI] [PubMed] [Google Scholar]
- 9.Damdinsuren B, Nagano H, Kondo M, Yamamoto H, Hiraoka N, Yamamoto T, et al. Expression of Id proteins in human hepatocellular carcinoma: relevance to tumor dedifferentiation. Int J Oncol. 2005;26(2):319–327. [PubMed] [Google Scholar]
- 10.Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA. 2001;98(1):130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desprez PY, Sumida T, Coppe JP. Helix-loop-helix proteins in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8(2):225–239. doi: 10.1023/A:1025957025773. [DOI] [PubMed] [Google Scholar]
- 12.Junankar S, Baker LA, Roden DL, Nair R, Elsworth B, Gallego-Ortega D, et al. ID4 controls mammary stem cells and marks breast cancers with a stem cell-like phenotype. Nat Commun. 2015;6:6548. doi: 10.1038/ncomms7548. [DOI] [PubMed] [Google Scholar]
- 13.Sharma P, Chinaranagari S, Patel D, Carey J, Chaudhary J. Epigenetic inactivation of inhibitor of differentiation 4 (Id4) correlates with prostate cancer. Cancer Med. 2012;1(2):176–186. doi: 10.1002/cam4.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey JP, Asirvatham AJ, Galm O, Ghogomu TA, Chaudhary J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer. 2009;9:173. doi: 10.1186/1471-2407-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian H, Ge C, Li H, Zhao F, Hou H, Chen T, et al. Ribonucleotide reductase M2B inhibits cell migration and spreading by early growth response protein 1-mediated phosphatase and tensin homolog/Akt1 pathway in hepatocellular carcinoma. Hepatology. 2014;59(4):1459–1470. doi: 10.1002/hep.26929. [DOI] [PubMed] [Google Scholar]
- 16.Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22(5):749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Li H, Ge C, Li M, Zhao FY, Hou HL, et al. Inhibitory effects of transcription factor Ikaros on the expression of liver cancer stem cell marker CD133 in hepatocellular carcinoma. Oncotarget. 2014;5(21):10621–10635. doi: 10.18632/oncotarget.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wennemers M, Bussink J, van den Beucken T, Sweep FC, Span PN. Regulation of TRIB3 mRNA and protein in breast cancer. PLoS ONE. 2012;7(11):e49439. doi: 10.1371/journal.pone.0049439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14(2):77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 21.Dell’Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G. ID4: a new player in the cancer arena. Oncotarget. 2010;1(1):48–58. doi: 10.18632/oncotarget.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon HM, Jin X, Lee JS, Oh SY, Sohn YW, Park HJ, et al. Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes Dev. 2008;22(15):2028–2033. doi: 10.1101/gad.1668708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branham MT, Marzese DM, Laurito SR, Gago FE, Orozco JI, Tello OM, et al. Methylation profile of triple-negative breast carcinomas. Oncogenesis. 2012;1:e17. doi: 10.1038/oncsis.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng W, Rushing EJ, Hartmann DP, Azumi N. Increased inhibitor of differentiation 4 (id4) expression in glioblastoma: a tissue microarray study. J Cancer. 2010;1:1–5. doi: 10.7150/jca.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren Y, Cheung HW, von Maltzhan G, Agrawal A, Cowley GS, Weir BA, et al. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Sci Transl Med. 2012;4(147):147ra12. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peretz Y, Wu H, Patel S, Bellacosa A, Katz RA. Inhibitor of DNA Binding 4 (ID4) is highly expressed in human melanoma tissues and may function to restrict normal differentiation of melanoma cells. PLoS ONE. 2015;10(2):e0116839. doi: 10.1371/journal.pone.0116839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson J, Berg T, Kurzejamska E, Pang MF, Tabor V, Jansson M, et al. MiR-155-mediated loss of C/EBPbeta shifts the TGF-beta response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene. 2013;32(50):5614–5624. doi: 10.1038/onc.2013.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang T, Cui M, Sun J, Ge C, Zhao F, Zhang L, et al. Orosomucoid 2 inhibits tumor metastasis and is upregulated by CCAAT/enhancer binding protein beta in hepatocellular carcinomas. Oncotarget. 2015;6(18):16106–16119. doi: 10.18632/oncotarget.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bein K, Leight H, Leikauf GD. JUN-CCAAT/enhancer-binding protein complexes inhibit surfactant-associated protein B promoter activity. Am J Respir Cell Mol Biol. 2011;45(2):436–444. doi: 10.1165/rcmb.2010-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dentesano G, Straccia M, Ejarque-Ortiz A, Tusell JM, Serratosa J, Saura J, et al. Inhibition of CD200R1 expression by C/EBP beta in reactive microglial cells. J Neuroinflammation. 2012;9:165. doi: 10.1186/1742-2094-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y, et al. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell. 2012;21(6):777–792. doi: 10.1016/j.ccr.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 32.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, et al. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71(9):3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- 33.Yin S, Li J, Hu C, Chen X, Yao M, Yan M, et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer. 2007;120(7):1444–1450. doi: 10.1002/ijc.22476. [DOI] [PubMed] [Google Scholar]


