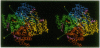Abstract
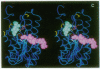
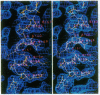
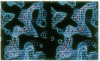
The x-ray structure of a short-chain dehydrogenase, the bacterial holo 3 alpha,20 beta-hydroxysteroid dehydrogenase (EC 1.1.1.53), is described at 2.6 A resolution. This enzyme is active as a tetramer and crystallizes with four identical subunits in the asymmetric unit. It has the alpha/beta fold characteristic of the dinucleotide binding region. The fold of the rest of the subunit, the quaternary structure, and the nature of the cofactor-enzyme interactions are, however, significantly different from those observed in the long-chain dehydrogenases. The architecture of the postulated active site is consistent with the observed stereospecificity of the enzyme and the fact that the tetramer is the active form. There is only one cofactor and one substrate-binding site per subunit; the specificity for both 3 alpha- and 20 beta-ends of the steroid results from the binding of the steroid in two orientations near the same cofactor at the same catalytic site.
Full text
PDF
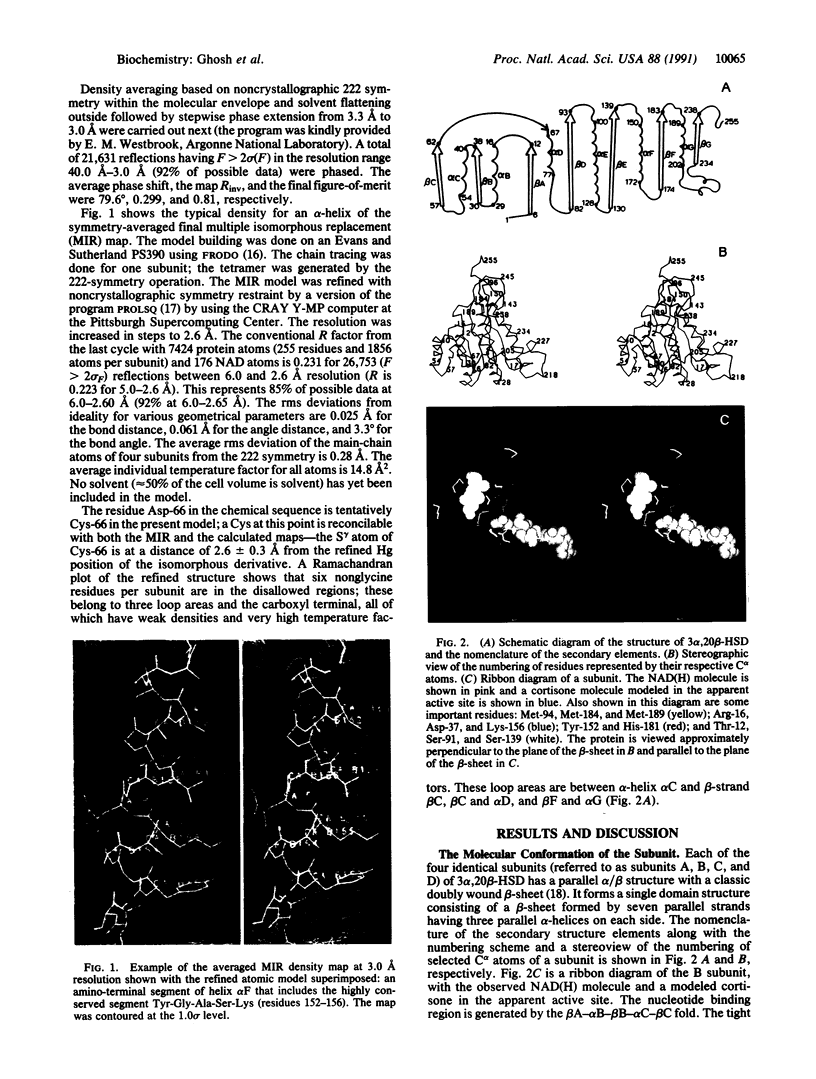


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
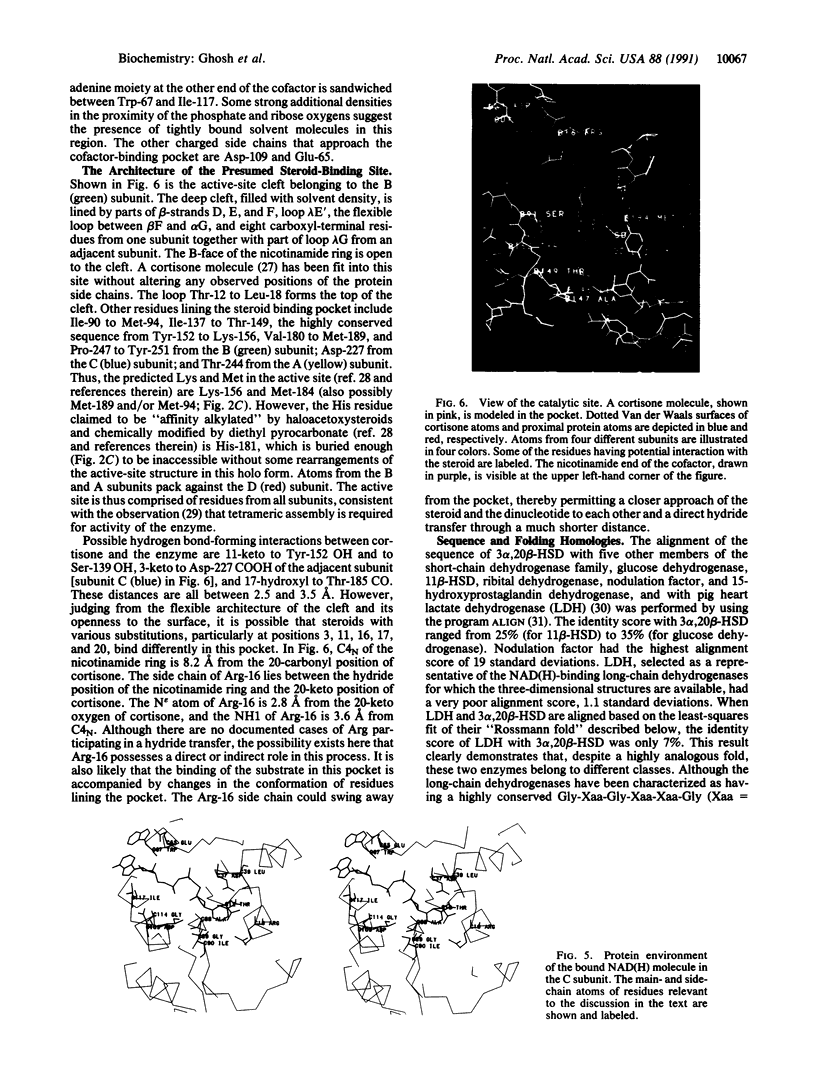
- Agarwal A. K., Monder C., Eckstein B., White P. C. Cloning and expression of rat cDNA encoding corticosteroid 11 beta-dehydrogenase. J Biol Chem. 1989 Nov 15;264(32):18939–18943. [PubMed] [Google Scholar]
- Benyajati C., Place A. R., Powers D. A., Sofer W. Alcohol dehydrogenase gene of Drosophila melanogaster: relationship of intervening sequences to functional domains in the protein. Proc Natl Acad Sci U S A. 1981 May;78(5):2717–2721. doi: 10.1073/pnas.78.5.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz G., Warren J. C. Reaction mechanism and stereospecificity of 20 beta-hydroxysteroid dehydrogenase. Arch Biochem Biophys. 1968 Dec;128(3):745–752. doi: 10.1016/0003-9861(68)90083-0. [DOI] [PubMed] [Google Scholar]
- Carrea G., Pasta P., Vecchio G. Effect of the lyotropic series of anions on denaturation and renaturation of 20 beta-hydroxysteroid dehydrogenase. Biochim Biophys Acta. 1984 Jan 18;784(1):16–23. doi: 10.1016/0167-4838(84)90167-5. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar K., McPherson A., Jr, Adams M. J., Rossmann M. G. Conformation of coenzyme fragments when bound to lactate dehydrogenase. J Mol Biol. 1973 Jun 5;76(4):503–518. doi: 10.1016/0022-2836(73)90488-9. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Debellé F., Sharma S. B. Nucleotide sequence of Rhizobium meliloti RCR2011 genes involved in host specificity of nodulation. Nucleic Acids Res. 1986 Sep 25;14(18):7453–7472. doi: 10.1093/nar/14.18.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Eklund H., Samama J. P., Jones T. A. Crystallographic investigations of nicotinamide adenine dinucleotide binding to horse liver alcohol dehydrogenase. Biochemistry. 1984 Dec 4;23(25):5982–5996. doi: 10.1021/bi00320a014. [DOI] [PubMed] [Google Scholar]
- Franklund C. V., de Prada P., Hylemon P. B. Purification and characterization of a microbial, NADP-dependent bile acid 7 alpha-hydroxysteroid dehydrogenase. J Biol Chem. 1990 Jun 15;265(17):9842–9849. [PubMed] [Google Scholar]
- Ghosh D., Punzi J. S., Duax W. L. Crystals of active tetramers of 3 alpha, 20 beta-hydroxysteroid dehydrogenase. J Biol Chem. 1986 Jan 25;261(3):1306–1308. [PubMed] [Google Scholar]
- Grau U. M., Trommer W. E., Rossmann M. G. Structure of the active ternary complex of pig heart lactate dehydrogenase with S-lac-NAD at 2.7 A resolution. J Mol Biol. 1981 Sep 15;151(2):289–307. doi: 10.1016/0022-2836(81)90516-7. [DOI] [PubMed] [Google Scholar]
- Krook M., Marekov L., Jörnvall H. Purification and structural characterization of placental NAD(+)-linked 15-hydroxyprostaglandin dehydrogenase. The primary structure reveals the enzyme to belong to the short-chain alcohol dehydrogenase family. Biochemistry. 1990 Jan 23;29(3):738–743. doi: 10.1021/bi00455a021. [DOI] [PubMed] [Google Scholar]
- Makino Y., Negoro S., Urabe I., Okada H. Stability-increasing mutants of glucose dehydrogenase from Bacillus megaterium IWG3. J Biol Chem. 1989 Apr 15;264(11):6381–6385. [PubMed] [Google Scholar]
- Marekov L., Krook M., Jörnvall H. Prokaryotic 20 beta-hydroxysteroid dehydrogenase is an enzyme of the 'short-chain, non-metalloenzyme' alcohol dehydrogenase type. FEBS Lett. 1990 Jun 18;266(1-2):51–54. doi: 10.1016/0014-5793(90)81504-h. [DOI] [PubMed] [Google Scholar]
- Matthews D. A., Alden R. A., Freer S. T., Xuong N., Kraut J. Dihydrofolate reductase from Lactobacillus casei. Stereochemistry of NADPH binding. J Biol Chem. 1979 May 25;254(10):4144–4151. [PubMed] [Google Scholar]
- Morris H. R., Williams D. H., Midwinter G. G., Hartley B. S. A mass-spectrometric sequence study of the enzyme ribitol dehydrogenase from Klebsiella aerogenes. Biochem J. 1974 Sep;141(3):701–713. doi: 10.1042/bj1410701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy M. R., Garavito R. M., Johnson J. E., Rossmann M. G. Structure of lobster apo-D-glyceraldehyde-3-phosphate dehydrogenase at 3.0 A resolution. J Mol Biol. 1980 Apr 25;138(4):859–872. doi: 10.1016/0022-2836(80)90069-8. [DOI] [PubMed] [Google Scholar]
- Pasta P., Mazzola G., Carrea G. Chemical modification of 3 alpha,20 beta-hydroxysteroid dehydrogenase with diethyl pyrocarbonate. Evidence for an essential, highly reactive, lysyl residue. Biochemistry. 1987 Mar 10;26(5):1247–1251. doi: 10.1021/bi00379a007. [DOI] [PubMed] [Google Scholar]
- Piontek K., Chakrabarti P., Schär H. P., Rossmann M. G., Zuber H. Structure determination and refinement of Bacillus stearothermophilus lactate dehydrogenase. Proteins. 1990;7(1):74–92. doi: 10.1002/prot.340070108. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Adams M. J., Buehner M., Ford G. C., Hackert M. L., Liljas A., Rao S. T., Banaszak L. J., Hill E., Tsernoglou D. Letter: Molecular symmetry axes and subunit interfaces in certain dehydrogenases. J Mol Biol. 1973 Jun 5;76(4):533–537. doi: 10.1016/0022-2836(73)90491-9. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Skarzyński T., Moody P. C., Wonacott A. J. Structure of holo-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus at 1.8 A resolution. J Mol Biol. 1987 Jan 5;193(1):171–187. doi: 10.1016/0022-2836(87)90635-8. [DOI] [PubMed] [Google Scholar]
- Sweet F., Samant B. R. Bifunctional enzyme activity at the same active site: study of 3 alpha and 20 beta activity by affinity alkylation of 3 alpha, 20 beta-hydroxysteroid dehydrogenase with 17-(bromoacetoxy)steroids. Biochemistry. 1980 Mar 4;19(5):978–986. doi: 10.1021/bi00546a023. [DOI] [PubMed] [Google Scholar]
- Vrielink A., Lloyd L. F., Blow D. M. Crystal structure of cholesterol oxidase from Brevibacterium sterolicum refined at 1.8 A resolution. J Mol Biol. 1991 Jun 5;219(3):533–554. doi: 10.1016/0022-2836(91)90192-9. [DOI] [PubMed] [Google Scholar]
- Webb L. E., Hill E. J., Banaszak L. J. Conformation of nicotinamide adenine dinucleotide bound to cytoplasmic malate dehydrogenase. Biochemistry. 1973 Dec 4;12(25):5101–5109. doi: 10.1021/bi00749a013. [DOI] [PubMed] [Google Scholar]
- Westbrook E. M., Piro O. E., Sigler P. B. The 6-A crystal structure of delta 5-3-ketosteroid isomerase. Architecture and location of the active center. J Biol Chem. 1984 Jul 25;259(14):9096–9103. [PubMed] [Google Scholar]