Abstract
In the study of allosteric proteins, understanding which effector/protein interactions contribute to allosteric activation is important both for designing allosteric drugs and for understanding allosteric mechanisms. The anti-hyperglycemic target, human liver pyruvate kinase (hL-PYK), binds its allosteric activator, fructose-1,6-bisphosphate (Fru-1,6-BP), such that the 1’-phosphate interacts with side chains of Arg501 and Trp494 and the 6’-phosphate interacts with Thr444, Thr446, Ser449 (i.e. the 444–449 loop), and Ser531. Additionally backbone atoms from the 527–533 loop interact with a sugar ring hydroxyl and the two effector phosphate moieties. An effector analogue series indicates that only one phosphate on the sugar is required for activation. However, singly phosphorylated sugars, including Fru-1-P and Fru-6-P, bind with a Kix in the rage of 0.07 to 1 mM. The second phosphate of Fru-1,6-BP causes tight effector binding, since this native effector has a Kix of 0.061 µM. Glucose-1,6-bisphosphate and ribulose-1,5-bisophosphate bind in the 0.07 to 1mM range. The contrast with higher Fru-1,6-BP binding indicates specificity for the fructose sugar conformation. Site-directed random mutagenesis at each residue that contacts bound Fru-1,6-BP identified that a negative charge introduced at 531 mimics allosteric activation, even in the absence of Fru-1,6-BP. Collectively, analogue and mutagenesis studies are consistent with the 527–533 loop playing a key role in allosteric function. Deletion mutations that shortened the 527–533 loop were expected to prevent formation of hydrogen bonds between backbone atoms on the loop and Fru-1,6-BP. Indeed, Fru-1,6-BP did not activate these loop-shortened mutant proteins. Previous structural comparisons of M1-PYK and M2-PYK indicate that the 527–533 loop makes interactions across a subunit interface when activator is not present. Mutating the hL-PYK subunit interface interactions among Trp527, Arg528 and Asp499 mimics allosteric activation. Considered with published structures, these results are consistent with 1) the two phosphates of Fru-1,6-BP dock to Arg501/Trp494 and the 444–449 loop, respectively; 2) the formation of hydrogen bonds among Fru-1,6-BP and backbone atoms of the 527–533 loop pulls this loop away from the subunit interface and results in breaking of the Trp527/Arg528/Asp499 interactions to elicit an allosteric response.
Keywords: Allosteric regulation, allostery, binding, activation
Graphical abstract
Introduction
Allosteric drugs that activate human liver pyruvate kinase (hL-PYK) are an attractive goal to counteract the hyperglycemia associated with diabetes. hL-PYK catalyzes the transfer of a phosphate moiety from phosphoenolpyruvate (PEP) to ADP to result in ATP and pyruvate as the last step in glycolysis. As an anti-hyperglycemic target, the hepatic glucose production associated with diabetes could be modulated via activation of hL-PYK to create a futile energy-burning cycle that is not expected to contribute to weight gain. One approach to design allosteric drugs is to create compounds that mimic physiological allosteric activators. To that end, it is imperative to realize that allosteric function can be localized to a limited number of the total protein/effector interactions (1–10). These allosterically important interactions can be distinct from those protein/effector interactions that make the largest contributions to effector binding. Therefore, the goal of this study was to distinguish the interactions between hL-PYK and the physiological allosteric activator, fructose-1,6-bisphosphate (Fru-1,6-BP), that contribute to allosteric activation.
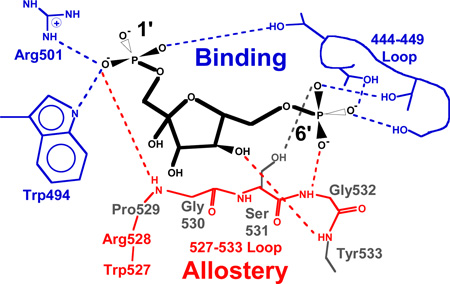
The Fru-1,6-BP binding site of pyruvate kinase isozymes was originally identified in a co-crystallized structure of yeast PYK (11). With the exception of the location of the hydroxyl moiety at the anomeric carbon (carbon 2), Fru-1,6-BP has a 2-fold rotational axis (C2-symmetry). This near symmetry has led to perceived disagreements in the binding orientation when bound to PYK isozymes (11–13). However, there are no differences in the actual electron densities for any of the co-crystallized structures. We recently solved a 1.85Å structure of hL-PYK with Fru-1,6-BP bound (14). The improved resolution confirms that Fru-1,6-BP binds to hL-PYK with the 1’-phosphate directed towards Arg501 of hL-PYK (Figure 1), the same binding orientation found in yeast-PYK, human M2-PYK, and human R-PYK.
Figure 1.
A schematic of the coordination of Fru-1,6-BP to human L-PYK (27). This schematic is intentionally designed to mimic that used in the initial characterization of Fru-1,6-BP binding to yeast PYK (11). Fru-1,6-BP is in black and protein is in gray. Interactions between Fru-1,6-BP and the protein are indicated with dashed lines).
To accomplish our goal of defining which interactions between hL-PYK and Fru-1,6-BP contribute to allostery, the study reported here employed both an effector analogue series and mutations in the activator binding site. In addition to facilitating future rational drug designs, the overall conclusions provided insights into the molecular mechanisms that give rise to allostery in hL-PYK.
Materials and Methods
Materials
The potassium salts of ADP and PEP were purchased from Chem-Impex International, Inc. NADH was from Sigma. L-lactate dehydrogenase (Type III bovine heart) was purchased from Calzyme Laboratories, Inc. Other buffer components were from Fisher Scientific and Sigma. The sodium salt of 2-deoxyribose 5-phosphate, the dipotassium salt of D-glucose 6-phosphate, The sodium salt of D-ribulose-1,5-bisphosphate, the dipotassium salt of α-D-glucose 1-phosphate and the disodium salt of D-fructose 6-phosphate were purchased from Sigma-Aldrich. DL-glyceraldehyde 3-phosphate, the sodium salt of D-fructose 1-phosphate, the dibarium salt of 2,5-anhydro-D-mannitol-1,6-bisphosphate, 2,5-anhydro-D-glucitol-1,6-bisphosphate and the cyclohexylammonium salt of α-D-glucose-1,6-bisphosphate were purchased from Santa Cruz Biotechnology. A table showing the structure of each analogue is included in the Supplemental Material.
Mutagenesis and Protein Expression and Purification
Mutagenesis of the human L-PYK gene was performed with Quikchange (Stratagene). Many of the included mutations were created using site-directed random mutagenesis via primers that were degenerate at the target codon. Other mutations were generated with specifically designed primers. Wild type and mutant proteins were expressed in the FF50 strain of Escherichia coli (15). Wild type protein used for analogue studies was purified using the cell lysis, ammonium sulfate fractionation and DEAE-cellulose column as previously described (15). Mutant proteins were partially purified using ammonium sulfate fractionation followed by dialysis (16). Estimates of ligand binding/affinity and allostery were equivalent whether evaluated using purified or ammonium sulfate partially purified protein (Supplemental Figure S1). Therefore, mutant proteins were only partially purified before evaluation, a method that allowed an assessment of considerably more mutations than would have been possible if purification of each was required.
Kinetic Assays and data analysis
Activity measurements were at 30°C, using a lactate dehydrogenase coupled assay in either HEPES or bicine buffer, pH 7.5 (17). As previously described, reaction conditions contained 50 mM HEPES or bicine, 10 mM MgCl2, 2 mM (K)ADP, 0.1 mM EDTA, 0.18 mM NADH, and 19.6 U/mL lactate dehydrogenase. PEP and effector concentrations were varied. The rate of the decrease in A340 due to NADH utilization was recorded at each concentration of PEP and these initial velocity rates as a function of PEP concentration were used to evaluate Kapp-PEP at any one effector concentration. In turn, Kapp-PEP evaluated over a concentration range of effector was used to evaluate allosteric parameters as noted in data fits below. All assays were completed in 96-well plates, such that 12 wells with 12 PEP concentrations were used to evaluate Kapp-PEP at each concentration of effecter. In turn, the 8 rows in the 96-well plate included 8 unique concentrations of effector. The enzymatic reaction was initiated with PEP and monitored at 340 nm over time in a UV-Star flat bottom 96-well plate (Greiner bio-one) containing a total reaction volume of 350 lL. All activity readings were collected using a Molecular Devices Spectramax Plus384 spectrophotometer.
Mutations were assayed in the exact conditions previously described (17). Commercially available analogues typically used either sodium or potassium as the counter-ion. To prevent a large percent change in either Na+ or K+ concentration, NaCl and KCl were added for a final concentration of 100 mM and 150 mM, respectively. Under these conditions, the highest concentration of effector that could be used before observing non-specific effects was determined to be 0.5 mM (see Supplemental Figure S2). Glucose-1,6-BP and 5-anhydro-D-mannitol-1,6-BP were purchased as the barium and cyclohexylammonium salt, respectively. Therefore, in assays with barium, the total Ba2+ concentration was held constant at 20 mM by BaCl2 additions and in assays with cyclohexylammonium, the total C6H14N+ concentration was held constant at 30 mM by additions of cyclohexylammonium chloride (in addition to 150 mM Na+ and 100 mM K+). We previously indicated that complete replacement of the monovalent cation from Na+ to K+ alters kinetic and allosteric parameters (17). Therefore, it was not surprising that addition of Na+, Ba2+, and/or C6H14N+ altered parameters as compared to those previously reported for the K+-only assay condition (Supplemental Figure S3). The Fru-1,6-BP activation of mutant proteins was assayed in the 100 mM K+ conditions previously reported (17). For each analogue, comparisons were only made to the wild type/Fru-1,6-BP assayed in the same condition.
Data fitting was with the nonlinear least-squares analysis of Kaleidagraph (Synergy) software. As previously noted, the dependency of enzymatic activity on PEP concentration is biphasic (15, 17). Kapp-PEP values were obtained by fitting initial rates obtained from kinetic assay to:
| Equation 1 |
Where Vmax is the maximum velocity associated with the low PEP phase, Kapp-PEP is the concentration of substrate that yields a rate equal to one-half the Vmax, nH is the Hill coefficient associated with the low PEP (i.e. high PEP affinity) phase, and c is the Vmax/Kapp-PEP for the low affinity PEP phase for which a maximal velocity is not observed within working PEP concentration range. Although nH values may influence fit parameters associated with PEP affinity, we previously demonstrated (15) that nH values from PEP titrations have minimum influence on allosteric parameters as evaluated below.
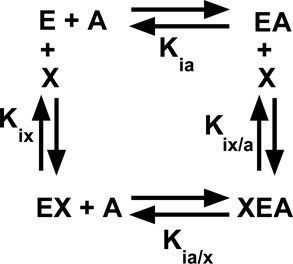
Our evaluation of allostery is based on a linked-function based energy cycle between substrate (A) and effector (X) binding to the enzyme (E). This energy cycle is represented in Reaction 1.
Reaction 1.
In addition, to the dissociation constants defined in Reaction 1,
| Equation 2 |
Since pyruvate kinase is considered a rapid equilibrium system, Kapp-PEP values obtained from initial velocity reactions were used as a proxy for PEP dissociation constant values. When evaluating allosteric regulation, Qax was determined by fitting a plot of the Kapp-PEP values as a function of effector concentration to equation 3 (18):
| Equation 3 |
where Ka = Kapp-PEP when [Effector] = 0; Kix = the dissociation constant for effector (X) binding to the protein in the absence of substrate (A); and Qax is the allosteric coupling constant discussed in detail elsewhere (18). Data for all analogue studies were replicated at least twice by two different researchers; average fit parameters are included in figures along with errors propagated from error estimates obtained from fitting individual data sets. Data for mutated proteins is from single assays. However, duplicate data collection for two independent protein predations was completed before concluding that a mutated enzyme lacks activity.
Results
The overall design of this project was to modify either a chemical moiety of the effector (analogue series) or the chemical nature of a residue side-chain of the protein (mutations) and determine if allosteric function and/or effector affinity were altered relative to the wild type/Fru-1,6-BP reference. The approach for quantifying allosteric function has previously been detailed (17). Briefly, the apparent affinity (Kapp-PEP) for substrate, PEP, was first determined by following initial velocity as a function of PEP concentration. Kapp-PEP as a function of activator concentration was fit to Equation 3 (see Materials and Methods) to obtain fit parameters for substrate affinity in the absence of effector (Ka-PEP), effector binding in the absence of substrate (Kix), and the allosteric coupling between the activator and PEP (Qax). Increased Ka-PEP and Kix indicate decreased binding affinity for the respective ligands, but Qax approaches unity as allosteric coupling in the system is reduced.
Binding of Fru-1,6-BP analogues
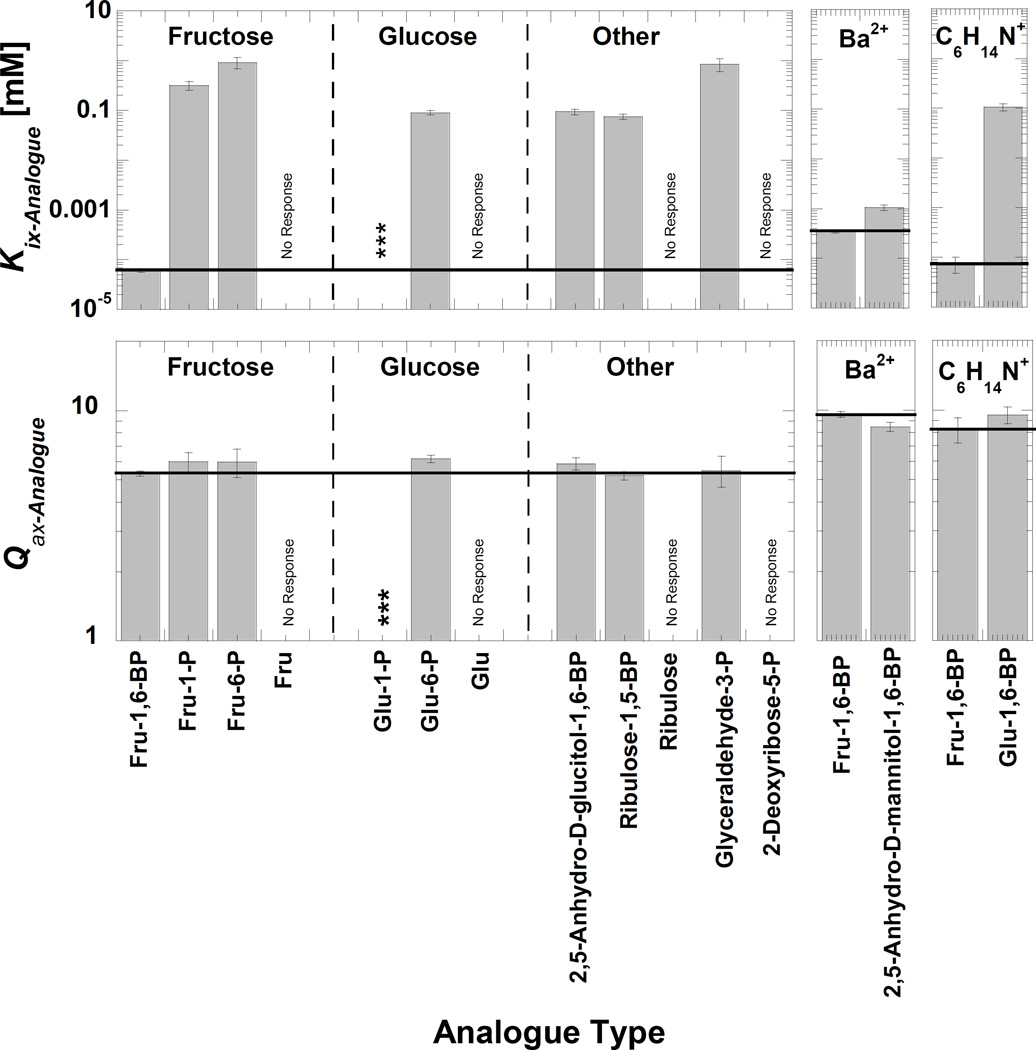
Fit parameters for activation by Fru-1,6-BP analogues are recorded in Figure 2 and Supplemental Table S2. Overall the effector binding affinity (Kix) was more sensitive to the varied chemical nature of the analogues than was the allosteric coupling (Qax). Analogue binding can be roughly classed into three groups: 1) one analogue that binds similarly to Fru-1,6-BP, 2) analogues that bind with lower affinity (higher values of Kix), and 3) the group of analogues that show no binding/activation.
Figure 2.
Binding (Kix) and allosteric coupling (Qax) of analogues of Fru-1,6-BP. The left panel is for data collected in the 100mM Na+/150 mM K+ condition. Glu-1-P (***) binding was sufficiently weak that within the concentration limit saturation was not possible, preventing a fit to Equation 3. There was no shift in Kapp-PEP (i.e. “No Response”) upon additions of Fru, Glu, ribulose, or 2-deoxyribose-5-P. Far right: Glucose-1,6-BP was purchased as a cyclohexyammonium salt and 2,5-Anhydro-D-Mannitol-1,6-BP (Mannitol-1,6-BP) was purchased as a barium salt. Therefore, Fru-1,6-BP controls assayed with an equivalent concentration of alternative counterion are included for comparison. In all panels, horizontal bars representing the value of the assay-matched Fru-1,6-BP control to aid comparison. Ligand abbreviations are: Fru-1,6-BP (fructose-1,6-bisphosphate), Fru-1-P (fructose-1-phosphate), Fru-6-P (fructose-6-phosphate), Fru-(fructose), Glu-1-P (glucose-1-phosphate), Glu-6-P (glucose-6-phosphate), Glu (glucose), 2,5-anhydro-D-glucitol-1,6-BP (2,5-anhydro-D-glucitol-1,6-bisphosphate), 2,5-anhydro-D-manitol-1,6-BP (2,5-anhydro-D-mannitol-1,6-bisphosphate), and Glu-1,6-BP (glucose-1,6-bisphosphate). Glyceraldehyde-3-P used in this study was a D/L mixture; the Kix estimate is based on the total concentration.
2,5-Anhydro-D-mannitol-1,6-BP was the only analogue that binds with an affinity close to that of Fru-1,6-BP. The hydroxyl at the anomeric #2 carbon is the only moiety that prevents Fru-1,6-BP from having 2-fold rotational symmetry (C2-symmetry). 2,5-Anhydro-D-mannitol-1,6-BP and 2,5-anhydro-D-glucitol-1,6-BP lack this anomeric hydroxyl, resulting in a sugar ring that is locked into one of two anomeric conformation. 2,5-Anhydro-D-mannitol-1,6-BP has C2-symmetry. Consistent with one previous study of yeast PYK (19) (but not the other (20)), only 2,5-anhydro-D-mannitol-1,6-BP mimics the binding and allosteric responses elicited by Fru-1,6-BP. This is, in turn, consistent with the β-anomer being the activating form of Fru-1,6-BP, an observation that is also consistent with structural data (11–13). Also, the relatively subtle effect of removing the anomeric hydroxyl (i.e. compare 2,5-anhydro-D-mannitol-1,6-BP with Fru-1,6-BP assayed in the same condition) implies that the anomeric hydroxyl provides only slight selection in the binding orientation of Fru-1,6-BP (see more below).
Excepting 2,5-anhydro-D-mannitol-1,6-BP, all analogues that elicit a response bind with a Kix value in the 0.07 to 1 mM range. This range is 3 to 4 orders of magnitude weaker than binding of Fru-1,6-BP. Since the group of analogues with low binding contains includes molecules with only one phosphate, clearly one phosphate is sufficient for weak binding and allosteric activation. Fru-1-P and Fru-6-P are included in the analogue group with Kix values in the 0.07 to 1 mM range. By contrasting these binding affinities with that of Fru-1,6-BP, we conclude that the second phosphate of Fru-1,6-BP must causes tight effector binding. In contrast to Fru-1,6-BP, glucose-1,6-bisphosphate and ribulose-1,5-bisophosphate bind in the 0.07 to 1mM range. Therefore, we can then suggest that the other bisphosphate examples are likely binding through interactions made with only one of the two phosphates and that Fru-1,6-BP can access some unique shape that is important for proper positioning of the two phosphates for binding. This fructose specific conformation is then the likely source of effector specificity.
Within the group of analogues that activate, but do so with greatly reduced binding (compared to Fru-1,6-BP), Fru-1-P and Fru-6-P deserve special consideration. With regards to which interactions contribute to binding, it is very clear that removal of either phosphate moiety from Fru-1,6-BP (i.e. Fru-1-P and Fru-6-P) greatly reduces binding. However given the minimal selectivity that is provided by the presence of the anomeric hydroxyl (compare Kix for Fru-1,6-BP with that for 2,5-anhydro-D-mannitol-1,6-BP), it seems likely that Fru-1-P and Fru-6-P can bind in the same orientation in the effector binding site (e.g. the phosphate of Fru-1-P and the phosphate of Fru-6-P might both interact with the 444–449 loop). In fact, the very slight difference in binding of Fru-1-P vs. Fru-6-P is reminiscent of the slight difference between binding of Fru-1,6-BP vs. 2,5-anhydro-D-mannitol-1,6-BP. This interpretation can be extrapolated to suggest that Fru-1,6-BP can bind in two possible orientations, with only a slight binding preference for the orientation represented in protein structures. More important to our overall goal of this study, the possibility that Fru-1-P and Fru-6-P can bind in the same orientation prevents an interpretation of the analogue series regarding which protein/effector interactions are most relevant to allosteric regulation.
2-Deoxyribose-5-P was the only phosphorylated analogue included in the group of mutations that showed no activation (Figure 2). (The binding of Glu-1-P was sufficiently low that saturation with this analogue was not obtained within the 0.5 mM maximum concentration, but Glu-1-P did activate.) Other analogues that did not activate were the non-phosphorylated sugars (fructose, glucose, and ribulose). This lack of response does not distinguish between binding without allostery vs. loss of binding. However, given the drastic decrease in binding caused by removing either of the ligand phosphate moieties, the lack of response by non-phosphorylated sugars likely reflects a complete lack of binding.
Allosteric activation by Fru-1,6-BP analogues
Considering only analogues that elicited an allosteric response, the magnitude of the allosteric coupling (Qax) was less sensitive than Kix to chemical differences among the analogues (Figure 2). With the exception of 2-deoxyribose-5-P and Glu-1-P discussed above, once sufficient concentrations of each of the phosphorylated analogues were added, the degree to which PEP affinity was increased (Qax) was approximately the same for all phosphorylated analogues. Since many of the activating analogues contained only one phosphate (including Fru-1-P and Fru-6-P), we conclude that the presence of only one phosphate on the sugar is necessary to elicit the allosteric response. However, as discussed in the next two sections, some component of the sugar ring is also required for allosteric function. Therefore, a single phosphate is necessary, but not sufficient for allostery.
Phosphate as an effector analogue
If the interaction of only one phosphate moiety from the activator is required for activation, then we asked if phosphate alone can cause increased PEP affinity. Phosphate was used cautiously, realizing that it might bind competitively with either PEP and/or ADP in the active site to result in inhibition. To our surprise, phosphate neither increased nor decreased PEP affinity, nor caused reduced Vmax. Instead, the presence of phosphate removed the previously reported (15) biphasic response to increasing PEP concentrations; phosphate caused the Vmax of the first phase to increase to a level equal to Vmax of the second phase, so at maximal phosphate only one phase is observed (Supplemental Figure S4). Therefore, the second phase observed only at high PEP concentrations may be a result of Vmax activation due to phosphate contamination in PEP stocks. We have already provided a number of other potential reasons for the biphasic response (15, 17). Since the second phase occurs only at very high PEP concentrations, rigorously describing the mechanism of this response is not a current priority. More important to the purpose of this work, phosphate ion concentrations up to 1 mM did not cause an increase in the affinity of hL-PYK for PEP. This is consistent with previous observations (21). As described above, the absence of a response does not distinguish between whether the phosphate ion binds in the Fru-1,6-BP binding site with no allosteric response vs. a failure to bind.
Minimal requirement for binding and allostery
Up to this point, we can conclude that neither fructose nor phosphate activate hL-PYK. However, phosphorylated sugars act as allosteric activators. Given these observations we can return to the analogue series (Figure 2) to consider the minimal chemical structure requirements that result in sufficient binding affinity to observe allosteric regulation. The smallest analogue that continues to elicit an increase in PEP affinity is glyceraldehyde-3-P. Therefore, the combined sugar-phosphate is required for activation and the contribution of the sugar can be further localized to the smaller glyceraldehyde-like substructure. A sugar ring hydroxyl is a likely candidate for the chemical moiety on the sugar that is required for allostery, especially given that one sugar ring hydroxyl appears to make a hydrogen bond with a backbone atom from the 527–533 loop (Figure 1).
Single Point Mutations of Arg501 and Trp494
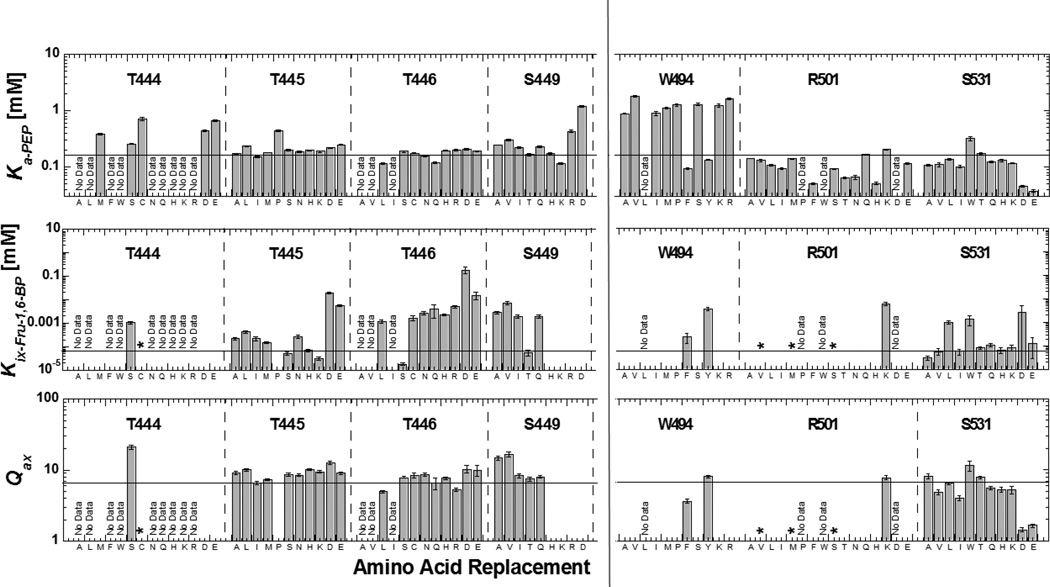
To further clarify which activator/protein interactions were required for allosteric function, we next turned to mutagenesis of the hL-PYK allosteric site. Each of the residues with side-chains that directly interact with Fru-1,6-BP (Figure 1) were mutated. In any mutational study, one challenge is discriminating whether altered properties are due to 1) the removal of the original side-chain or 2) addition of the substitute side-chain. One approach to resolve this issue is to make a series of mutations, in order to determine which side-chain properties do and do not modify function (i.e. Ka, Kix and Qax in this study). Furthermore, any single substitution at a given residue position may cause multiple perturbations in the protein and these various perturbations can compensate each other in a given protein function (22). This potential compensation can mask the identification of residues that participate in a function when screened by a mutagenesis study. Multiple substitutions introduced at a given position increase the likelihood of identifying substitutions that perturb function. For these reasons, a series of substitutions was introduced at each position in the Fru-1,6-BP binding site. Positional-random mutagenesis (using primers degenerate at the codon of interest) was used to generate a substitution series at each position, such that there was no control over which substitutions were obtained for each probed position. The nature of the substitution was determined by DNA sequencing. Each mutated gene was isolated and introduced into FF50 E. coli for protein expression. The allosteric activation by Fru-1,6-BP was determined using the same approach described for characterization of the analogues (above). Fit parameters for activation of mutated proteins by Fru-1,6-BP are recorded in Figure 3 and Table S3 in Supplemental Material.
Figure 3.
Ka-PEP, Kix and Qax determined for mutant proteins. Mutated residues are included in the respective section of the graph, sections dedicated to each residue position are divided by vertical dashed lines, and replacement residues at the respective positions are listed on the x-axis. For each position, replacement residues are roughly ordered from smallest to greatest hydrophobic, smallest to largest hydrophilic, positive charge and negative charge from left to right. In each panel, a line is included to indicate the wild type value. Several mutations were not active and are noted with “No Data”. Examples that show some response to Fru-1,6-BP, but with sufficiently decreased affinity to prevent saturation with the activator are indicated by an asterisk (*). The absence of any data/symbol in graphs of Kix and Qax indicate no allosteric response.
In the crystal structure, Arg501 clearly makes a charge-charge interaction with the 1’-phosphate of Fru-1,6-BP (Figure 1). Most mutations of Arg501 retain enzymatic activity, but show either no allosteric response or greatly decreased Fru-1,6-BP binding such that data fitting was not possible (Figure 3). R501K is the one exception that retains allosteric properties close to that of the wild type enzyme. This highlights the role of the positive charge of the 501 side-chain in activator binding/activation.
Trp494 might interact with the sugar ring of the allosteric activator, although a hydrogen bond interaction between the indole ring nitrogen and the 1’-phosphate has also been indicated (11, 12). Trp494 also potentially contributes to creating a local hydrophobic environment that influences other ionic and hydrogen bonding and proper folding in the local area of the protein. Consistent with this possible role of the hydrophobic nature of Trp494, Tyr and Phe were the only replacements at position 494 that retained effector binding and allosteric functions (Figure 3). In contrast, smaller hydrophobic (Met, Ile, Val, Ala) or hydrophilic (Ser, Lys, Arg) replacements caused loss of response to Fru-1,6-BP. Notably, many mutations at the 494 position caused reduced PEP affinity (see below).
Single Point Mutations in the 444–449 loop
Thr444, Thr446, and Ser449 interact with the 6’-phosphate of Fru-1,6-BP (Figure 1), whereas the side-chain of Thr445 is orientated towards the 1’-phosphate of the activator. Since most substitutions at position 444 abolished detectible enzymatic activity, we can speculate that this position is important to proper protein folding. Of the exceptions that retained activity, both T444E and T444D lacked an allosteric response to Fru-1,6-BP. These negatively charged residues are expected to cause charge repulsion with the effector phosphate moieties, thereby preventing Fru-1,6-BP binding.
Mutations introduced at positions 445, 446, and 449 resulted in common data trends. Altering the side-chain chemistry at these positions modified Fru-1,6-BP binding, but caused only limited perturbations of the allosteric coupling (Qax). In fact, some of the largest reductions in Kix occurred when a negatively charged side-chain was substituted at either the 445 or 446 positions. This likely reflects charge repulsion between the replacement acidic side-chain and ligand phosphates. Overall, Thr445, Thr446, and Ser449 appear to contribute to effector binding, but are not required for allosteric regulation.
Mutations in the 527–533 loop
The side-chain of the Ser531 residue is the only side-chain on loop 527–533 that appears to directly interact with Fru-1,6-BP and it does so via an interaction with the 6’-phosphate of the activator. In general, mutations at Ser531 followed a similar trend as mutations introduced at 445, 446, and 449, albeit with less impact on Fru-1,6-BP binding. However, acidic substitutions at position 531 caused considerable differences. Not only were values of Ka-PEP greatly decreased (increased affinity), but also the allosteric activation (Qax) caused by Fru-1,6-BP was reduced. Both observations indicate that the S531D and S531E proteins are constitutively in a more activated state (even in the absence of Fru-1,6-BP; Ka-PEP below 0.5mM) compared to the wild type enzyme. The addition of Fru-1,6-BP activated both S531D and S531E, although with a decreased allosteric response compared to the wild type protein. Therefore, we can envision that Fru-1,6-BP may displace the putative interaction between the introduced negative charge at 531 and some other region of the activator binding site. Fru-1,6-BP activation of S531D and S531E also implies that the carboxylic acid of a side-chain at position 531 can only partially fulfill the role of a negatively charged ligand phosphate. The need for Fru-1,6-BP to displace a preexisting interaction would also suggest that Fru-1,6-BP should have reduced binding affinity to the S531D and S531E proteins. As predicted the S531D (but not S531E) shows the expected increase in the Kix value (Supplemental Figure S5).
Double and Multiple Mutations
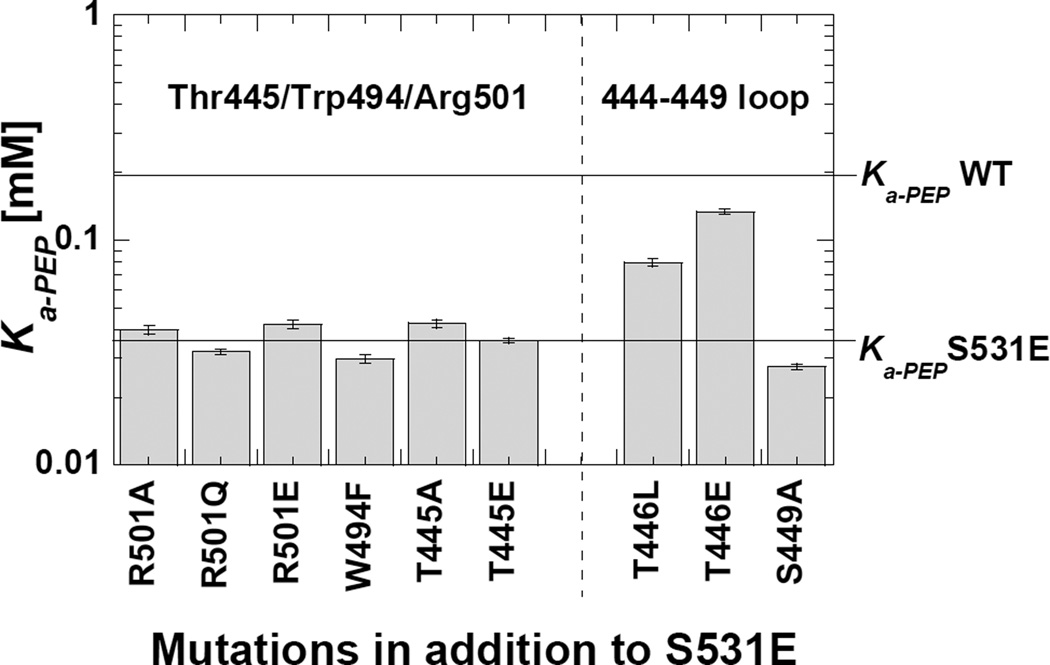
We combined S531E with other mutations to identify how the introduced native charge at 531 interacts within the Fru-1,6-BP binding site. We were not surprised that none of the mutant proteins in this series responded to Fru-1,6-BP. Therefore, to evaluate these mutations we exclusively considered whether Ka-PEP is below 0.05 mM like the Fru-1,6-BP activated enzyme or closer to 0.2 mM, similar to the non-activated wild type enzyme (Figure 4). Adding a negative charge at 446 appears to counteract the S531E activation based on a Ka-PEP value near 0.2 mM. Charge repulsion between the negative charged side-chains likely indicates that S531E binds to the 444–449 loop. In contrast, adding mutations at Thr445, Arg501 or Trp494 (in addition to S531E) resulted in Ka-PEP values close to that of the S531E protein. We can then propose that S531E interacts with the 444–449 loop.
Figure 4. A double mutant study with S531E. The goal of this design was to identify residues that likely interact with the introduced negative charge from the S531E mutation. For the second substitution, only mutations (when introduced as the single mutation in the protein; see Figure 3) that caused minimal changes on Ka-PEP were included. In the presence of S531E, modification of residues in the 444–449 loop cause the Ka-PEP value to approach that for the wild type enzyme. Therefore, S531E likely interacts with this 444–449 loop to mimic activation.
With regards to allostery, we considered that the introduced negative charge from S531E could place a negative charge in a position to interact with the 444–449 loop and, thus elicit allostery by causing some change in this 444–449 loop. However, mutational probing of the 444, 446, and 449 positions did not yield clues for which side-chain might play that role in allostery (Figure 3). We then questioned if multiple interactions with the 444–449 loop (or alternatively Arg501/Trp494) are redundant in their role to elicit allostery; i.e. either of two interactions might be sufficient to cause a conformational shift in the 444–449 loop. However, combining mutation of residues in either of the phosphate binding sites did not provide new insights into a side-chain residue that might be required to elicit allostery (Supplemental Table S4).
527–533 loop shortening deletions
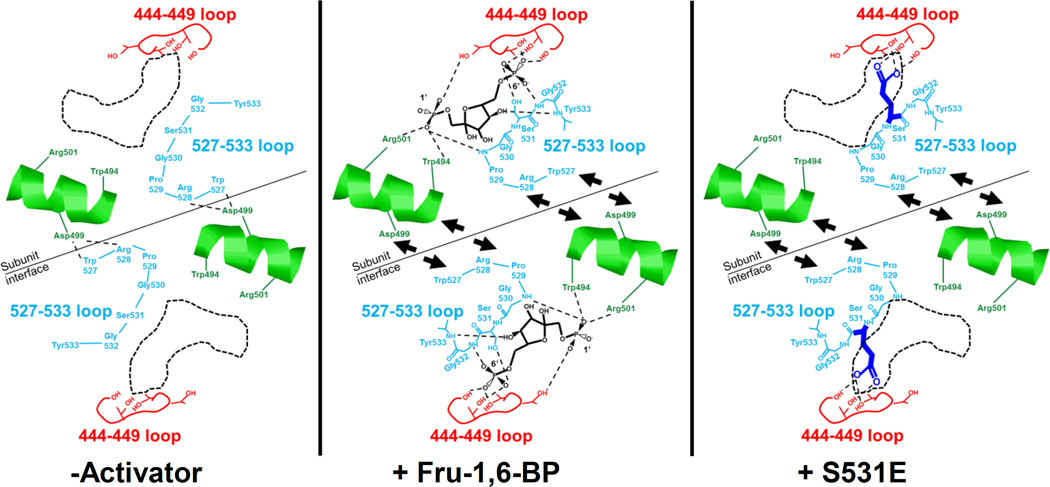
In contrast to the S531E–introduced negative charge causing changes in the 444–449 loop, we next considered whether S531E mimics activation by its influence on the 527–533 loop. The binding of Fru-1,6-BP with the formation of interactions with the 527–533 backbone atoms could cause the 527–533 loop to shift towards the Fru-1,6-BP binding site (Figure 5). Similarly, the introduced negative charge of S531E could interact with the 444–449 loop to cause the 527–533 loop to shift towards the Fru-1,6-BP binding site. In both cases this shift in the 527–533 loop would be in comparison to the location of the same loop in the wild type protein in the absence of activator.
Figure 5.
A schematic of proposed allosterically relevant structural changes that are based on available PYK structures and that could be consistent with data in this study. In the absence of activator, Arg528 and Trp527 from the 527–533 loop of one subunit make contacts with Asp499 from the neighboring subunit. Upon Fru-1,6-BP binding or in the presence of the S531E mutation, the 527–533 loop is pulled away from the subunit interface. Interruption of the Trp527/Arg528/Asp499 interactions appears to be the primary requirement for allosteric activation. Movement of the helix containing Trp494, Asp499, and Arg501 may further relay the presence/absence of the Trp527/Arg528/Asp499 interactions to modify PEP affinity.
The suggested conformational shifting of the 527–533 loop as just described was also previously proposed based on structural comparisons of M1-PYK with M2-PYK (12, 23). In fact the earlier of the two previous structural studies labeled the 527–533 loop as the “FBP activating loop.” No testing of this structural comparison-based proposed role for the 527–533 loop has been reported. Therefore, although the 527–533 loop shift has previously been proposed, more extensive probing of this potential shift as a contributor to the allosteric mechanism is needed.
In our attempt to dissect how each effector/protein interaction contributes to allostery, we can first revisit the influence of various side-chain substitutions introduced at Ser531 (Figure 3). Mutations of Ser531 (except those that introduced negative charges) did not influence allosteric function. Therefore, the interaction of Fru-1,6-BP with the native Ser at position 531 does not appear to be required for allostery. Since the interactions between Fru-1,6-BP and the backbone atoms of the 527–533 loop are the interactions that were not probed in the random mutagenesis study, those backbone interactions are then implicated for primary importance for a role in allostery.
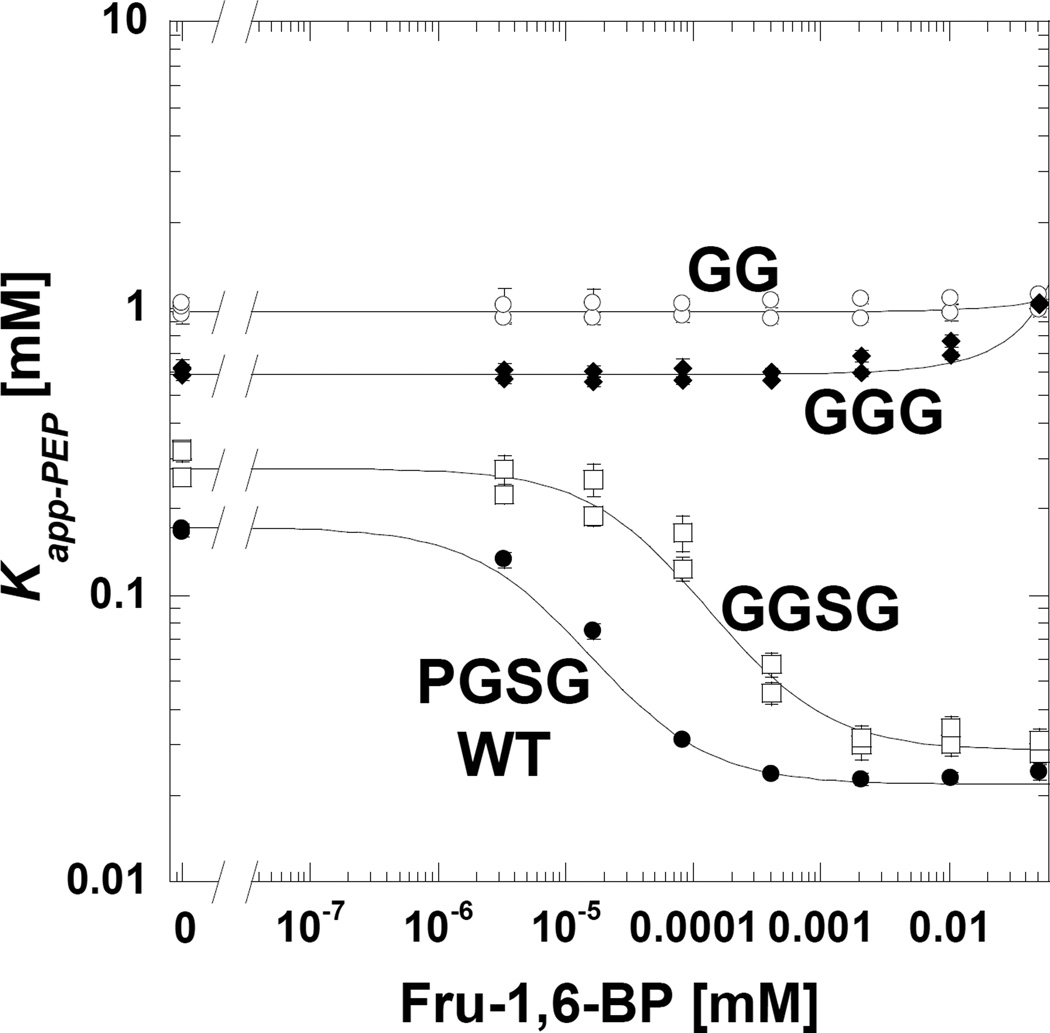
Mutational probing of backbone interactions is obviously challenging. We chose to shorten the 527–533 loop by deleting residues. We predicted that the shortened loop would not make interactions with bound Fru-1,6-BP. Therefore even when Fru-1,6-BP is present, the shortened 527–533 loop is predicted to remain in the same location that it resides in when Fru-1,6-BP is absent. Initially the proline in the 527-WRPGSGY-533 loop was replaced with glycine (with minimal effect on Qax; Figure 6) and then the loop was sequentially shortened to WRGGGY and then WRGGY. Consistent with predictions based on the proposed 527–533 loop-shift mechanism, shortening the 527–533 loop removed an allosteric response to Fru-1,6-BP (Figure 6).
Figure 6.
Allosteric activation of wild type protein and proteins with deletions that shorten the 527–533 loop. In this data presentation, Qax is the distance between the upper plateau (in the absence of Fru-1,6-BP) and the lower plateau (at high Fru-1,6-BP). The sequence of the 527–533 loop is 527-WRPGSGY-533. The 529-PGSG-532 portion of this loop is modified, first to remove substitute the proline with glycine, and then shortened sequentially to GGG and GG. The shortened loop prevents a response to Fru-1,6-BP, as predicted. When error bars are not apparent, they are smaller than data point symbols.
Mutating Trp527/Arg528/Asp499 interactions across the subunit interface
A structural change in the 527–533 loop was initially described by the Mesecar laboratory (12). However, a most recent structural comparison of the M1-PYK and M2-PYK isozymes emphasized that in the absence of activator, residues equivalent to Trp527 and Arg528 in hL-PYK form an interaction across the subunit interface with Asp499 of the neighboring subunit (23). A Fru-1,6-BP-dependent shift in the 527–533 loop would interrupt interactions among Trp527, Arg528, and Asp499 across the subunit interface. If the Fru-1,6-BP binding interrupts the Trp527/Arg528/Asp499 interactions to elicit allostery, then we predicted that mutations at 527, 528, and 499 would also likely mimic allosteric activation (i.e. increase PEP affinity).
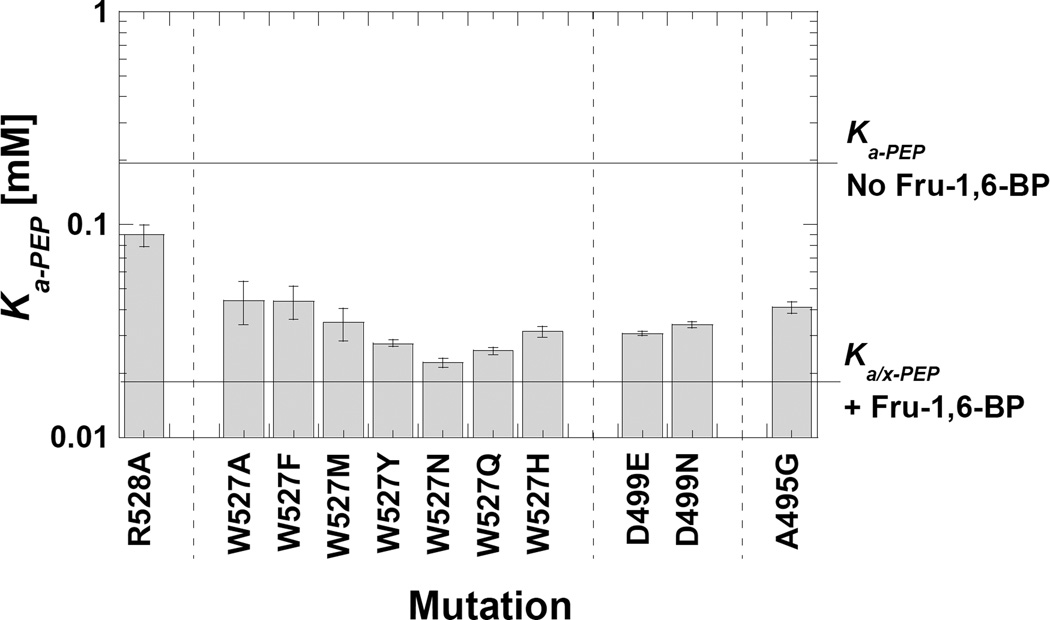
Removal of the Arg528 (R528A) side-chain slightly increased PEP affinity, but overall the effects of this mutation were minor (Figure 7). In contrast, modifying Trp527 resulted in the expected increased PEP affinity associated with Fru-1,6-BP activation (i.e. Ka-PEP below 0.05 mM). This is true for mutations that conserve hydrophobic properties (i.e. W527F, W527Y and W527M) and mutations that conserve hydrogen bonding properties (i.e. W527N, W527Q, W527Y, and W527H). Therefore, Trp at the 527 position appears to serve a very specialized role in allosteric activation. Similarly, modifying Asp499 results in Ka-PEP values close to that of the Fru-1,6-BP activated wild type enzyme. These observations are consistent with the idea that any modification that disrupts or perturbs the Trp527-Asp499 interaction results increased binding of PEP.
Figure 7.
Ka-app values for point mutations introduced with the intent of interrupting the Trp527/Arg528/Asp499 subunit interface interaction. The Ka-app values for the wild type proteins, both in the presence and absence of Fru-1,6-BP are indicated by horizontal reference lines. V498G and D499T mutant proteins were not activity and are not represented in the graph. Modification of Trp527 and Asp499 both caused increased affinity for PEP, similar to the allosteric activation by Fru-1,6-BP.
The mechanism by which Trp494 mutations modify PEP affinity
At the onset of this study, we focused primarily on interpreting Qax as the fit parameter associated with allosteric coupling between PEP and Fru-1,6-BP binding. However, mutations that mimic activation can directly modify PEP affinity (Ka-PEP) in the absence of activator. This direct effect on Ka-PEP was exemplified by S531E and mutations at Trp527 and Asp499. We noted earlier that several substitutions at the Trp494 position modified PEP affinity. The mechanism included in Figure 5 offers a highly plausible explanation for this observation. Removing the large aromatic side-chain from the 494 position likely perturbs the helix that also contains Asp499. Therefore, this helix may be energetically coupled to PEP binding. To test that possibility, we replaced alanine residues in the relevant helix with glycine residues. Consistent with potential energetic coupling with PEP binding, interrupting the helix containing Trp494, Asp499, and Arg501 resulted in a Ka-PEP value that is shifted toward the Ka/x-PEP value observed for the wild type protein when Fru-1,6-BP is bound (see A495G in Figure 7). Therefore, our working hypothesis is that the influence of mutations at the Trp494 position on Ka-PEP are due to an influence on the properties of this helix. However since mutations at the subunit interface (Trp527/Arg528/Asp499) are sufficient to mimic an allosteric effect, we do not anticipate that the interaction of Trp494 with the 1’-phosphate of Fru-1,6-BP contributes to the allosteric mechanism in the wild type protein.
Discussion
The goal of this study was to determine which interactions between Fru-1,6-BP and its binding site on hL-PYK are responsible for eliciting the allosteric increase in substrate PEP affinity. Using a series of Fru-1,6-BP analogues, we concluded the following: 1) Both phosphates of Fru-1,6-BP contribute substantially to activator binding. 2) The hydroxyl moiety at the anomeric carbon contributes only moderately to binding affinity and, therefore, there is only a moderate selection for one of two similar binding orientations of the nearly symmetrical Fru-1,6-BP molecule (either 1’-phosphate or 6’-phosphate interacting with the 444–449 loop). 3) Only one phosphate is necessary (but not sufficient) for activation. 4) At least a small portion of the sugar is required for binding/activation, since glyceraldehyde-3-P activates, but phosphate alone does not. Importantly, our data are consistent with the previous analogue studies of L-PYK (21, 24–26).
From mutational probes, we concluded several additional key points: 1) The 444–449 loop interaction with the 6’-phosphate contributes to effector binding, but not eliciting the allosteric response. 2) The Trp494/Arg501 interaction with the 1’-phosphate of the activator contribute to effector binding. Trp494 may also contribute to allostery. However, we find it more likely that mutations at this location influence the helix that contains Trp494, Arg501, and Asp499 and this mutation-dependent helix perturbation influences PEP affinity. 3) The acidic side-chain introduced by S531E likely interacts with the 444–449 loop. We can speculate that this interaction can pull the 527–533 loop towards the Fru-1,6-BP binding site and disrupting the Trp527/Arg528/Asp499 interactions. 4) Formation of interactions between Fru-1,6-BP and backbone atoms from the 527–533 loop is also speculated to interrupt the Trp527/Arg528/Asp499 interactions. 5) Activation seems to result from any modification that disrupts the intersubunit interaction between Asp499 and Trp494. Importantly, a large number of positions were mutated with no influence on allostery and these serve as an important internal negative control, indicating that not all positions in the protein are allosterically sensitive to modification.
In summary, the speculative picture emerges that for activation of hL-PYK by Fru-1,6-BP 1) the Thr444/Thr446/Ser449 interaction with the 6’-phosphate of the activator and the Arg501/1’-phosphate interaction are the major determinants of activator binding, 2) the backbone interactions between the 527–533 loop with one of two phosphates and a single hydroxyl of Fru-1,6-BP contribute to disrupting the intersubunit Trp527-Asp499 interaction, and 3) any disruption of the intersubunit Trp527-Asp499 interaction results in increased PEP affinity. We can also speculate that the helix containing Trp494, Asp499 and Arg501 plays an important role allosterically coupling the PEP and Fru-1,6-BP binding events.
Supplementary Material
Acknowledgments
We thank Cassandra Field for her aid in initiating the studies with Fru-1,6-BP analogues. This work was supported by NIH grant DK78076.
Footnotes
Supporting Information
Supplemental material include: 1) a table of the structures of activators used in this study; 2) a comparison of data collected with purified wild type protein and that collected with partially purified wild type protein; 3) an evaluation of the effect of very high activator concentration as a means of determining concentrations that result in non-specific effect; these were used to determine the upper concentration limit used in throughout this work; 4) a comparison of the effects of including various ions in the assay; 5) the response of activity to PO4, and 4) the allosteric activation of S531D and S531E by Fru-1,6-BP, 4) tables of values represented in figures. Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Williams R, Holyoak T, McDonald G, Gui C, Fenton AW. Differentiating a Ligand’s Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase. Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 2.Alontaga AY, Fenton AW. Effector analogues detect varied allosteric roles for conserved protein-effector interactions in pyruvate kinase isozymes. Biochemistry. 2011;50:1934–1939. doi: 10.1021/bi200052e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frieden C. Glutamic dehydrogenase. II. The effect of various nucleotides on the association-dissociation and kinetic properties. J Biol Chem. 1959;234:815–820. [PubMed] [Google Scholar]
- 4.Cheng A, Fitzgerald TJ, Bhatnagar D, Roskoski R, Jr, Carlson GM. Allosteric nucleotide specificity of phosphorylase kinase: correlation of binding, conformational transitions, and activation. Utilization of lin-benzo-ADP to measure the binding of other nucleoside diphosphates, including the phosphorothioates of ADP. J Biol Chem. 1988;263:5534–5542. [PubMed] [Google Scholar]
- 5.Brown PH, Beckett D. Use of binding enthalpy to drive an allosteric transition. Biochemistry. 2005;44:3112–3121. doi: 10.1021/bi047792k. [DOI] [PubMed] [Google Scholar]
- 6.Abraham DJ, Kister J, Joshi GS, Marden MC, Poyart C. Intrinsic activity at the molecular level: E. J. Ariens’ concept visualized. J Mol Biol. 1995;248:845–855. doi: 10.1006/jmbi.1995.0265. [DOI] [PubMed] [Google Scholar]
- 7.Abraham DJ, Safo MK, Boyiri T, Danso-Danquah RE, Kister J, Poyart C. How allosteric effectors can bind to the same protein residue and produce opposite shifts in the allosteric equilibrium. Biochemistry. 1995;34:15006–15020. doi: 10.1021/bi00046a007. [DOI] [PubMed] [Google Scholar]
- 8.Khasawneh FT, Huang JS, Turek JW, Le Breton GC. Differential mapping of the amino acids mediating agonist and antagonist coordination with the human thromboxane A2 receptor protein. J Biol Chem. 2006;281:26951–26965. doi: 10.1074/jbc.M507469200. [DOI] [PubMed] [Google Scholar]
- 9.Su KL, Chang KY, Hung HC. Effects of structural analogues of the substrate and allosteric regulator of the human mitochondrial NAD(P)+-dependent malic enzyme. Bioorg Med Chem. 2009;17:5414–5419. doi: 10.1016/j.bmc.2009.06.052. [DOI] [PubMed] [Google Scholar]
- 10.Garuti L, Roberti M, Bottegoni G. Non-ATP competitive protein kinase inhibitors. Current medicinal chemistry. 2010;17:2804–2821. doi: 10.2174/092986710791859333. [DOI] [PubMed] [Google Scholar]
- 11.Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 12.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 13.Valentini G, Chiarelli LR, Fortin R, Dolzan M, Galizzi A, Abraham DJ, Wang C, Bianchi P, Zanella A, Mattevi A. Structure and function of human erythrocyte pyruvate kinase. Molecular basis of nonspherocytic hemolytic anemia. J Biol Chem. 2002;277:23807–23814. doi: 10.1074/jbc.M202107200. [DOI] [PubMed] [Google Scholar]
- 14.Holyoak T, Zhang B, Deng J, Tang Q, Prasannan CB, Fenton AW. Energetic coupling between an oxidizable cysteine and the phosphorylatable N-terminus of human liver pyruvate kinase. Biochemistry. 2013 doi: 10.1021/bi301341r. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenton AW, Hutchinson M. The pH dependence of the allosteric response of human liver pyruvate kinase to fructose-1,6-bisphosphate, ATP, and alanine. Arch Biochem Biophys. 2009;484:16–23. doi: 10.1016/j.abb.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenton AW, Tang Q. An activating interaction between the unphosphorylated n-terminus of human liver pyruvate kinase and the main body of the protein is interrupted by phosphorylation. Biochemistry. 2009;48:3816–3818. doi: 10.1021/bi900421f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenton AW, Alontaga AY. The impact of ions on allosteric functions in human liver pyruvate kinase. Methods Enzymol. 2009;466:83–107. doi: 10.1016/S0076-6879(09)66005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 19.Wurster B, Hess B. Tautomeric and anomeric specificity of allosteric activation of yeast pyruvate kinase by D-fructose 1, 6-bisphosphate and its relevance in D-glucose catabolism. FEBS Lett. 1976;63:17–21. doi: 10.1016/0014-5793(76)80185-8. [DOI] [PubMed] [Google Scholar]
- 20.Fishbein R, Benkovic PA, Benkovic SJ. The Anomeric Specificity of Yeast Pyruvate Kinase toward Activation by D-Fructose 1,6-Bisphosphate. Biochemistry. 1975;14:4060–4063. [Google Scholar]
- 21.Koster JF, Hulsmann WC. The influence of inorganic phosphate and phosphorylated hexoses on the activity of pyruvate kinase. Arch Biochem Biophys. 1970;141:98–101. doi: 10.1016/0003-9861(70)90111-6. [DOI] [PubMed] [Google Scholar]
- 22.Fenton AW. Allostery: an illustrated definition for the ‘second secret of life’. Trends Biochem Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan HP, O’Reilly FJ, Wear MA, O’Neill JR, Fothergill-Gilmore LA, Hupp T, Walkinshaw MD. M2 pyruvate kinase provides a mechanism for nutrient sensing and regulation of cell proliferation. Proc Natl Acad Sci U S A. 2013;110:5881–5886. doi: 10.1073/pnas.1217157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggleston LV, Woods HF. Activation of liver pyruvate kinase by fructose-1-phosphate. FEBS Lett. 1970;6:43–45. doi: 10.1016/0014-5793(70)80038-2. [DOI] [PubMed] [Google Scholar]
- 25.Balinsky D, Cayanis E, Bersohn I. The effects of various modulators on the activities of human pyruvate kinases isolated from normal adult and foetal liver and hepatoma tissue. International Journal of Biochemistry. 1973;4:489–501. doi: 10.1021/bi00729a013. [DOI] [PubMed] [Google Scholar]
- 26.Koster JF, Slee RG, Staal GE, van Berkel TJ. The influence of glucose I,6-diphosphate on the enzymatic activity of pyruvate kinase. Biochim Biophys Acta. 1972;258:763–768. doi: 10.1016/0005-2744(72)90177-5. [DOI] [PubMed] [Google Scholar]
- 27.Holyoak T, Zhang B, Deng J, Tang Q, Prasannan CB, Fenton AW. Energetic coupling between an oxidizable cysteine and the phosphorylatable N-terminus of human liver pyruvate kinase. Biochemistry. 2013;52:466–476. doi: 10.1021/bi301341r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.