ABSTRACT
Recently, linear ubiquitin assembly complex (LUBAC)-mediated linear ubiquitination has come into focus due to its emerging role in activation of NF-κB in different biological contexts. However, the role of LUBAC in LMP1 signaling leading to NF-κB and interferon regulatory factor 7 (IRF7) activation has not been investigated. We show here that RNF31, the key component of LUBAC, interacts with LMP1 and IRF7 in Epstein-Barr virus (EBV)-transformed cells and that LUBAC stimulates linear ubiquitination of NEMO and IRF7. Consequently, LUBAC is required for LMP1 signaling to full activation of NF-κB but inhibits LMP1-stimulated IRF7 transcriptional activity. The protein levels of RNF31 and LMP1 are correlated in EBV-transformed cells. Knockdown of RNF31 in EBV-transformed IB4 cells by RNA interference negatively regulates the expression of the genes downstream of LMP1 signaling and results in a decrease of cell proliferation. These lines of evidence indicate that LUBAC-mediated linear ubiquitination plays crucial roles in regulating LMP1 signaling and functions.
IMPORTANCE We show here that LUBAC-mediated linear ubiquitination is required for LMP1 activation of NF-κB but inhibits LMP1-mediated IRF7 activation. Our findings provide novel mechanisms underlying EBV-mediated oncogenesis and may have a broad impact on IRF7-mediated immune responses.
KEYWORDS: LUBAC, LMP1, IRF7, ubiquitination
INTRODUCTION
Epstein-Barr virus (EBV) is clearly an oncogenic virus in humans and is quite frequently involved in AIDS-related malignancies. EBV-encoded latent membrane protein 1 (LMP1), a member of the tumor necrosis factor receptor (TNFR) superfamily, is essential for altered cell growth, survival, adhesion, and invasive potential. LMP1 shares many features with CD40 and Toll-like receptors (TLRs) in signaling transduction, including the activation of NF-κB, mitogen-activated protein kinases (MAPKs), and phosphatidylinositol 3-kinase (PI3K)/Akt (1–5).
Interferon regulatory factor 7 (IRF7) is the “master” regulator of type I interferon (IFN) production in antiviral innate immunity. IRF7 also plays a role in EBV latency maintenance and oncogenesis, among other potential functions (6). IRF7 is overexpressed in EBV-positive AIDS-related central nervous system lymphoma and causes anchorage-independent growth of NIH 3T3 cells that induce tumor formation in nude mice (7). In addition, IRF7 targets the BamHI-A rightward transcript (BART) P1 promoter (8), and BART-derived microRNAs (miRNAs) play a role in EBV oncogenesis (9, 10). Furthermore, we and others have presented vast evidence that IRF7 is induced, as well as activated, by LMP1 (11–15). Thus, tight regulation of IRF7 activity is of great importance in EBV latent infection, not only for shaping EBV latency by refining type I IFN signaling, but also for EBV oncogenesis.
Ubiquitination is a pervasive mechanism important to the activation of NF-κB and other transcription factors, including IRFs, and is crucial to coordinating complex aspects of many fundamental cellular activities. DNA tumor viruses, especially EBV, are well known to manipulate the host ubiquitin (Ub) machinery to facilitate their latent persistence and oncogenesis (16–20). We have provided solid evidence showing that IRF7 is activated by LMP1 through a TRAF6/RIP-dependent, K63-linked ubiquitination pathway (11, 12). We have further shown that TNFAIP3 regulates this process as a deubiquitinating enzyme (DUB) (21).
More recently, LUBAC (linear ubiquitin assembly complex)-mediated linear ubiquitination has been shown to be crucial for canonical NF-κB activation in diverse biological contexts (22–27), including apoptotic and immune stimuli, such as tumor necrosis factor alpha (TNF-α) (28, 29), interleukin 1β (IL-1β) (30), genotoxic stress (31), CD40 (32), TLRs (33), NOD2 (34), and NLRP3 (35). However, whether LUBAC is required for NF-κB activation in the setting of EBV infection has not been examined. LUBAC is a ternary ubiquitin ligase complex composed of HOIP (RNF31), HOIL1L (RNF54), and Sharpin and is constitutively formed under normal physiological conditions (28, 29, 36). RNF31 is likely the central component of the complex (37). LUBAC is abundantly expressed in thymus and spleen, implying its potential role in lymphocytes (25). Originally, LUBAC was shown to target TAB2/3 for degradation and therefore negatively regulates TNF-α- or IL-1β-stimulated NF-κB activation (30, 36). Recent reports clearly show that LUBAC specifically activates the canonical NF-κB pathway, but not the JNK pathway, by conjugating linear polyubiquitin chains onto NEMO and RIP1 (25, 38). More recently, the deubiquitinase Gumby (OTULIN), CYLD, and TNFAIP3 were shown to negatively regulate LUBAC-mediated NF-κB activation through different mechanisms in different biological contexts (34, 39–42). These intriguing findings have advanced our current understanding of the novel functions of ubiquitination in signal transduction pathways and may provide novel paradigms for the treatment of human cancers and immune diseases (43).
In this study, we provide evidence that LUBAC is involved in LMP1 activation of NF-κB but negatively regulates LMP1-promoted IRF7 transcriptional activity. Since LUBAC activation of NF-κB has been shown in other contexts, we focus on LUBAC regulation of IRF7, which has never been investigated in any context.
RESULTS
RNF31 physically interacts with LMP1 and IRF7.
LUBAC is crucial for canonical NF-κB activation in diverse biological contexts (22–27). However, whether LUBAC is required for NF-κB activation by LMP1 has never been reported. Furthermore, the relationship between LUBAC and IRF7 has never been investigated. To assess if LUBAC participates in LMP1 signaling transduction to NF-κB and IRF7 activation, we first determined if LMP1 interacts with RNF31. To this end, hemagglutinin (HA)-LMP1 and Flag-RNF31 expression plasmids were transfected into 293T cells, and the cell lysates were subjected to immunoprecipitation (IP). The results showed that LMP1 and RNF31 clearly interact (Fig. 1A).
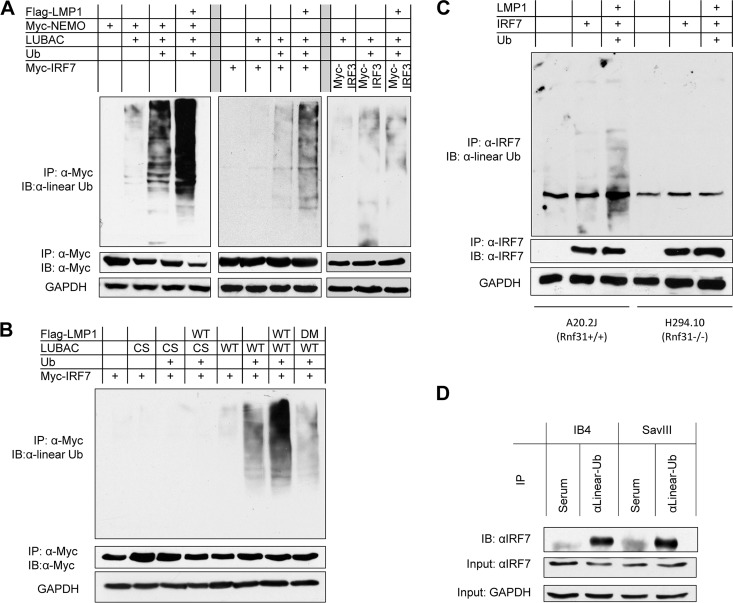
FIG 1.
RNF31 interacts with LMP1 and IRF7. (A to C) 293T cells in 60-mm dishes were transfected with 1 μg Flag-RNF31 or Myc-RNA31; 0.5 μg HA-LMP1, Myc-LMP1, or Flag-LMP1 and its mutants; and 0.5 μg HA-IRF7 using Effectene transfection reagent. Cells were collected 48 h after transfection, and cell lysates in NP-40 lysis buffer were subjected to immunoprecipitation with anti-Flag M2 (Sigma) and protein A/G beads (Santa Cruz). After extensive washes, the beads were subjected to immunoblotting (IB) with anti-HA HA7 (Roche). (D and E) Cell lysates were prepared with NP-40 lysis buffer from IB4, JiJoye, and SavIII cells and were then subjected to preclearing with rabbit (D) or mouse (E) serum before immunoprecipitation with rabbit anti-RNF31 (Abcam) (D) or mouse anti-LMP1 (Dako) (E) or corresponding serum as a control. After extensive washes, the beads were subjected to immunoblotting with the indicated antibodies. Inputs were 5% of total cell lysates.
To check if the interaction between RNF31 and LMP1 is specific, we used a panel of LMP1 deletion mutants for IP assays (Fig. 1B). Our results indicated that LMP1 CTAR2 specifically interacts with RNF31.
Of special note and novelty, we further show that RNF31 and IRF7 interact when both are transiently expressed in 293T cells and that their interaction is much stronger in the presence of LMP1, indicating that LMP1 promotes their interaction (Fig. 1C).
We further checked their endogenous interaction in the context of EBV latent infection. A panel of EBV-transformed cells were subjected to immunoprecipitation with anit-RNF31 and then to immunoblotting with anti-LMP1 and anti-IRF7. The results showed that endogenous RNF31 interacts with both LMP1 and IRF7 (Fig. 1D). Furthermore, we showed that LMP1 interacts with the other two LUBAC components, Sharpin and HOIL1L, in EBV-transformed cells (Fig. 1E).
Taken together, these results demonstrate that RNF31, as a component of LUBAC (37), physically interacts with IRF7 and LMP1 in latently EBV-infected cells.
LUBAC stimulates linear ubiquitination of IRF7 and NEMO downstream of LMP1 signaling.
LUBAC promotes NF-κB activation mainly by conjugating linear ubiquitin chains to NEMO (25, 38). Thus, we sought to evaluate if LUBAC can promote NEMO linear ubiquitination downstream of LMP1 signaling. On the other hand, we have shown that LMP1 stimulates K63-linked polyubiquitination of IRF7 (11, 12). Here, we asked if LMP1 also stimulates linear ubiquitination of IRF7, as we have already shown that IRF7 and RNF31 interact (Fig. 1). To this end, 293T cells were transfected with expression plasmids for Flag-LMP1, Myc-NEMO (or Myc-IRF7), Ub, and LUBAC, which includes Flag-RNF31, HA-HOIL1L, and Flag-Sharpin. Cell lysates were collected 2 days after transfection and denatured before immunoprecipitation. Our results indicated that LUBAC promotes linear ubiquitination of both NEMO and IRF7 and that LMP1 potentiates their ubiquitination (Fig. 2A). LMP1 specifically promotes IRF7 linear ubiquitination, as LMP1 did not enhance IRF3 ubiquitination (Fig. 2A). However, LUBACcs, which consists of the Flag-RNF31 E3-ligase-dead mutant Flag-RNF31 (C699/702S C871/874S) (36), the HA-HOIL1L E3-ligase-dead mutant HA-HOIL1 (C282/285S C447/450S) (36), and the Flag-Sharpin Ub-binding-dead mutant Flag-Sharpin (T358L F359V) (44), did not stimulate ubiquitination of IRF7 (Fig. 2B) or of NEMO (not shown) under all conditions.
FIG 2.
LUBAC promotes linear ubiquitination of NEMO and IRF7. (A and B) 293T cells in 60-mm dishes were transfected with 0.5 μg Flag-LMP1, 0.5 μg Myc-NEMO or Myc-IRF7, 0.5 μg Ub, and 0.5 μg LUBAC (mixed with its three components). Cell lysates were prepared after 48 h with NP-40 lysis buffer. IP was performed with 1.5 μg anti-Myc 9E10 (Roche) and protein A/G-Sepharose beads (Santa Cruz) for 4 h. The beads were washed four times in NP-40 lysis buffer and two times with low-salt buffer (20 mM Tris, pH 7.4, 25 mM NaCl, 1 mM dithiothreitol [DTT]) and then denatured in 50 μl of 1% SDS. After dilution with buffer A (20 mM Tris, pH 7.4, 250 mM NaCl, 1 mM DTT, 1 mM sodium orthovanadate, 2 mM EDTA, 1% Triton X-100), the proteins were subjected to a second IP with 1 μg anti-Myc and protein A/G beads overnight. The beads were washed four times with buffer A and two times with low-salt buffer before immunoblotting with the linear ubiquitin antibody 1E3 (Millipore). (C) A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−) cells were transfected with LMP1, IRF7, and Ub expression plasmids using a Nucleofector kit, following the manufacturers' instructions. Denatured immunoprecipitation was performed with anti-IRF7 G-8 (Santa Cruz), and then the immunoprecipitate was subjected to immunoblotting with the linear Ub antibody. (D) Cell lysates from IB4 and SavIII cells were denatured, precleared with rabbit serum, and then subjected to IP with rabbit anti-linear Ub or rabbit serum (control). After extensive washes, the beads were probed with rabbit anti-IRF7 (H-246). WT, wild type.
To ensure that LMP1 does stimulate IRF7 linear ubiquitination, we used a pair of mouse B cell lines, A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−). We transfected the cells with LMP1, IRF7, and Ub expression plasmids, and cell lysates were collected after 48 h and subjected to denatured immunoprecipitation with the IRF7 antibody clone G-8 (Santa Cruz) and then to immunoblotting with the linear Ub antibody. As expected, LMP1 promoted IRF7 linear ubiquitination in A20.2J (Rnf31+/+) but not in H294.10 (Rnf31−/−) cells (Fig. 2C).
To further verify LMP1-stimulated IRF7 linear ubiquitination, we performed IP with a linear Ub antibody and then probed with an IRF7 antibody. We clearly detected IRF7 that was pulled down by the linear Ub antibody (Fig. 2D) but did not detect IRF3 under our experimental conditions (data not shown).
Taken together, these results demonstrate that LMP1 promotes linear ubiquitination of NEMO and IRF7 via LUBAC, in which RNF31 is the central component (37).
LUBAC is required for full activation of NF-κB by LMP1.
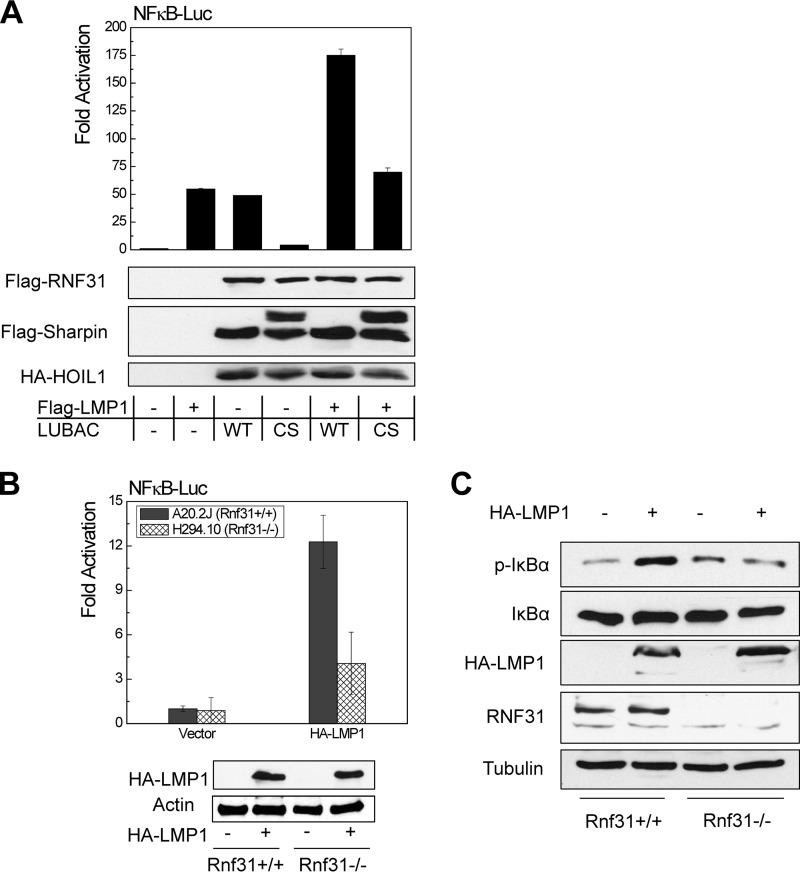
To check if LUBAC plays a potential role in LMP1 activation of NF-κB, we performed promoter-reporter assays by transfecting 293 cells with combinations of Flag-LMP1 and LUBAC (or LUBACcs) plus NF-κB promoter (NF-κBp)–Luc and Renilla luciferase. The results show that LUBAC, but not LUBACcs, significantly increased LMP1 activation of the NF-κB promoter construct (Fig. 3A).
FIG 3.
LUBAC is required for LMP1 full activation of NF-κB. (A) 293 cells in 24-well plates were transfected with 150 ng LUBAC (equal amounts of each component), 10 ng Flag-LMP1, 40 ng pGL3/NF-κB–Luc, and 10 ng Renilla luciferase. A dual-luciferase assay was performed 24 h after transfection, with a Dual Luciferase kit (Promega). The results are the averages and SE of duplicates. Representative results from at least three independent experiments are shown. The ability of the vector control to activate the promoter construct was set to 1. (B and C) A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−) cells were transfected with 3 μg HA-LMP1, 1 μg pGL3/NF-κB–Luc, and 0.5 μg Renilla luciferase using a Nucleofector kit. Dual-luciferase assays and immunoblotting were performed after 24 h. Experiments were repeated at least three times, and representative results are shown.
This finding was further confirmed with the mouse B cell lines A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−). In H294.10 (Rnf31−/−) cells, LMP1-stimulated NF-κB promoter activity was significantly lower than that in the parental A20.2J (Rnf31+/+) cells (Fig. 3B). Further, NF-κB activation was evaluated by IκBα phosphorylation at S32/36. The results showed that a significant increase of IκBα S32/36 phosphorylation was detected in A20.2J (Rnf31+/+) cells, but not in H294.10 (Rnf31−/−) cells, in the presence of LMP1 (Fig. 3C). These results indicate that LUBAC is required for full activation of NF-κB by LMP1.
LUBAC inhibits LMP1 activation of IRF7.
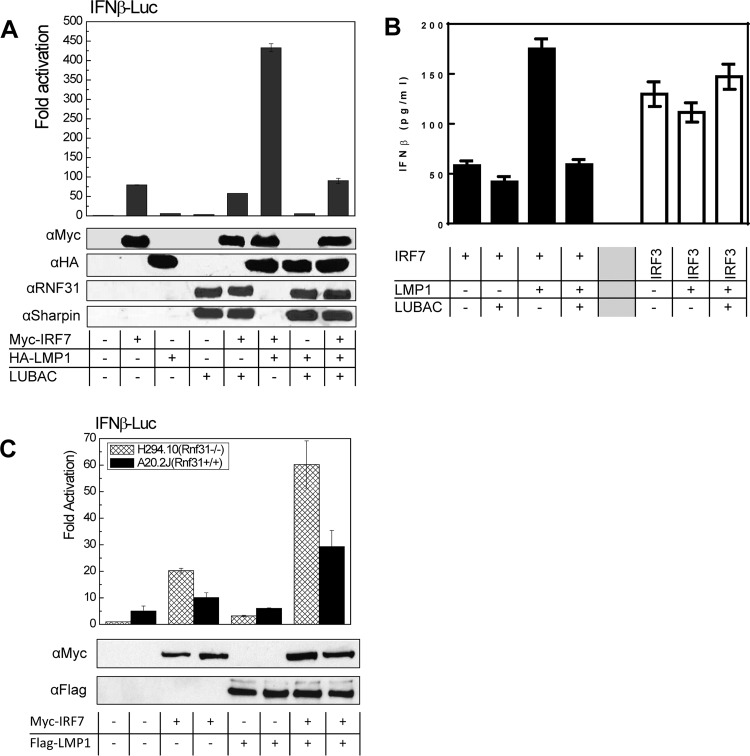
We then performed a promoter-reporter assay to check the effect of LUBAC on LMP1-stimulated IRF7 transcriptional activity. Surprisingly, our results showed that LUBAC dramatically inhibits LMP1-stimulated IRF7 activity (Fig. 4A) and consequently inhibits IFN-β production mediated by the LMP1/IRF7 pathway. However, LMP1 did not stimulate IRF3 activity, and LUBAC had no significant effect on the basal IRF3-mediated IFN-β production (Fig. 4B). These data indicate that LUBAC-mediated linear ubiquitination specifically inhibits LMP1 activation of IRF7. We also confirmed the finding in A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−) cells, and the results showed that LMP1-promoted IRF7 activity was much higher in H294.10 (Rnf31−/−) cells (Fig. 4C).
FIG 4.
LUBAC inhibits LMP1-promoted IRF7 transcriptional activity. (A) 293 cells in 24-well plates were transfected with 150 ng LUBAC (equal amounts of each component), 10 ng Flag-LMP1, 50 ng Myc-IRF7, 40 ng pGL3/IFN-β–Luc, and 10 ng Renilla luciferase. A dual-luciferase assay was performed 24 h after transfection. (B) 293 cells in 24-well plates were transfected with 150 ng LUBAC (equal amounts of each component), 10 ng Flag-LMP1, and 50 ng IRF7 or IRF3. IFN-β production in the medium was measured 48 h after transfection with a human IFN-β enzyme-linked immunosorbent assay (ELISA) kit following the manufacturer's instructions (PBL Assay Science). (C) A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−) cells were transfected with 1 μg HA-LMP1, 2 μg Myc-IRF7, 1 μg pGL3/IFN-β–Luc, and 0.5 μg Renilla luciferase using a Nucleofector kit. Dual-luciferase assays and immunoblotting were performed after 24 h. Experiments were repeated at least three times, and representative results are shown.
LUBAC modulates the expression of LMP1 target genes.
To assess the role of LUBAC in regulation of LMP1 target gene expression, we knocked down the endogenous RNF31 in IB4 cells by lentivirus-mediated transfection of RNF31-specific short hairpin RNAs (shRNAs). As shown in Fig. 5A, we reached high knockdown efficiencies by two selected RNF31 shRNA constructs. After selection of the cells with puromycin, we performed immunoblotting for IRF7, IRF4, A20, and SOCS1, all of which are known to be upregulated by LMP1 signaling (7, 45, 46). Our results showed that expression of these genes was significantly downregulated in RNF31 shRNA-expressing cells compared with control shRNA-expressing cells (Fig. 5A). We further confirmed the regulation of IRF4 and SOCS1 and evaluated expression of the IRF7 target genes, Tap2 and ISG56, by quantitative real-time (qRT)-PCR. Our results showed that mRNA levels of Tap2 and ISG56 were elevated in RNF31-deficient IB4 cells (Fig. 5A).
FIG 5.
LUBAC regulates LMP1 target gene expression. (A) IB4 cells were infected with lentivirus expressing GIPz/shRNF31 (or control), and stable cells were selected with 1 μg/ml puromycin. Immunoblotting was performed for cell lysates with antibodies against IRF7, IRF4, A20, SOCS1, RNF31, and GAPDH. mRNAs from the cells were also used for real-time PCR for detection of selected targets and Rnf31 with the following primers: Rnf31, F (5′-CAGATGCCTATGCGTTGTT-3′) and R (5′-TGGTATTCTGGGTCGTTCAT-3′); Irf4, F (5′-TTAATTCTCCAAGCGGATGC-3′) and R (5′-AAGGAATGAGGAAGCCGTTC-3′); Socs1, F (5′-GTGGCAGCCGACAATGCAGT-3′) and R (5′-CGAGGCCATCTTCACGCTAAGG-3′); Tap2, F (5′-ACGGCTGAGCTCGGATACCAC-3′) and R (5′-CCTCGGCCCCAAAACTGC-3′); Isg56, F (5′-TCTCAGAGGAGCCTGGCTAAG-3′) and R (5′-CCACACTGTATTTGGTGTCTAGG-3′). (B and C) A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−) cells were transfected with LMP1 or vector control (B) or LMP1 and/or IRF7 (C), using a Nucleofector kit. (B) Immunoblotting was performed after 48 h with antibodies against IRF4, IRF7, RNF31, and GAPDH. (C) Signals were quantitated with Quantity One software (Bio-Rad). Cells were also collected for real-time PCR for IFN-β mRNA expression using the following primers: F (5′-AACCTCACCTACAGGGCGGACTTCA-3′) and R (5′-TCCCACGTCAATCTTTCCTCTTGCTTT-3′). Experiments were repeated at least three times, and representative results are shown. **, P < 0.01; ***, P < 0.001.
We then evaluated the ability of LMP1 to induce its targets in the mouse B cell lines A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−). Our results showed that LMP1 can induce expression of IRF4 and IRF7 in A20.2J (Rnf31+/+) cells but not in H294.10 (Rnf31−/−) cells (Fig. 5B). However, IFN-β mRNA, which was shown to be induced by LMP1 through IRF7 upon superinfection (47), was significantly induced in H294.10 (Rnf31−/−) cells, but not in A20.2J (Rnf31+/+) cells, in the presence of LMP1 and IRF7 (Fig. 5C). These results indicated that LUBAC is required for LMP1 induction of IRF7 expression but inhibits LMP1 activation of IRF7.
RNF31 correlates with LMP1 in EBV-transformed cells, and its depletion decreases cell proliferation.
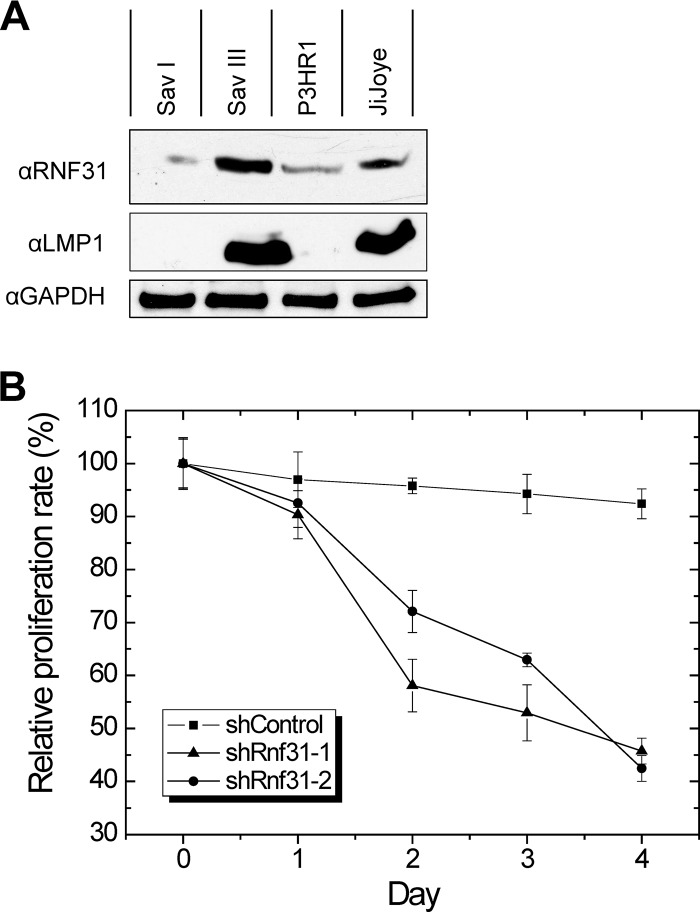
We next evaluated RNF31 expression in EBV latency by immunoblotting. Our results showed that RNF31 is correlated with LMP1 in EBV latency, that is, its expression is not detectable in SavI cells (latency 1) but is readily detectable in SavIII cells (latency 3). Both P3HR1 and JiJoye cell lines are latency 3. However, P3HR1 lacks LMP1 expression due to lack of the EBNA2 gene. Correspondingly, RNF31 is expressed at a very low level in P3HR1 cells but at a considerably higher level in JiJoye cells (Fig. 6A).
FIG 6.
RNF31 is correlated with LMP1 in EBV latency, and its depletion results in a decrease of cell proliferation. (A) Cell lysates were prepared from SavI, SavIII, P3HR1, and JiJoye cells for immunoblotting with RNF31 and LMP1 antibodies. (B) IB4 cells were infected with lentivirus expressing RNF31 shRNAs; 48 h later, the cells were subjected to MTT cell proliferation assays using a kit from R&D Systems, following the manufacturer's instructions. The absorbance at a wavelength of 570 nm was recorded, using a 96-well plate reader (Turner Biosystems). RNF31 depletion significantly decreased cell proliferation 48 h after infection (P < 0.0001 and P = 0.0058 for shRNA-1 and shRNA-2, respectively; unpaired t test). Experiments were repeated at least three times, and representative results are shown. The error bars indicate SE.
To assess the functional consequences of LUBAC regulation of LMP1 signaling, we evaluated the proliferation of IB4 cells infected with lentivirus expressing RNF31 shRNAs. The results showed that depletion of RNF31 significantly decreased cell proliferation (by 25%) after 48 h (Fig. 6B).
These results indicate that RNF31 is critical for LMP1 oncogenic functions and for maintenance of EBV latency.
DISCUSSION
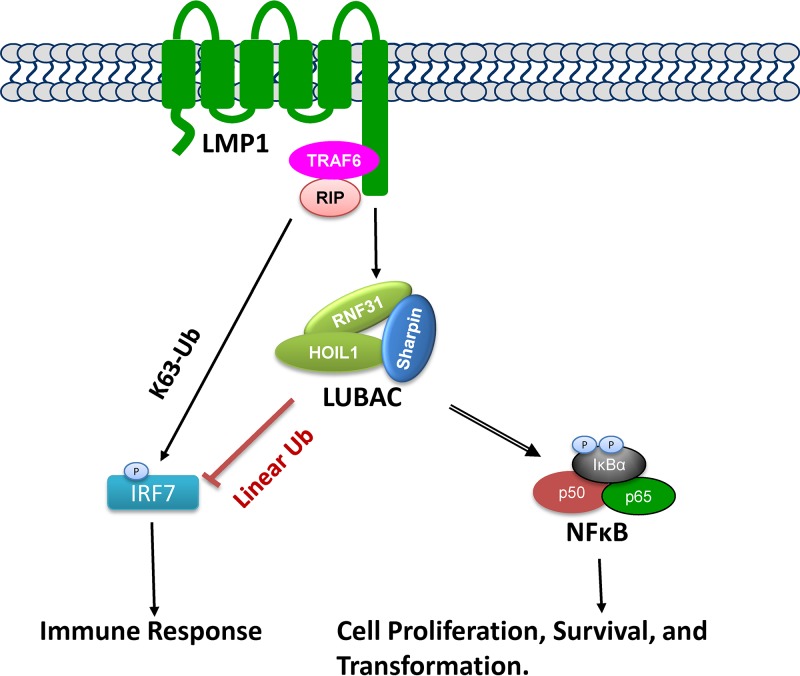
In this study, we revealed a close link between RNF31 and EBV latent infection (Fig. 7). We have shown that RNF31 interacts with LMP1 and IRF7 and stimulates linear ubiquitination of NEMO and IRF7. Consequently, LUBAC is required for LMP1 functions by regulating NF-κB and IRF7 activation.
FIG 7.
Diagram showing the involvement of LUBAC in LMP1 signaling transduction. LUBAC is required for LMP1 activation of NF-κB by promoting NEMO linear ubiquitination. LMP1 is known to activate IRF7 through TRAF6/RIP-dependent, K63-linked polyubiquitination. However, LUBAC negatively regulates LMP1 activation of IRF7 by promoting IRF7 linear ubiquitination.
Posttranslational modifications (PTMs) of proteins by ubiquitin and ubiquitin-like proteins are a pervasive theme important to the activation of NF-κB and other transcription factors in many fundamental cellular processes (48, 49). Many pathogens, such as DNA tumor viruses, encode proteins that mimic, block, or redirect the activity of the ubiquitin-proteasome machinery to modify the cellular environment and protect infected cells from host immune attack (50). EBV encodes several proteins that exploit the host ubiquitin system to regulate its latency and persistence in the host cell (20, 50–53). For example, EBNA1 resistance to ubiquitination-dependent degradation creates a perfect camouflage to prevent recognition by the host immune system (50). EBNA3C, another latent antigen, targets the tumor suppressor retinoblastoma protein (Rb) for proteasome-dependent degradation through the well-known SCFSkp2 ubiquitin ligase in different systems, including B cells (54). The EBV deubiquitinating enzyme BPLF1 targets TRAF6 and inhibits NF-κB activation during lytic replication (55).
On the other hand, EBV exploits the host ubiquitin system to benefit its latency and oncogenesis. For example, EBNA1 competes with p53 to bind to HAUSP and consequently protect the cell from apoptotic challenge by reducing p53 levels (56). LMP1 controls cellular p53 protein levels by modulating its ubiquitination (57, 58). LMP1 also induces expression of a pool of ubiquitin-related proteins, including A20, which we have shown to inhibit LMP1 activation of IRF7 by functioning as a deubiquitinase (21). In this study, we have shown LMP1 utilizes the host LUBAC-mediated linear ubiquitination to regulate NF-κB and IRF7 activation. In fact, a recent report has shown that TRAF1, which participates in LMP1 CTAR1 signaling to NF-κB activation, is a target for LUBAC and that TRAF1 linear ubiquitination contributes to LMP1-mediated cell growth and survival (59). Thus, LMP1 may employ LUBAC for its benefits via different mechanisms. Our results also show that RNF31 is associated with LMP1 in expression levels. Thus, RNF31 is likely a transcriptional target downstream of LMP1 signaling. We will investigate the mechanism underlying RNF31 expression by LMP1.
Here, we have shown novel and exciting findings that LUBAC interacts with IRF7 and inhibits IRF7 transcriptional activity, although further investigation is needed to elucidate the mechanism. IRF7 is the “master” regulator of type I IFN production in innate immune responses, required not only for early-stage IFN priming, but also for late-stage IFN amplification. Thus, our findings suggest a potential role for LUBAC in modulation of IRF7-mediated IFN response in innate immunity. In fact, in addition to NF-κB activation downstream of pathogen-associated recognition receptors, LUBAC negatively regulates RIG-I-mediated innate immune responses by targeting RIG-I, TRIM25, and IRF3 for degradation (60, 61) and by disrupting the TRAF3-MAVS complex (62). LUBAC also promotes IRF3 linear ubiquitination at two specific sites to activate the RLR-induced IRF-3-mediated pathway of apoptosis (63). Thus, LUBAC seems to be an important player in modulation of innate immune responses through different mechanisms. We will investigate the role of LUBAC in regulating IRF7-mediated IFN responses.
Our results have defined key roles for LUBAC in controlling LMP1 signaling and functions. The findings broaden our knowledge of the interaction between EBV and the host ubiquitin system in shaping its latency and oncogenesis. The role of linear chains in regulating numerous biological functions will advance our current understanding of the novel functions of ubiquitination in signal transduction and may provide novel paradigms for the treatment of human cancers.
MATERIALS AND METHODS
Constructs, antibodies, and reagents.
LUBAC constructs and mutants were gifts from Kazuhiro Iwai (28, 36). LUBAC is a complex that includes Flag-RNF31, HA-HOIL1L, and Flag-Sharpin. LUBACcs is a complex that includes the Flag-RNF31 E3-ligase-dead mutant Flag-RNF31 (C699/702S C871/874S), the HA-HOIL1L E3-ligase-dead mutant HA-HOIL1 (C282/285S C447/450S), and the Flag-Sharpin Ub-binding-dead mutant Flag-Sharpin (T358L F359V). The Flag-IRF7 expression construct and its mutants (64, 65) and pGL3/IFNA4p-Luc (66) were described previously. 3×Flag-tagged LMP1 and its mutants were described in our recent paper (67). Other constructs with different tags and deletion and point mutants were generated by subcloning or site-directed mutagenesis (Stratagene) and verified by sequencing. Rabbit linear-Ub monoclonal antibody (clone 1E3) was purchased from Millipore. Mouse anti-IRF7 (clone G-8) for immunoprecipitation, rabbit anti-IRF7 (clone H-246) for immunoblotting, goat anti-mouse IgG-horseradish peroxidase (HRP), mouse anti-rabbit IgG-HRP, and mouse anti-goat IgG-HRP were purchased from Santa Cruz. Goat and rabbit polyclonal anti-RNF31 antibodies from Abcam were used for immunoblotting and immunoprecipitation, respectively. Mouse anti-Sharpin, rabbit anti-HOIL1L, and rabbit anti-RNF31 from Sigma were used for immunoblotting. Flag (clone M2) and Myc (clone 9E10) antibodies were from Sigma and Roche, respectively.
Cell lines.
The mouse B cell lines A20.2J (Rnf31+/+) and H294.10 (Rnf31−/−) were gifts from Bruce S. Hostager and were described previously (32). 293 and 293T are human kidney epithelial cell lines. IB4, SavIII, JiJoye, and LCL00045 are human B cell lines transformed with EBV. Epithelial cells were cultured with Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS) and antibiotics, and B cells were cultured with RPMI 1640 medium plus 10% FBS and antibiotics. All cell culture supplies were purchased from Life Technologies.
Transfection.
For knockdown of RNF31 in B cell lines, a set of RNF31 shRNAs (4 in total) cloned in pGIPZ/GFP (green fluorescent protein) were purchased from Open Biosystems. We chose two of them that had the highest knockdown efficiencies for our experiments. Their insert sequences were as follows: 5′-TAAACTTGACACCACGCCA-3′ (shRnf31-1) and 5′-GACACATCACCTCCGTGCT-3′ (shRnf31-2). Lentiviral packing, preparation, infection, and selection of stable cells with puromycin were performed as detailed in our previous publication (68). For other transfections of B cells, the Nucleofector kit for human B cells (Lonza) was used. 293 and 293T cells were transfected with Effectene (Qiagen).
Promoter-reporter assays.
293 cells were transfected with expression plasmids as indicated, together with NF-κB–Luc (or IFN-β–Luc) and Renilla luciferase as an internal transfection control. Empty vector was used to equalize the total amounts of DNA in all transfections. Cells were collected 24 h after transfection. Luciferase activity was measured with equal amounts (10% of the total for each sample) of protein lysates with the use of a Dual Luciferase Assay kit (Promega) on a multimode microplate reader (Turner Biosystems). The results are shown as the mean ± standard error (SE) of duplicates for each sample. At least three consistent results were obtained from independent experiments, and representative results are shown. The ability of the empty-vector controls to activate the promoter constructs was set to 1.
Immunoprecipitation and immunoblotting.
For endogenous protein interaction, 1 × 107 cells were used for each IP. For interaction between transiently expressed proteins, 293T cells in 60-mm dishes were collected 48 h after transfection. The cells were lysed with NP-40 lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris, pH 8.0, plus protease inhibitors), and cell lysates were subjected to immunoprecipitation with 1.5 μg of the indicated antibodies overnight and then incubated with 40 μl protein A/G beads (Santa Cruz) for 1 h. For ubiquitination detection, a second immunoprecipitation was performed with anti-Myc after denaturing the immunoprecipitated proteins in 1% SDS at 95°C for 5 min. After three washes, proteins on beads were denatured before separation by SDS-PAGE. Immunoblotting was carried out with the indicated antibodies, and signals were detected with an enhanced chemiluminescence (ECL) kit following the manufacturer's protocol (Amersham Pharmacia Biotech).
Real-time quantitative PCR.
Quantitative PCR (qPCR) was performed with the use of SYBR green (Applied Biosystems) on a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Inc.). All reactions were run in duplicate. Mean cycle threshold (CT) values were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase), yielding a normalized CT (ΔCT). The ΔΔCT value was calculated by subtracting the respective control from the ΔCT, and the expression level was then calculated by 2 raised to the power of the respective −ΔΔCT value. The results are the average ± SE of duplicates for each sample.
MTT proliferation assay.
The 3-(4,5-methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) growth assay was used to measure the cell proliferation rate using a Cell Titer 96 nonradioactive cell proliferation assay kit (R&D Systems) according to the manufacturer's recommendations. Basically, 5 × 104 live cells were placed into 85 μl RPMI medium plus 10% FBS in a 96-well plate and incubated with retrovirus expressing RNF31 shRNA or an shRNA control cloned in GIPz/GFP for 0 to 4 days; 15 μl dye solution was added to each well, and the cells were further incubated at 37°C for 4 h. Solubilization solution/stop mix (100 μl) was added, and the cells were incubated for another hour. The absorbance at a wavelength of 570 nm was recorded using a 96-well plate reader (Turner Biosystems). The results are the averages ± SE of duplicates for each sample. The results obtained consistently from at least three independent experiments are shown.
Statistical analysis.
Two-tailed t tests were executed using GraphPad Prism (version 5). A P value of <0.01 was considered significant, and a P value of <0.001 was considered very significant. The data are expressed as means ± SE of duplicate samples, and representative results from at least three independent experiments are shown.
ACKNOWLEDGMENTS
This work was supported by an NIH NIDDK grant to Z.Q.Y. and J.P.M. (R01DK093526), an NIH NIAID grant to Z.Q.Y. and J.P.M. (R01AI114748), an American Society of Hematology Scholar Award to S.N., and in part by NIH grant C06RR0306551.
We thank Bruce S. Hostager for the mouse B cell lines. This publication is the result of work supported with resources of and the use of facilities at the James H. Quillen Veterans Affairs Medical Center.
The contents of this publication do not represent the views of the Department of Veterans Affairs or the United States Government.
We declare that we have no competing interests.
REFERENCES
- 1.Dawson CW, Port RJ, Young LS. 2012. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin Cancer Biol 22:144–153. doi: 10.1016/j.semcancer.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Kieser A. 2007. Signal transduction by the Epstein-Barr virus oncogene latent membrane protein 1 (LMP1). Signal Transduct 7:20–33. doi: 10.1002/sita.200600116. [DOI] [Google Scholar]
- 3.Li HP, Chang YS. 2003. Epstein-Barr virus latent membrane protein 1: structure and functions. J Biomed Sci 10:490–504. doi: 10.1007/BF02256110. [DOI] [PubMed] [Google Scholar]
- 4.Middeldorp JM, Pegtel DM. 2008. Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin Cancer Biol 18:388–396. doi: 10.1016/j.semcancer.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Soni V, Cahir-McFarland E, Kieff E. 2007. LMP1 TRAFficking activates growth and survival pathways. Adv Exp Med Biol 597:173–187. doi: 10.1007/978-0-387-70630-6_14. [DOI] [PubMed] [Google Scholar]
- 6.Ning S, Pagano J, Barber G. 2011. IRF7: activation, regulation, modification, and function. Genes Immun 12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Zhang J, Lambert Q, Der CJ, Del Valle L, Miklossy J, Khalili K, Zhou Y, Pagano JS. 2004. Interferon regulatory factor 7 is associated with Epstein-Barr virus-transformed central nervous system lymphoma and has oncogenic properties. J Virol 78:12987–12995. doi: 10.1128/JVI.78.23.12987-12995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Huang J, Wu FY, Liao G, Hutt-Fletcher L, Hayward SD. 2005. Regulation of expression of the Epstein-Barr virus BamHI-A rightward transcripts. J Virol 79:1724–1733. doi: 10.1128/JVI.79.3.1724-1733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo AKF, To KF, Lo KW, Lung RWM, Hui JWY, Liao G, Hayward SD. 2007. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A 104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquitz AR, Raab-Traub N. 2012. The role of miRNAs and EBV BARTs in NPC. Semin Cancer Biol 22:166–172. doi: 10.1016/j.semcancer.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huye LE, Ning S, Kelliher M, Pagano JS. 2007. IRF7 is activated by a viral oncoprotein through RIP-dependent ubiquitination. Mol Cell Biol 27:2910–2918. doi: 10.1128/MCB.02256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning S, Campos AD, Darnay B, Bentz G, Pagano JS. 2008. TRAF6 and the three C-terminal lysine sites on IRF7 are required for its ubiquitination-mediated activation by the tumor necrosis factor receptor family member Latent Membrane Protein 1. Mol Cell Biol 28:6536–6546. doi: 10.1128/MCB.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song YJ, Izumi KM, Shinners NP, Gewurz BE, Kieff E. 2008. IRF7 activation by Epstein-Barr virus latent membrane protein 1 requires localization at activation sites and TRAF6, but not TRAF2 or TRAF3. Proc Natl Acad Sci U S A 105:18448–18453. doi: 10.1073/pnas.0809933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Wu L, Hong K, Pagano JS. 2001. Intracellular signaling molecules activated by Epstein-Barr virus for induction of interferon regulatory factor 7. J Virol 75:12393–12401. doi: 10.1128/JVI.75.24.12393-12401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Pagano JS. 2000. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J Virol 74:1061–1068. doi: 10.1128/JVI.74.3.1061-1068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavorgna A, Harhaj E. 2014. Regulation of HTLV-1 tax stability, cellular trafficking and NF-κB activation by the ubiquitin-proteasome pathway. Viruses 6:3925. doi: 10.3390/v6103925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minor M, Slagle B. 2014. Hepatitis B virus HBx protein interactions with the ubiquitin proteasome system. Viruses 6:4683. doi: 10.3390/v6114683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wimmer P, Schreiner S. 2015. Viral mimicry to usurp ubiquitin and SUMO host pathways. Viruses 7:4854–4872. doi: 10.3390/v7092849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan J, Qiao N, Strahan R, Zhu C, Liu L, Verma SC, Wei F, Cai Q. 2016. Manipulation of ubiquitin/SUMO pathways in human herpesviruses infection. Rev Med Virol 26:435–445. doi: 10.1002/rmv.1900. [DOI] [PubMed] [Google Scholar]
- 20.Shackelford J, Pagano J. 2007. Role of the ubiquitin system and tumor viruses in AIDS-related cancer. BMC Biochem 8:S8. doi: 10.1186/1471-2091-8-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ning S, Pagano J. 2010. The A20 deubiquitinase activity negatively regulates LMP1 activation of IRF7. J Virol 84:6130–6138. doi: 10.1128/JVI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwai K, Fujita H, Sasaki Y. 2014. Linear ubiquitin chains: NF-kappaB signalling, cell death and beyond. Nat Rev Mol Cell Biol 15:503–508. doi: 10.1038/nrm3836. [DOI] [PubMed] [Google Scholar]
- 23.Rieser E, Cordier SM, Walczak H. 2013. Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem Sci 38:94–102. doi: 10.1016/j.tibs.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Tokunaga F. 2013. Linear ubiquitination-mediated NF-kappaB regulation and its related disorders. J Biochem 154:313–323. doi: 10.1093/jb/mvt079. [DOI] [PubMed] [Google Scholar]
- 25.Tokunaga F, Iwai K. 2012. Linear ubiquitination: a novel NF-kappaB regulatory mechanism for inflammatory and immune responses by the LUBAC ubiquitin ligase complex. Endocr J 59:641–652. doi: 10.1507/endocrj.EJ12-0148. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu Y, Taraborrelli L, Walczak H. 2015. Linear ubiquitination in immunity. Immunol Rev 266:190–207. doi: 10.1111/imr.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda F. 2015. Linear ubiquitination signals in adaptive immune responses. Immunol Rev 266:222–236. doi: 10.1111/imr.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJL, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I. 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata Si Tanaka K, Nakano H, Iwai K. 2011. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature 471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 30.Tian Y, Zhang Y, Zhong B, Wang YY, Diao FC, Wang RP, Zhang M, Chen DY, Zhai ZH, Shu HB. 2007. RBCK1 negatively regulates Tumor Necrosis Factor- and Interleukin-1-triggered NF-kappaB activation by targeting TAB2/3 for degradation. J Biol Chem 282:16776–16782. doi: 10.1074/jbc.M701913200. [DOI] [PubMed] [Google Scholar]
- 31.Niu J, Shi Y, Iwai K, Wu ZH. 2011. LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J 30:3741–3753. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hostager BS, Kashiwada M, Colgan JD, Rothman PB. 2011. HOIL-1L interacting protein (HOIP) is essential for CD40 signaling. PLoS One 6:e23061. doi: 10.1371/journal.pone.0023061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zak DE, Schmitz F, Gold ES, Diercks AH, Peschon JJ, Valvo JS, Niemista A, Podolsky I, Fallen SG, Suen R, Stolyar T, Johnson CD, Kennedy KA, Hamilton MK, Siggs OM, Beutler B, Aderem A. 2011. Systems analysis identifies an essential role for SHANK-associated RH domain-interacting protein (SHARPIN) in macrophage Toll-like receptor 2 (TLR2) responses. Proc Natl Acad Sci U S A 108:11536–11541. doi: 10.1073/pnas.1107577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, Rieser E, Rickard JA, Bankovacki A, Peschel C, Ruland J, Bekker-Jensen S, Mailand N, Kaufmann T, Strasser A, Walczak H, Silke J, Jost PJ, Gyrd-Hansen M. 2012. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell 46:746–758. doi: 10.1016/j.molcel.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Rodgers MA, Bowman JW, Fujita H, Orazio N, Shi M, Liang Q, Amatya R, Kelly TJ, Iwai K, Ting J, Jung JU. 2014. The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J Exp Med 211:1333–1347. doi: 10.1084/jem.20132486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emmerich CH, Schmukle AC, Walczak H. 2011. The emerging role of linear ubiquitination in cell signaling. Sci Signal 4:re5. doi: 10.1126/scisignal.2002187. [DOI] [PubMed] [Google Scholar]
- 38.Tokunaga F, Sakata Si Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. 2009. Involvement of linear polyubiquitylation of NEMO in NF-[kappa]B activation. Nat Cell Biol 11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 39.Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D, Hofmann K, Komander D. 2013. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivkin E, Almeida SM, Ceccarelli DF, Juang YC, MacLean TA, Srikumar T, Huang H, Dunham WH, Fukumura R, Xie G, Gondo Y, Raught B, Gingras AC, Sicheri F, Cordes SP. 2013. The linear ubiquitin-specific deubiquitinase Gumby regulates angiogenesis. Nature 498:318–324. doi: 10.1038/nature12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takiuchi T, Nakagawa T, Tamiya H, Fujita H, Sasaki Y, Saeki Y, Takeda H, Sawasaki T, Buchberger A, Kimura T, Iwai K. 2014. Suppression of LUBAC-mediated linear ubiquitination by a specific interaction between LUBAC and the deubiquitinases CYLD and OTULIN. Genes Cells 19:254–272. doi: 10.1111/gtc.12128. [DOI] [PubMed] [Google Scholar]
- 42.Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, Kamei K, Ma A, Iwai K, Nureki O. 2012. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NFκB regulation. EMBO J 31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang WL, Zhang X, Lin HK. 2010. Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene 29:4493–4503. doi: 10.1038/onc.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. 2012. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep 13:840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu D, Zhao L, Del Valle L, Miklossy J, Zhang L. 2008. Interferon regulatory factor 4 is involved in Epstein-Barr virus-mediated transformation of human B lymphocytes. J Virol 82:6251–6258. doi: 10.1128/JVI.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fries KL, Miller WE, Raab-Traub N. 1996. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol 70:8653–8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu D, Brumm K, Zhang L. 2006. The latent membrane protein 1 of Epstein-Barr Virus (EBV) primes EBV latency cells for type I interferon production. J Biol Chem 281:9163–9169. doi: 10.1074/jbc.M511884200. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Chen ZJ. 2013. Regulation of NF-κB by ubiquitination. Curr Opin Immunol 25:4–12. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harhaj EW, Dixit VM. 2012. Regulation of NF-kappaB by deubiquitinases. Immunol Rev 246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masucci MG. 2004. Epstein-Barr virus oncogenesis and the ubiquitin-proteasome system. Oncogene 23:2107–2115. doi: 10.1038/sj.onc.1207372. [DOI] [PubMed] [Google Scholar]
- 51.Dantuma NP, Masucci MG. 2003. The ubiquitin/proteasome system in Epstein-Barr virus latency and associated malignancies. Semin Cancer Biol 13:69–76. doi: 10.1016/S1044-579X(02)00101-3. [DOI] [PubMed] [Google Scholar]
- 52.Shackelford J, Pagano JS. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem 41:139–156. [DOI] [PubMed] [Google Scholar]
- 53.Shackelford J, Pagano JS. 2004. Tumor viruses and cell signaling pathways: deubiquitination versus ubiquitination. Mol Cell Biol 24:5089–5093. doi: 10.1128/MCB.24.12.5089-5093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knight JS, Sharma N, Robertson ES. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci U S A 102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito S, Murata T, Kanda T, Isomura H, Narita Y, Sugimoto A, Kawashima D, Tsurumi T. 2013. Epstein-Barr virus deubiquitinase down-regulates TRAF6-mediated NF-kappaB signaling during productive replication. J Virol 87:4060–4070. doi: 10.1128/JVI.02020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, Arrowsmith CH, Frappier L. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1: IMplications for EBV-mediated immortalization. Mol Cell 18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Li W, Xiao L, Xu J, Chen X, Tang M, Dong Z, Tao Q, Cao Y. 2012. Viral oncoprotein LMP1 disrupts p53-induced cell cycle arrest and apoptosis through modulating K63-linked ubiquitination of p53. Cell Cycle 11:2327–2336. doi: 10.4161/cc.20771. [DOI] [PubMed] [Google Scholar]
- 58.Husaini R, Ahmad M, Soo-Beng Khoo A. 2011. Epstein-Barr virus latent membrane protein LMP1 reduces p53 protein levels independent of the PI3K-Akt pathway. BMC Res Notes 4:551. doi: 10.1186/1756-0500-4-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greenfeld H, Takasaki K, Walsh MJ, Ersing I, Bernhardt K, Ma Y, Fu B, Ashbaugh CW, Cabo J, Mollo SB, Zhou H, Li S, Gewurz BE. 2015. TRAF1 coordinates polyubiquitin signaling to enhance Epstein-Barr virus LMP1-mediated growth and survival pathway activation. PLoS Pathog 11:e1004890. doi: 10.1371/journal.ppat.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU. 2011. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell 41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, Chen DY, Zhai ZH, Shu HB. 2008. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res 18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 62.Belgnaoui SM, Paz S, Samuel S, Goulet ML, Sun Q, Kikkert M, Iwai K, Dikic I, Hiscott J, Lin R. 2012. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe 12:211–222. doi: 10.1016/j.chom.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel Jaime L, Sen Ganes C. 2016. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity 44:1151–1161. doi: 10.1016/j.immuni.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ning S, Huye LE, Pagano JS. 2005. Regulation of the transcriptional activity of the IRF7 promoter by a pathway independent of interferon signaling. J Biol Chem 285:12262–12270. [DOI] [PubMed] [Google Scholar]
- 65.Ning S, Hahn AM, Huye LE, Joseph PS. 2003. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. J Virol 77:9359–9368. doi: 10.1128/JVI.77.17.9359-9368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. 2006. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem 281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Ren J, Li G, Moorman JP, Yao ZQ, Ning S. 7 November 2016. LMP1 signaling pathway activates IRF4 in EBV latency and a positive circuit between PI3K and Src is required. Oncogene. doi: 10.1038/onc.2016.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Toomey NL, Diaz LA, Walker G, Ramos JC, Barber GN, Ning S. 2011. Oncogenic IRFs provide a survival advantage for EBV- or HTLV1-transformed cells through induction of BIC expression. J Virol 85:8328–8337. doi: 10.1128/JVI.00570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]