Abstract
BACKGROUND
Activator Protein-1 (AP-1) family (cJun, JunB, JunD, cFos, FosB, Fra1, and Fra2) plays a central role in the transcriptional regulation of many genes that are associated with cell proliferation, differentiation, migration, metastasis, and survival. Many oncogenic signaling pathways converge at the AP-1 transcription complex. Transforming growth factor beta (TGF-β) is a multifunctional regulatory cytokine that regulates many aspects of cellular function, including cellular proliferation, differentiation, migration, apoptosis, adhesion, angiogenesis, immune surveillance, and survival.
METHODS
This study investigated, the role of FOS proteins in TGF-β signaling in prostate cancer cell proliferation, migration, and invasion. Steady state expression levels of FOS mRNA and proteins were determined using RT-PCR and western blotting analyses. DU145 and PC3 prostate cancer cells were exposed to TGF-β1 at varying time and dosage, RT-PCR, western blot, and immunofluorescence analyses were used to determine TGF-β1 effect on FOS mRNA and protein expression levels as well as FosB subcellular localization. Transient silencing of FosB protein was used to determine its role in cell proliferation, migration, and invasion.
RESULTS
Our data show that FOS mRNA and proteins were differentially expressed in human prostate epithelial (RWPE-1) and prostate cancer cell lines (LNCaP, DU145, and PC3). TGF-β1 induced the expression of FosB at both the mRNA and protein levels in DU145 and PC3 cells, whereas cFos and Fra1 were unaffected. Immunofluorescence analysis showed an increase in the accumulation of FosB protein in the nucleus of PC3 cells after treatment with exogenous TGF-β1. Selective knockdown of endogenous FosB by specific siRNA did not have any effect on cell proliferation in PC3 and DU145 cells. However, basal and TGF-β1- and EGF-induced cell migration was significantly reduced in DU145 and PC3 cells lacking endogenous FosB. TGF-β1- and EGF-induced cell invasion were also significantly decreased after FosB knockdown in PC3 cells.
CONCLUSION
Our data suggest that FosB is required for migration and invasion in prostate cancer cells. We also conclude that TGF-β1 effect on prostate cancer cell migration and invasion may be mediated through the induction of FosB.
Keywords: AP-1, FosB, TGF-β, prostate cancer, cell migration, cell invasion
INTRODUCTION
Prostate cancer is the most diagnosed and the second leading cause of cancer deaths among American men. According to American Cancer Society, 180,896 men will be diagnosed and 26,120 men will die of prostate cancer in US in 2016. Early stage prostate cancer which is localized in the prostate gland is treatable by surgery and radiation therapy and the prognosis in these patients is very good [1]. However, the prostate cancers in later stages of disease metastasize to other tissues and bone and pose a significant problem for treatment [1]. Death from prostate cancer results when cancer cells become metastatic after invading the lymph nodes and blood vessels and migrate to bone [2,3]. Current treatments for metastatic disease are hormonal therapy and chemotherapy. Hormonal therapies are based on inhibition of biosynthesis and/or action of androgens [4,5]. However, the cancer cells develop resistance to these treatments resulting in development of castration resistant or hormone refractory prostate cancers. There is no effective therapy for these cancers which are responsible for mortality in majority of patients.
In mammals, TGF-β is the prototype of a family of secreted polypeptide growth factors. To date, up to 33 TGF-β related genes have been identified which include TGF-β itself, bone morphogenic proteins (BMPs), activins/inhibin, growth and differentiation factors, nodal, and anti-müllerian hormone [6]. TGF-β signaling inhibits cell proliferation in a multitude of cell types, including normal endothelial, epithelial, hematopoietic, and neural cells, certain types of mesenchymal cells, and especially many primary cancer cells [7]. However, in a different cellular context, TGF-β can act as a tumor promoter because it is able to induce changes in transcriptional activities that re-program epithelial cells into mesenchymal cells, thereby facilitating tumor metastasis and invasion [6,8,9]. Previous studies from several laboratories have investigated the role of TGF-β secreted by the epithelial and stromal cells in the development and progression of prostate cancer [10–12]. Despite these studies, the molecular mechanisms and the intracellular effectors surrounding these differential effects of TGF-β1 during different stages of cancer progression are not well understood [13].
Activator protein-1 (AP-1) was one of the first transcription factors to be identified, but its physiological functions are still being unraveled [14]. AP-1 activity is induced by a plethora of physiological stimuli and environmental insults [14] such as growth factors, cytokines, tumor-promoters, and UV-irradiation [15]. AP-1 transcription factor consists of a variety of dimers composed primarily of members of the Fos and Jun families of proteins [16,17]. While the Fos proteins (FosB, cFos, Fra1, and Fra2) can only heterodimerize with members of Jun family, the Jun proteins (JunB, cJun, and JunD) can both homo- and heterodimerize with Fos members to form transcriptionally active complexes. AP-1 has been implicated in a variety of biological processes including cell differentation, proliferation, apoptosis, and oncogenic transformation [18]. AP-1 activity is modulated by interactions with other transcriptional regulators and is further controlled by upstream kinases that link AP-1 proteins to various signal transduction pathways [18]. AP-1 activity converts extra-cellular signals into changes in gene expression patterns through the binding of the AP-1 dimers to specific target sequences, the TPA-response element (TREs, TGAC/GTCA), in the promoter and enhancer regions of target genes [19,20]. Dimerization serves as a prerequisite for DNA binding as well as an enhancer of nuclear translocation. In vitro studies have determined that, in many situations, JUN–FOS heterodimers are more stable and have stronger DNA binding activities than JUN–JUN homo-dimers [21]. Although AP-1 proteins share a high level of sequence and function homology, they exhibit distinct expression patterns and differ in their transcriptional and biological activities [19]. Previous studies have shown that AP-1 family proteins are differentially expressed in human cancers and have been suggested to play specific roles in cancer progression. The available evidence suggests higher proliferation rate, malignant transformation, and enhanced aggressiveness is accompanied by changes in AP-1 complex compositions [22]. For most processes, the precise composition of the AP-1 complexes and the critical target genes remain to be defined. Furthermore, the role of each individual AP-1 family member in prostate cancer development and progression and in TGF-β1 signaling have not been investigated. To address this, we have studied the expression of FOS family members in prostate epithelial and prostate cancer cells in vitro and utilized knockdown approaches to determine the role of individual FOS family members in prostate cancer cell proliferation, migration, and invasion.
MATERIALS AND METHODS
Chemical and Reagents
Recombinant human TGF-β1 (Catalog # HEK293 100-21) was purchased from Peprotech (Rocky Hill, NJ). The antibodies against FosB (Catalog# 2251S), cFos (Catalog # SC-52), Fra1 (Catalog # SC-605), and Fra2 (Catalog # SC-604) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). The anti-β-Actin (Catalog # A5441) antibody was purchased from Sigma–Aldrich (St. Louis, MO). The anti-mouse and anti-rabbit immunoglobulins coupled with horseradish peroxidase (IgG-HRP) were obtained from Promega (Madison, WI). Cell lysis buffer was purchased from Cell Signaling (Danvers, MA). TRIzol was purchased from Invitrogen (Carlsbad, CA).
Human Prostate Cell Lines and Treatments
All cell lines were obtained from American Type Culture Collection. These include immortalized prostate luminal epithelial cell line (RWPE1) and prostate cancer cell lines (LNCaP, DU145, and PC3).
Expression of FOS family members
Prostate cells (RWPE1, LNCaP, DU145, and PC3) were cultured using established procedures [23–25]. To determine the basal expression of Fos mRNA and protein levels, RWPE1, LNCaP, DU145, and PC3 cells were seeded at a density 1.0 × 106 cells/dish in a 10 cm petri dish in the appropriate growth media and cultured at 37°C for 48 h. After 48 hr, the cells were washed with ice-cold phosphate buffered saline (PBS) and lyzed in cell lysis buffer containing 20 mM Tris–HCl (pH 7.5), 150 mm NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, and 1× protease inhibitor cocktail (Calbiochem, San Diego, CA). Protein concentrations were determined by the Lowry HS assay using the Bio-Rad DC Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA) according to the instructions provided by the manufacturer. The total RNAs were isolated from parallel experiments and were also used for RT-PCR as described below.
To determine the effects of TGF-β1 on FosB, cFos, Fra1, and Fra2 expression, prostate cancer cells were seeded in six-well plates at a density of 3.0 × 105 cells per well. Before each experiment DU145 and PC3 cells were incubated with media supplemented with 1% serum for 2 hr followed by treatment with different doses of TGF-β1 (0, 1, 5, 10 ng/ml) for specific time periods. RNA and proteins were isolated and quantified.
RNA Isolation, cDNA Synthesis, and RT-PCR
Total RNAs were isolated from prostate cells using TRIzol (Invitrogen) and the resulting RNA samples were quantified by optical density reading at 260 nm as described previously [26]. Total RNAs (2 μg) were reverse transcribed in a 50 μl reaction mixture containing 0.5 mM dNTP (Fisher Scientific, Pittsburgh, PA), 0.5 mM dithiotreitol (Bio-Rad), 0.5 μg of oligo dT, and 400 U of M-MLV Reverse Transcriptase (Promega) at 37°C for 1.5 hr. The reaction was terminated by heating the samples at 65°C for 5 min and subsequently cooled to 4°C. PCR was performed to detect mRNA levels of FosB1, FosB2, cFos, Fra1, Fra2, and L-19. The PCR mixture was composed of 0.1 mM deoxynucleotide triphosphates, 0.5 U Taq DNA polymerase, 10× PCR Buffer with 3 mM MgCl2, and 25 pM of the specific primers in a total volume of 15 μl. Primer information and the size of specific amplicons for individual genes are shown in Table I. L-19 (a ribosomal protein) was used as a loading control. RNA samples processed without RT and PCR amplified were used as negative controls. Amplification was performed at 1, initial denaturant 94°C for 2 min; 2, 94°C for 15 sec; 3, 58°C for 15 sec; 4, 72°C for 30 sec, 5, repeating steps two and four for 35 cycles for FosB1, FosB2, cFos, Fra1, Fra2, and L-19; 5, final extension 72°C for 2 min. The PCR products were separated on 1.0–2.0% agarose gels, and viewed under UV.
TABLE I.
Gene-Specific Primers Used for RT-PCR Amplification
| Gene | Primers | Sequence 5′-3′ | Product size (bp) |
|---|---|---|---|
| Full length FosB | Sense | AGCAGCAGCTAAATGCAGGA | 131 |
| Antisense | GACGTTCCTTCTCCTTTTGGA | ||
| Delta FosB | Sense | CGAGAGGAGACGGAGACAGA | 139 |
| Antisense | CTTCGTAGGGGATCTTGCAG | ||
| cFos | Sense | AGTGCCAACTTCATTCCCAC | 265 |
| Antisense | CCCTTCGGATTCTCCTTTTC | ||
| Fra1 | Sense | TCTTCACCTACCCCAGCACTC | 220 |
| Antisense | GCTGGAGTTGGATGTGGGATAC | ||
| Fra2 | Sense | CTGCTGGATCAAGTGCTTTCTC | 150 |
| Antisense | GATTCAACAGACAACCAGGAATGG | ||
| L-19 | Sense | GAAATCGCCAATGCCAACTC | 405 |
| Antisense | TCTTAGACCTGCGAGCCTCA |
Western Blot Analysis
Total cellular proteins were prepared from different prostate cell lines and were analyzed by Western blot as described previously [26]. Briefly, cell lysates were mixed with Laemmeli's buffer (62.5 mM Tris, pH 6.8, 2% SDS, 5% β–mercaptoethanol, and 10% glycerol). Individual samples containing 30–35 μg protein were subjected to SDS–PAGE in 10% gels and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corp., Bedford, MA). After blocking the membranes with 5% fat free milk in TBST (50 mM Tris, pH 7.5, containing 0.15 M NaCl, and 0.05% Tween–20) (cFos, Fra1, Fra2) or 5% bovine serum albumin (BSA) in TBST (FosB), for 1 hr at room temperature, the membranes were incubated with appropriate dilutions of specific primary antibodies (1:500 for FOS proteins and 1:5,000 for β-actin) overnight at 4°C. After washing, the blots were incubated with secondary anti-rabbit (for FOS proteins) and anti-mouse (for β-actin) IgG HRPs (1:20,000) for 1 hr. The blots were developed in ECL mixture (Thermo Fisher Scientific, Inc., Rockford, IL), and placed inside the Syngene PXi/PXi Touch darkroom imaging (high resolution, multi-application image analysis systems) (Frederick, MD) according to the manufacturer's directions and the density of specific protein bands were determined using ImageJ image processing and analysis software and values were normalized using β-actin.
Immunofluorescence of FosB
PC3 cells were seeded at a density of 8.0 × 104 cells/well into six-well plates containing sterile glass coverslips. The cells were incubated at 37°C and allowed to attach for 48 hr. The media were replaced with fresh media containing 1% FBS for 2 hr before treatment with TGF-β1 (10 ng/ml) for 4 hr. At the end of the treatment, media were aspirated and cells were fixed in 3.7% paraformaldehyde for 20 min at room temperature. The fixative was aspirated and cells were rinsed three times with 1.0 ml 1× PBS and permeabilized using 0.1% Triton at room temperature. The cover slips were transferred to an aluminum wrapped 20 cm Petri dish and outlined using a hydrophobic marker. The cells on the cover slips were blocked in blocking buffer containing 10% normal goat serum in 1× PBS for 1 hr. Blocking solution was aspirated and primary antibody (1:1,000) for FosB was added overnight at 4°C in 1× PBS with 2% normal goat serum. After washing five times in 1× PBS for 10 min each, secondary antibody containing green fluorochrome (Alexa Fluor 488 goat anti-rabbit IgG, Life Technologies, Carlsbad, CA) was added at room temperature for 1 hr in 1× PBS (light sensitive). After washing, DAPI (3 μg/ml) was added to cells for 20 min at room temperature to stain the nuclei. The cells were rinsed and the cover slips were then mounted on slides. Slides were kept at room temperature in the dark for 2 hr before viewing under inverted florescence microscope. Images were captured using 40× magnification with an Axiovision camera of a Carl Zeiss zoom inverted florescence microscope (Carl Zeiss, Thornwood, NY).
Transfection With FosB siRNA
To knockdown endogenous FosB expression, DU145 and PC3 cells were seeded in six-well plates at the density 1.5 × 105 cells per well in 1.0 ml antibiotic-free normal growth medium supplemented with 5% FBS. The cells were cultured at 37°C to 60–80% confluence. Control (scrambled) and FosB specific siRNAs were transfected into DU145 and PC3 cells according to the manufacturers’ instructions. Briefly, transfection complex were mixed together in a 1:1 ratio (2.5 μl for FosB) of siRNA to transfection reagent in 200 μl of antibiotic-free normal growth medium. The mixture was allowed to incubate at room temperature in the dark for 20 min. During this time the cells were washed once with 1 ml of siRNA transfection medium, after which the antibiotic-free medium was mixed with 1% FBS and added to the transfection reagent mixture. The transfection reagent siRNA duplex was overlaid onto the washed cells, and the cells were incubated overnight. The medium containing the transfection complex was replaced with complete medium containing 5% FBS and incubated for 48 hr. Cells were trypsinized (0.25% Trypsin/2.21 mM EDTA) for 1 min and trypsin was neutralized with 3.0 ml of complete medium. Cells were centrifuged at 1,000 RPM 4°C for 5 min and re-plated for biological assays. Western blot analyses were used to confirm FosB protein knockdown.
Cell Proliferation Assay
After transfection, the cells were counted and seeded into 96-well plates at a density of 5 × 103 cells/well and treated with 10 ng/ml of TGF-β1 in the presence of 1% FBS for 72 hr. Cell viability was measured using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. MTT assays were performed using Cell Titer 96 Non-radioactive Cell proliferation assay (Promega) following the manufacturer's instructions.
Cell Migration Assay
After the transfections, in vitro cell migration assays were performed using 24-well transwell inserts (8 μm) as previously described [27,28]. Chemoattractant solutions were made by diluting TGF-β1 (10 ng/ml) or EGF (10 ng/ml) into MEM for DU145 and PC3 cells supplemented with 0.2% BSA. The results were expressed as migration index defined as: the average number of cells per field for test substance/the average number of cells per field for the medium control. The experiments were conducted at least three times using independent cell preparations.
Cell Invasion Assay
After transfection, the invasive behavior of PC3 cells was measured using the BD BioCoat Matrigel Invasion inserts [29]. Cell culture inserts (VWR International, Bridgeport, NJ) were coated with 50 μl of 1:4 Matrigel/Medium dilutions (BD Sciences, San Jose, CA) and allowed to solidify at 37°C for 1 hr. Cells were resuspended (5.0 × 104 cells/ml) in MEM and 0.1% FBS and 500 μl of cell suspension was added to each insert. Chemoattractant solutions were made as described above with TGF-β1 and EGF into MEM supplemented with 0.1% FBS. Matrigel and non-invading cells were removed by scrubbing. Invading cells in the membrane were fixed in 3.7% paraformaldehyde and stained with DAPI. Pictures were taken from five different fields for average number of invading cells to be determined. The results were expressed as an invasive index defined as: the average number of cells per field for test substance/the average number of cells per field for the medium control. The experiments were conducted at least three times using independent cell preparations.
Statistical Analysis
All experiments were repeated at least three times. One way analysis of variance (ANOVA), Duncan's modified multiple range and Student–Newman–Keuls tests were employed to assess the significance of differences between treatment groups.
RESULTS
Expression of FOS Family Members in Prostate Cancer Cell Lines
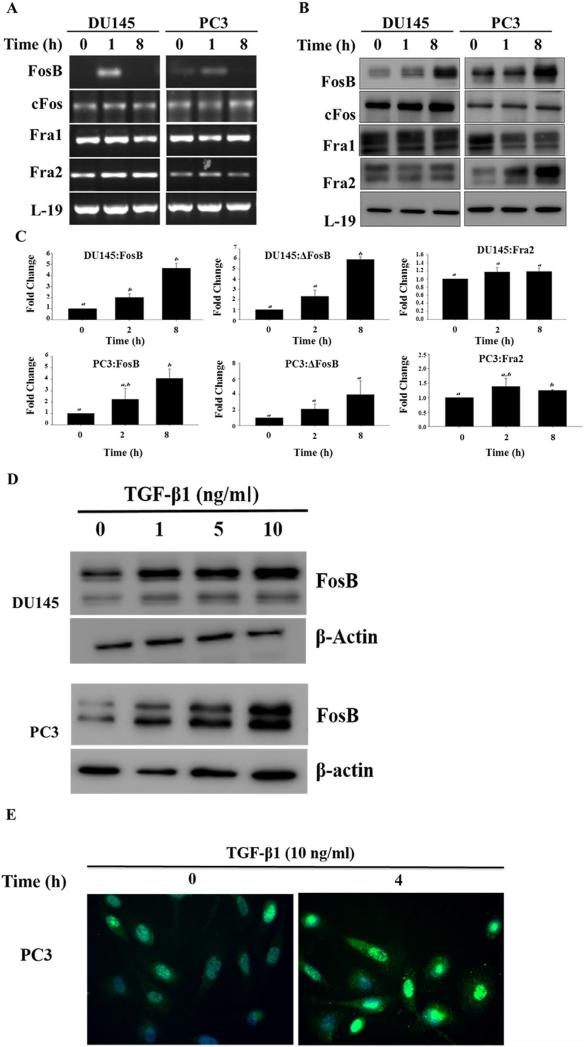
We initially screened four human prostate cell lines, RWPE1 (normal prostate epithelial cells), LNCaP, DU145, and PC3 (prostate cancer cell lines) for expression of FOS family members. Levels of mRNA for FosB, cFos, Fra1, and Fra2 were determined using RT-PCR, with L-19 used as control (Fig. 1A). Fra2 mRNA was robust in all four cell lines, Fra1 and FosB mRNA were highly expressed in all cell lines except in LNCaP, which also had very low mRNA levels of cFos. ΔFosB was highly expressed in all cells with only moderate expression in RWPE1 cells (Fig. 1A). Western blot analyses was performed to determine the relative protein abundance of FosB, cFos, Fra1, and Fra2 (Fig. 1B). Fra1 protein levels were very low in all prostate cancer cell lines (LNCaP, DU145, and PC3) with slightly higher levels in RWPE1 cells (Fig. 1B). Fra2 protein levels were relatively high in RWPE1, DU145, and PC3 but were not detectable in LNCaP cells. cFos showed moderate expression in RWPE1, DU145, and PC3 cells but was very low in LNCaP cells. FosB and ΔFosB protein levels were high in RWPE1 and PC3 cells but low to moderate in LNCaP and DU145 cells, respectively, (Fig. 1B).
Fig. 1.
FOS family basal expression. Steady state mRNA levels of FOS (FosB, cFos, Fra1, and Fra2) mRNA and protein expression. (A) Total RNA's were isolated and semi quantitative RT-PCR was performed to determine the mRNA levels of FosB, cFos, Fra1, and Fra2 in prostate cells. L-19 was used as an internal control. (B) Western blot analysis of FosB, cFos, Fra1, and Fra2 in prostate cells. β-actin was used as a loading control.
TGF-β1 Effects on FOS Protein Expression and Nuclear Accumulation in Prostate Cancer Cells
DU145 and PC3 cells were treated with exogenous TGF-β1 (10 ng/ml) for 0, 1, 2, and 8 hr. TGF-β1 induced an increase in the mRNA levels of FosB in both cell lines but had little effect on the mRNA levels of the other FOS family members (Fig. 2A). At the protein levels, TGF-β1 induced an increase in the levels of FosB protein in both cell lines and an increase in Fra2 protein levels was observed only in PC3 cells. TGF-β1 had no effect on the levels of cFos and Fra1 in both cell lines (Fig. 2B). Spot densitometry analysis confirmed TGF-β1 effects on FosB and Fra2 protein levels in DU145 and PC3 cells in a time-dependent manner (Fig. 2C). TGF-β1 induction of FosB was dose dependent (Fig. 2D). Immunofluorescence of FosB in PC3 cells showed that treatment with TGF-β1 induced increased expression and nuclear localization of FosB (Fig. 2E).
Fig. 2.
TGF-β1 effects on expression of FOS family members. (A) RT-PCR analysis of FosB, cFos, Fra1, and Fra2 mRNA levels in DU145 and PC3 prostate cancer cells after exposure to exogenous TGF-β (10 ng/ml) for different times. (B) Western blot analysis of FosB, the FosB antibody used recognizes both full length FosB (higher molecular weight band) and ΔFosB (lower molecular weight band), cFos, Fra1, and Fra2 protein levels DU145 and PC3 prostate cancer cells after exposure to exogenous TGF-β1 (10 ng/ml) for different times. (C) Band density analysis of FosB and Fra2 in DU145 and PC3 cells after treatment with TGF-β1 for 2 and 8 hr. Each band density was normalized by density of β-actin bands. Each bar represents the Mean ± SD from three independent experiments. “a and b” denote significant differences (P < 0.05) from untreated controls. (D) The Dose dependent effects of TGF-β1 on expression of FosB; Western blot analysis of FosB in prostate cancer cells DU145 and PC3 after treatment with varying concentrations of exogenous TGF-β1 (0, 1, 5, 10 ng/ml) for 4 hr. (E) Immunofluorescence, TGF-β1 activation of FosB in PC3. Cells were treated with exogenous TGF-β1 (10 ng/ml) for 0 and 4 hr.
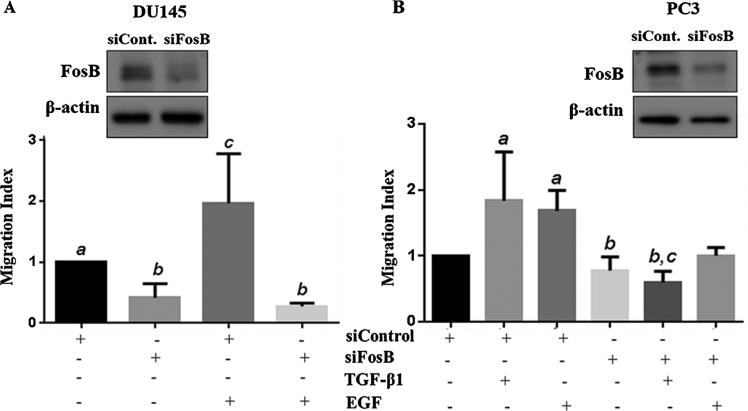
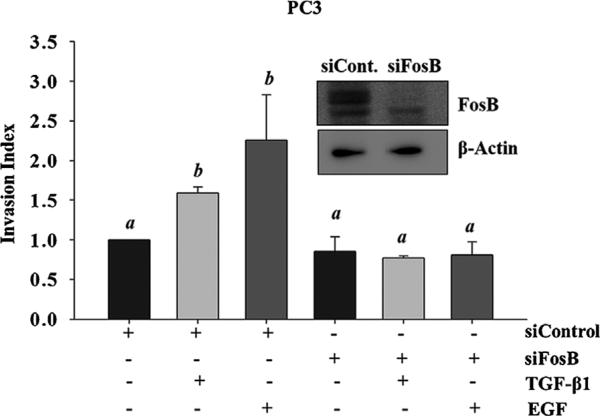
FosB Knockdown Has No Effect on TGF-β1-Mediated Cell Proliferation But Abrogates TGF-β1 and EGF-Mediated Cell Migration and Invasion
Next, we determined the role of FosB in TGF-β1-induced cell proliferation, migration and invasion in prostate cancer cells. A transient knock down of FosB was carried out using siRNA specific to FosB, followed by a MTT proliferation (Fig. 3A and B), trans-well inserts migration assays (Fig. 4A and B), and matrigel invasion assays (Fig. 5). Knock down of FosB had no effect on cell proliferation in DU145 and PC3 cells (Fig. 3A and B); however, there was a significant decrease in cell migration (DU145, PC3) and cell invasion (PC3) in response to TGF-β1 and EGF in these cells (Fig. 4A, B and Fig. 5). Our data also suggests that FosB knockdown reduced both TGF-β1- and EGF-induced cell invasion but had no significant effect on the basal invasive potential of these cells (Fig. 5).
Fig. 3.
Effects of FosB knock down on TGF-β1-induced cell proliferation. DU145 (A) and PC3 (B) cells were transfected with siRNA to transiently silence FosB followed by an in vitro proliferation assay.
Fig. 4.
Effects of FosB knock down on cell migration. Prostate cancer cells DU145 (A) and PC3 (B) were pretreated with siRNA against FosB for 72 hr. Western blots were used to confirm knock down of endogenous FosB (inserts). DU145 and PC3 cells were pretreated with siRNA against FosB, followed by treatment with 10 ng/ml of exogenous TGF-β1 and 10 ng/ml EGF migratory behavior were measured using transwell insert migration assay. Each bar represents Mean ± SEM from three independent experiments. Different letters designate statistically significant (P < 0.05) differences among different treatments.
Fig. 5.
Effects of FosB knock down on cell invasion. PC3 cells were pretreated with siRNA against FosB, followed by treatment with 10 ng/ml of exogenous TGF-β1 and 10 ng/ml EGF. Invasive behavior was measured using a matrigel in vitro invasion assay. Insert shows western blot used to confirm FosB knock down. Each bar represents Mean ± SEM from three independent experiments. Different letters designate statistically significant (P < 0.05) differences among different treatments.
DISCUSSION
In this study, we demonstrate for the first time the role of Fos transcription factors in migration and invasion of prostate epithelial cancer cells. We performed two types of experiments: first, the effect of TGF-β1 on the Fos family expression levels were determined by western blot analysis using different doses and varying time of exposure to TGF-β1; the second part of this study, the influence of forced knock down of FosB transcription factor was used to determine the role of FosB on the proliferation, migration, and invasion of two prostate cancer cell lines of different proliferative, migratory, and invasive potential. The key findings in this study are that (i) TGF-β1 induces and increased expression of FosB in prostate epithelial cancer cells; (ii) FosB is essential for migration and invasion to occur in prostate cancer cells; and (iii) FosB is required for TGF-β1- and EGF-induced cell migration and invasion.
AP-1 family of transcription factors are a part of the complex immediate early genomic response of a variety of cells to transmembrane signaling agents [30]. Additional complexity results from the variety of possible Jun dimers and the Jun–Fos heterodimers and from potential dimer formation between Jun or Fos and other leucine zipper proteins [30]. Jun and Fos proteins share extensive homology within the leucine zipper and basic domains. However, despite their homology, these proteins display different transcriptional activity [31]. The Fos proteins contribute distinct functions toward the activity of the AP-1 heterodimers. For example, c-Fos can both activate and repress transcription [32], the full-length FosB is a transcriptional activator [33] and a naturally occurring short form of FosB inhibits AP-1 transactivation [34]. The Fos-related antigens, Fra-1 and Fra-2 lack functional transactivation domains and are poor transcriptional activators [34]. We believed that an alteration in the composition of AP-1 either directly or indirectly regulates cell growth and motility, which in turn pushes the normal cell into pre-malignant or malignant state. Therefore, we analyzed the effect of TGF-β1 on Fos mRNA and protein expression in both normal as well as different prostate cancer cell lines. The most interesting observation was an immediate increase in FosB expression both at the mRNA and the protein levels in prostate cancer cells.
TGF-β super family signaling is well known as a key regulator of many biological processes [35] including differential effects on cell proliferation and migration in prostate cancer cells. These differential effects of TGF-β during different stages of cancer progression presumably depend on selective loss or acquisition of specific intracellular signals that are required to elicit different biological effects to TGF-β. A loss of TGF-β receptors and/or Smad proteins has been shown to result in TGF-β resistance in cancer cells [12,23,36]. However, most cancer cells retain classical TGF-β signaling components throughout cancer progression but modify or recruit additional signaling pathways to exert novel or different biological effects [23]. Our data shows that TGF-β1 increases FosB expression in prostate cancer cells, which, in turn, mediates its effects on migration and invasion but does not play a role in TGF-β1 effects on cell proliferation. Thus TGF-β1 induction of FosB may represent a shift in intracellular signaling involved in the escape from inhibition of proliferation to the stimulation of more migratory and invasive behavior in advanced stages of prostate cancer.
While essential to normal development and homeostasis, the process of cellular migration is also a trait essential for metastasis. Enhanced migration is key across the metastatic cascade and is involved in the initial scattering of cells and migration from the primary tumor [37]. Numerous proteins and pathways have been implicated in altering the migratory potentials of cancer cells and therefore their aggressive nature. Given its essential role in cancer progression, treatments that inhibit cell migration or such proteins/pathways involved in enhancing cellular motility represent an attractive strategy for controlling meta-static dissemination [37]. Because Fra1 and Fra2 exhibit a lack of trans-activating domain as seen in cFos and FosB, they might exert inhibitory functions on tumor growth. However, recent data points to positive effects of Fra1 and partly Fra2, on tumor progression in many tumor types [20,38]. In contrast, to the bulk of data on the function of cFos and Fra1, far less is known about the role of FosB and its smaller splice variant ΔFosB which is often expressed more strongly than Fra1 in clinical cancer tissues [20,39]. Although, the Fos family of proteins has been extensively studied as immediate early genes, the role of FosB in cancer cell proliferation and migration has not been previously investigated. Our data shows that transient silencing of FosB with or without the presence of TGF-β1 has no effect on prostate cancer cell proliferation but significantly reduces cell migration and invasion. Numerous studies have demonstrated that TGF-β1 induces the migration and invasion of prostate cancer cells; however, we show in this study that TGF-β1 is unable to induce prostate cancer cell migration and invasion without FosB. The data also suggests that epidermal growth factor (EGF), a potent mitogenic factor that plays an important role in the growth, proliferation and differentiation of numerous cell types is unable to induce migration and invasion in prostate cancer cells in the absence of FosB; further confirming that FosB does indeed have a major role in migration and invasion of prostate cancer cells. Thus the differences in migratory and invasive behavior observed in different stages of prostate cancer progression can be due to AP-1 specifically FosB activation. FosB may have a role in the aggressive phenotype observed in prostate cancer, thus inhibition of FosB activity may serve as a therapeutic tool in the management of prostate cancer.
CONCLUSIONS
In conclusion, our results obtained using human prostate cancer cell lines suggest that the transcription factor FosB may be an important regulator of TGF-β1 effects on migration and invasion in human prostate cancer cells. The further study of the role of FosB in prostate cancer carcinogenesis, especially in vivo, will be of great importance and will probably open new perspectives for therapy.
ACKNOWLEDGMENTS
We are grateful to Ms. Chelesie Leath and Ms. Jasmine Mosley for technical assistance. This study was supported by grants from NIH/NIMHD/RCMI grant #G12MD007590 and NIH/NIMHD #5P20MD002285.
Footnotes
Conflict of interest: The authors have nothing to disclose.
REFERENCES
- 1.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Arnold JT, Isaacs JT. Mechanisms involved in the progression of androgen-independent prostate cancers: It is not only the cancer cell's fault. Endocr Relat Cancer. 2002;9:61–73. doi: 10.1677/erc.0.0090061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collazo J, Zhu B, Larkin S, Martin SK, Pu H, Horbinski C, Koochekpour S, Kyprianou N. Cofilin drives cell-invasive and metastatic responses to TGF-beta in prostate cancer. Cancer Res. 2014;74:2362–2373. doi: 10.1158/0008-5472.CAN-13-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong CM, Gao AC. Drug resistance in castration resistant prostate cancer: Resistance mechanisms and emerging treatment strategies. Am J Clin Exp Urol. 2015;3:64–76. [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrasekar T, Yang JC, Gao AC, Evans CP. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl Androl Urol. 2015;4:365–380. doi: 10.3978/j.issn.2223-4683.2015.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Zhang X, Xie F, Zhang Z, van Dam H, Zhang L, Zhou F. The regulation of TGF-beta/SMAD signaling by protein deubiquitination. Protein Cell. 2014;5:503–517. doi: 10.1007/s13238-014-0058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat Cell Biol. 2007;9:1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 9.Massague J. A very private TGF-beta receptor embrace. Mol Cell. 2008;29:149–150. doi: 10.1016/j.molcel.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Bhowmick NA, Moses HL. Tumor-stroma interactions. Curr Opin Genet Dev. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielpour D. Functions and regulation of transforming growth factor-beta (TGF-beta) in the prostate. Eur J Cancer. 2005;41:846–857. doi: 10.1016/j.ejca.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 12.Guo Y, Kyprianou N. Overexpression of transforming growth factor (TGF) beta1 type II receptor restores TGF-beta1 sensitivity and signaling in human prostate cancer cells. Cell Growth Differ. 1998;9:185–193. [PubMed] [Google Scholar]
- 13.Strong N, Millena AC, Walker L, Chaudhary J, Khan SA. Inhibitor of differentiation 1 (Id1) and Id3 proteins play different roles in TGFbeta effects on cell proliferation and migration in prostate cancer cells. Prostate. 2013;73:624–633. doi: 10.1002/pros.22603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 15.Gallo A, Cuozzo C, Esposito I, Maggiolini M, Bonofiglio D, Vivacqua A, Garramone M, Weiss C, Bohmann D, Musti AM. Menin uncouples Elk-1, JunD and c-Jun phosphorylation from MAP kinase activation. Oncogene. 2002;21:6434–6445. doi: 10.1038/sj.onc.1205822. [DOI] [PubMed] [Google Scholar]
- 16.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 17.Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 18.Shaulian E, Schreiber M, Piu F, Beeche M, Wagner EF, Karin M. The mammalian UV response: C-Jun induction is required for exit from p53-imposed growth arrest. Cell. 2000;103:897–907. doi: 10.1016/s0092-8674(00)00193-8. [DOI] [PubMed] [Google Scholar]
- 19.Yazgan O, Pfarr CM. Regulation of two JunD isoforms by Jun N-terminal kinases. J Biol Chem. 2002;277:29710–29718. doi: 10.1074/jbc.M204552200. [DOI] [PubMed] [Google Scholar]
- 20.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Renaud SJ, Kubota K, Rumi MA, Soares MJ. The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem. 2014;289:5025–5039. doi: 10.1074/jbc.M113.523746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milde-Langosch K, Kappes H, Riethdorf S, Loning T, Bamberger AM. FosB is highly expressed in normal mammary epithelia, but down-regulated in poorly differentiated breast carcinomas. Breast Cancer Res Treat. 2003;77:265–275. doi: 10.1023/a:1021887100216. [DOI] [PubMed] [Google Scholar]
- 23.Vo BT, Morton D, Jr, Komaragiri S, Millena AC, Leath C, Khan SA. TGF-beta effects on prostate cancer cell migration and invasion are mediated by PGE2 through activation of PI3K/AKT/mTOR pathway. Endocrinol. 2013;154:1768–1779. doi: 10.1210/en.2012-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ten Dijke P, Yamashita H, Ichijo H, Franzen P, Laiho M, Miyazono K, Heldin CH. Characterization of type I receptors for transforming growth factor-beta and activin. Sci. 1994;264:101–104. doi: 10.1126/science.8140412. [DOI] [PubMed] [Google Scholar]
- 25.Wieser R, Attisano L, Wrana JL, Massague J. Signaling activity of transforming growth factor beta type II receptors lacking specific domains in the cytoplasmic region. Mol Cell Biol. 1993;13:7239–7247. doi: 10.1128/mcb.13.12.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millena AC, Reddy SC, Bowling GH, Khan SA. Autocrine regulation of steroidogenic function of Leydig cells by transforming growth factor-alpha. Mol Cell Endocrinol. 2004;224:29–39. doi: 10.1016/j.mce.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Vo BT, Khan SA. Expression of nodal and nodal receptors in prostate stem cells and prostate cancer cells: Autocrine effects on cell proliferation and migration. Prostate. 2011;71:1084–1096. doi: 10.1002/pros.21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zigmond SH, Foxman EF, Segall JE. Chemotaxis assays for eukaryotic cells. Curr Protoc Cell Biol. 2001 doi: 10.1002/0471143030.cb1201s00. chapter 12. Section 12.1. [DOI] [PubMed] [Google Scholar]
- 29.Walker L, Millena AC, Strong N, Khan SA. Expression of TGFbeta3 and its effects on migratory and invasive behavior of prostate cancer cells: Involvement of PI3-kinase/AKT signaling pathway. Clin Exp Metastasis. 2013;30:13–23. doi: 10.1007/s10585-012-9494-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakabeppu Y, Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991;64:751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- 31.Finch S, Joseloff E, Bowden T. JunB negatively regulates AP-1 activity and cell proliferation of malignant mouse keratinocytes. J Cancer Res Clin Oncol. 2002;128:3–10. doi: 10.1007/s00432-001-0298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gius D, Cao XM, Rauscher FJ, 3rd, Cohen DR, Curran T, Sukhatme VP. Transcriptional activation and repression by Fos are independent functions: The C terminus represses immediate-early gene expression via CArG elements. Mol Cell Biol. 1990;10:4243–4255. doi: 10.1128/mcb.10.8.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobrazanski P, Noguchi T, Kovary K, Rizzo CA, Lazo PS, Bravo R. Both products of the fosB gene, FosB and its short form, FosB/SF, are transcriptional activators in fibroblasts. Mol Cell Biol. 1991;11:5470–5478. doi: 10.1128/mcb.11.11.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutberg SE, Saez E, Lo S, Jang SI, Markova N, Spiegelman BM, Yuspa SH. Opposing activities of c-Fos and Fra-2 on AP-1 regulated transcriptional activity in mouse keratinocytes induced to differentiate by calcium and phorbol esters. Oncogene. 1997;15:1337–1346. doi: 10.1038/sj.onc.1201293. [DOI] [PubMed] [Google Scholar]
- 35.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 37.Darnell JE., Jr Variety in the level of gene control in eukaryotic cells. Nat. 1982;297:365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- 38.Sommerville J. RNA polymerase I promoters and transcription factors. Nature. 1984;310(5974):189–190. doi: 10.1038/310189a0. [DOI] [PubMed] [Google Scholar]
- 39.Moquet-Torcy G Tolza C, Piechaczyk M, Jariel-Encontre I. Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res. 2014;42:11011–11024. doi: 10.1093/nar/gku814. [DOI] [PMC free article] [PubMed] [Google Scholar]