ABSTRACT
African green monkeys (AGM) and sooty mangabeys (SM) are well-studied natural hosts of simian immunodeficiency virus (SIV) that do not progress to AIDS when infected with their species-specific viruses. Natural hosts of SIV express very low levels of the canonical entry coreceptor CCR5, and recent studies have shown that CCR5 is dispensable for SIV infection of SM in vivo and that blocking of CCR5 does not prevent ex vivo infection of peripheral blood mononuclear cells (PBMC) from SM or vervet AGM. In both hosts, CXCR6 is an efficient entry pathway in vitro. Here we investigated the use of species-matched CXCR6 and other alternative coreceptors by SIVagmSab, which infects sabaeus AGM. We cloned sabaeus CD4 and 10 candidate coreceptors. Species-matched CXCR6, CCR5, and GPR15 mediated robust entry into transfected cells by pseudotypes carrying SIVagmSab92018ivTF Env, with lower-level entry through GPR1 and APJ. We cloned genetically divergent env genes from the plasma of two wild-infected sabaeus AGM and found similar patterns of coreceptor use. Titration experiments showed that CXCR6 and CCR5 were more efficient than other coreceptors when tested at limiting CD4/coreceptor levels. Finally, blocking of CXCR6 with its ligand CXCL16 significantly inhibited SIVagmSab replication in sabaeus PBMC and had a greater impact than did the CCR5 blocker maraviroc, confirming the use of CXCR6 in primary lymphocyte infection. These data suggest a new paradigm for SIV infection of natural host species, whereby a shared outcome of virus-host coevolution is the use of CXCR6 or other alternative coreceptors for entry, which may direct SIV toward CD4+ T cell subsets and anatomical sites that support viral replication without disrupting immune homeostasis and function.
IMPORTANCE Natural hosts of SIV do not progress to AIDS, in stark contrast to pathogenic human immunodeficiency virus type 1 (HIV-1)-human and SIVmac-macaque infections. Identifying how natural hosts avoid immunodeficiency can elucidate key mechanisms of pathogenesis. It is known that despite high viral loads, natural hosts have a low frequency of CD4+ cells expressing the SIV coreceptor CCR5. In this study, we demonstrate the efficient use of the coreceptor CXCR6 by SIVagmSab to infect sabaeus African green monkey lymphocytes. In conjunction with studies of SIVsmm, which infects sooty mangabeys, and SIVagmVer, which infects vervet monkeys, our data suggest a unifying model whereby in natural hosts, in which the CCR5 expression level is low, the use of CXCR6 or other coreceptors to mediate infection may target SIV toward distinct cell populations that are able to support high-level viral replication without causing a loss of CD4+ T cell homeostasis and lymphoid tissue damage that lead to AIDS in HIV-1 and SIVmac infections.
KEYWORDS: African green monkey, CXCR6, coreceptor, natural host, simian immunodeficiency virus, tropism
INTRODUCTION
Over 40 species of African nonhuman primates (NHP) have been identified as natural hosts of simian immunodeficiency virus (SIV), harboring endemic species-specific viruses that appear to have coevolved with their hosts and cause little if any disease (1). Sooty mangabeys (SM) and African green monkeys (AGM) are the best-studied natural hosts. Despite sustained high viral loads following infection and the short life span of infected cells (2–4), natural hosts typically maintain normal blood CD4+ T cell counts, lack chronic immune activation once acute infection resolves, maintain normal lymph node architecture and function, and remain healthy (5, 6). In stark contrast, infections of nonnatural hosts, including human immunodeficiency virus type 1 (HIV-1) infection of humans and SIV infection of macaques, are characterized by CD4+ T cell loss, chronic immune activation, and progressive inflammation, fibrosis, and dysfunction of lymphoid tissue (7). Understanding differences between natural and nonnatural hosts can highlight key elements in pathogenesis and may elucidate novel targets for intervention.
Several lines of evidence suggest that differential cell targeting may be a central feature underlying distinct outcomes of infections of natural and nonnatural hosts. One mechanism implicates CD4+ T cell populations critical for immune cell homeostasis, including T central memory (Tcm) and T stem cell memory (Tscm) cells, as studies have shown that these subsets are infected at a lower frequency in SM than in rhesus macaques (RM) (8, 9). Second, infected humans and RM show high levels of virus in lymphoid tissue, particularly in germinal center CD4+ T follicular helper (Tfh) cells, which are critical for humoral immunity, and robust lymphoid tissue infection is associated with progressive inflammation, fibrosis, and architectural damage (10, 11). In contrast, lymph nodes of chronically infected SM and AGM are marked by a lower viral burden, minimal fibrosis, and preserved function (11–14). Finally, CD4+ Th17 cells, which are required for gut immune barrier integrity, are maintained in natural host infection but lost in infection of nonnatural hosts (15–17). The loss of gut barrier integrity is linked to the translocation of bacterial products and chronic immune activation in nonnatural hosts, a key feature absent in infected natural hosts (18–21). These observations demonstrate the need to fully understand determinants of cell targeting by SIV.
HIV/SIV cell targeting is determined largely at entry by the expression of CD4 and a seven-transmembrane coreceptor. The established dogma has been that CCR5 is the coreceptor used by SIV in vivo, although several other molecules have long been known to support entry in transfected cells or cell lines in vitro (22–24). The level of CCR5 expression is very low on CD4+ T cells of natural host species, which led to the suggestion that restricted coreceptor availability may limit CD4+ T cell infection (8, 9, 25). However, the high levels of viremia and short life span of infected cells in natural hosts are comparable to those in nonnatural hosts (2–4), raising a paradox of robust SIV replication and infected-cell turnover in the face of exquisitely low CCR5 levels. Several years ago, our laboratory found that pathways other than CCR5 contributed to SIVsmm entry and cell targeting in SM in vivo when we identified a common CCR5 deletion allele in SM that abrogates surface CCR5 expression (26). SM that were homozygous for CCR5-null alleles were infected with SIVsmm at a frequency similar to, and had viral loads only slightly lower than, those of SM with wild-type CCR5. We then showed that SM CXCR6 was an efficient coreceptor for SIVsmm in vitro and, using blocking agents, that both CXCR6 and CCR5 were used by SIVsmm for infection of SM peripheral blood mononuclear cells (PBMC) ex vivo (27, 28). Subsequently, a study of vervet AGM (which inhabit East and Southern Africa) showed that CCR5 was dispensable for SIVagmVer infection of vervet PBMC, and vervet CXCR6 and GPR15 were robust coreceptors in vitro (29). While that study did not identify the non-CCR5 coreceptor(s) responsible for infection of vervet PBMC, it suggested that the use of alternative pathways to target and enter cells was shared by other natural hosts of SIV.
In this study, we wished to understand coreceptor use in another natural host infection, SIVagmSab infection of sabaeus AGM (which inhabit West Africa), and identify the pathways responsible for primary cell infection. Since previous studies of SIVagmSab coreceptor use employed human-derived CD4 and coreceptors, yet small amino acid differences can substantially impact coreceptor activity (30), we cloned sabaeus CD4 and 10 candidate coreceptors. Additionally, since most SIVagmSab isolates available have been passaged in human cell lines (31, 32), potentially altering patterns of coreceptor use, we employed the recently described transmitter/founder SIVagmSab (33) and cloned additional genetically diverse env genes from plasma of wild, naturally infected sabaeus monkeys. Since coreceptor function can be highly dependent on expression levels (34, 35), and also because AGM have a propensity to downregulate CD4 (36), we analyzed entry with a range of coreceptor and CD4 levels. Finally, we used blocking agents to confirm the pathways mediating entry into sabaeus PBMC. We demonstrate here that species-matched sabaeus CXCR6/CD4 is as efficient an entry pathway for SIVagmSab in transfected cells as CCR5/CD4 and that blocking of CXCR6 inhibits SIVagmSab infection of sabaeus PBMC, with an impact that is substantially greater than that of blocking of CCR5. These findings demonstrate a major role for CXCR6 in sabaeus SIVagmSab infection and support the emerging idea that entry mediated by non-CCR5 pathways, particularly CXCR6, is a common feature of SIV infection of natural hosts.
RESULTS
Cloning and sequence analysis of sabaeus African green monkey CD4 and candidate coreceptors.
Recent work from our laboratory and others has described the use of species-matched non-CCR5 coreceptors by the natural host viruses SIVsmm and SIVagmVer (27–29). Here we asked whether an SIV of another natural host, SIVagmSab of sabaeus AGM, also utilized species-matched alternative coreceptors in the context of species-matched CD4. We first cloned CD4 from cDNA of two sabaeus monkeys and identified two alleles that differed from each other by 5 amino acids (CD4-30 and CD4-31) (Table 1). These differences included several previously described AGM CD4 polymorphisms (37, 38), and CD4-30 also contained a novel C-terminal serine. Two polymorphisms fell within domain 1 of CD4 but not within residues implicated in human CD4/HIV-1 gp120 interactions (39). Like other reported AGM CD4 alleles, these sequences share ∼90% amino acid identity to human CD4 and are most closely related to the CD4 molecules of other African green monkey subspecies, including vervet and grivet (Table 1).
TABLE 1.
Sabaeus African green monkey CD4 moleculesa
| CD4 clone | % amino acid identity to CD4 from: |
||||
|---|---|---|---|---|---|
| Human | Rhesus macaque | Sooty mangabey | Grivet AGM (Chlorocebus aethiops) | Vervet AGM (Chlorocebus pygerythrus) | |
| AGM Sab 30 | 90.6 | 95.4 | 95.0 | 99.1 | 98.9 |
| AGM Sab 31 | 90.8 | 95.6 | 95.2 | 99.3 | 99.1 |
CD4 was cloned from PBMC cDNA of two individual animals, yielding two AGM CD4 clones that are 98.9% identical at the amino acid level (5 amino acids are different).
We then cloned APJ, CCR2b, CCR3, CCR4, CCR5, CCR8, CXCR4, CXCR6, GPR1, and GPR15 from genomic DNA (gDNA) or cDNA using primers that lie outside the open reading frame (ORF) of each gene (27). The sequences of most cloned coreceptors were identical to those of the genes described in the recently sequenced sabaeus genome (BioProject accession number PRJNA215854) (40). One exception was CCR2b, which contained a conservative V340A difference. In addition, sabaeus CCR5 is known to be polymorphic, and the clone generated here is unique to reported sequences but contains several previously described AGM polymorphisms (N57S and R163G) (24). We aligned the amino acid sequences of these 10 sabaeus AGM candidate coreceptors with those of human, rhesus macaque, and sooty mangabey. As shown in Table 2, the amino acid sequences of these coreceptors all vary from those of human, including in the N terminus and second extracellular loop (ECL2), the two domains expected to interact with Env based on studies of CCR5-Env interactions (41). These differences underscore the importance of testing the coreceptor usage of SIVs on their species-matched molecules.
TABLE 2.
Sabaeus African green monkey candidate coreceptorsa
| AGM Sab 7TMR | % amino acid identity to 7TMR from: |
No. of amino acids differing from human sequence |
|||
|---|---|---|---|---|---|
| Human | Rhesus macaque | Sooty mangabey | NTD | ECL2 | |
| APJ | 98.7 | 99.7 | 99.5 | 0 | 1 |
| CCR2b | 96.7 | 99.2 | 99.4 | 2 | 2 |
| CCR3 | 93.5 | 98 | 97.2 | 6 | 4 |
| CCR4 | 98.6 | 100 | 100 | 0 | 1 |
| CCR5 | 97.4 | 99.1 | 98.9 | 2 | 1 |
| CCR8 | 94.6 | 99.2 | 98.9 | 8 | 1 |
| CXCR4 | 98.6 | 100 | 99.7 | 3 | 1 |
| CXCR6 | 95.3 | 98.8 | 98.3 | 6 | 1 |
| GPR1 | 98 | 100 | 99.7 | 1 | 1 |
| GPR15 | 97.2 | 99.4 | 99.4 | 1 | 2 |
Seven-transmembrane receptors (7TMRs) were cloned from PBMC genomic DNA or cDNA, and the amino acid identity compared to human, rhesus macaque, and sooty mangabey molecules is shown. Also shown is the number of amino acids differing from the sequence of the human molecule in the N-terminal domain (NTD) and extracellular loop 2 (ECL2), which are implicated in coreceptor function based on studies with CCR5.
The sabaeus sequences were overall more similar to those of other nonhuman primates than to human sequences, and several were identical to those of other monkeys at the amino acid level, such as GPR1 and CXCR4 (100% amino acid identity between sabaeus monkeys and RM) and CCR4 (100% amino acid identity among sabaeus monkeys, RM, and SM). Of note, CXCR6 of macaques (rhesus, pigtail, and cynomolgus macaques) encodes an R at position 31, which results in poor coreceptor activity for this molecule (30), but sabaeus CXCR6 possesses the more common S31 residue that is seen in SM and most other primate CXCR6 sequences and is associated with efficient coreceptor function. The sabaeus CXCR6, GPR15, and CXCR4 amino acid sequences are identical to those reported previously for vervet (29), while vervet GPR1, APJ, CCR2b, CCR3, CCR4, and CCR8 sequences have not been reported.
SIVagmSab Envs use non-CCR5 coreceptors in vitro.
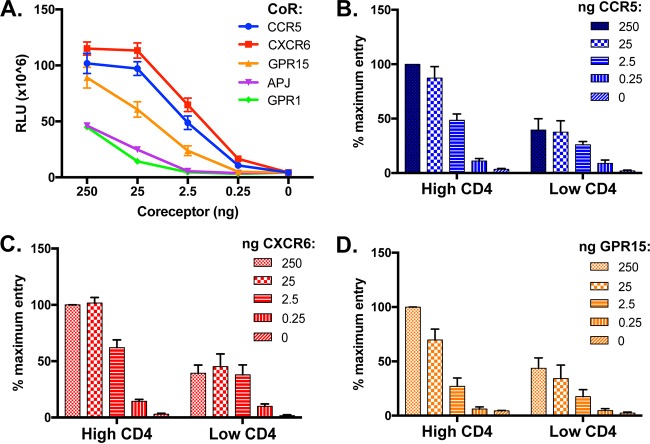
We then tested the ability of each candidate coreceptor to facilitate SIVagmSab entry in vitro and the impact of the two distinct CD4 alleles. Luciferase reporter pseudotype viruses were generated, carrying Env from SIVagmSab92018ivTF, an infectious molecularly cloned transmitted/founder virus derived from intravenous infection with an unpassaged SIVagmSab stock (33). 293T cells were transfected with each candidate coreceptor in conjunction with each of the two sabaeus CD4 alleles and infected with the pseudotype virus, and entry was measured by luciferase production 3 days later. SIVagmSab92018ivTF readily entered cells expressing species-matched CXCR6, CCR5, and GPR15 (Fig. 1). We saw lower levels of entry through sabaeus GPR1 and APJ. Both sabaeus CD4 alleles were used by SIVagmSab for entry, and there was no evidence that the specific CD4 allele altered the preference for any coreceptor tested. Of note, SIVagmSab has been reported to use human CXCR4 (33, 42), but we found no detectable entry through species-matched CXCR4 and CD4. Because we observed no functional difference between the two CD4 alleles, subsequent studies were done with CD4-31.
FIG 1.
Efficient use of alternative sabaeus coreceptors with two sabaeus CD4 molecules by prototype SIVagmSab92018ivTF Env pseudotypes. 293T cells were transfected with sabaeus candidate coreceptors in conjunction with each of the two sabaeus CD4 alleles. Cells were infected 48 h later with pseudotypes carrying Env from the transmitter/founder SIVagmSab92018ivTF. Cells were then lysed 72 h later, and luciferase content was measured as relative light units (RLU). Infections were carried out in triplicate, and data (means ± standard deviations) are representative of results from 4 replicate experiments.
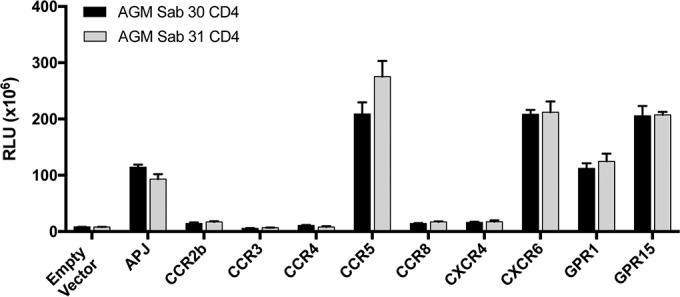
We then sought to expand this analysis to additional SIVagmSab envelopes. Aside from SIVagmSab92018ivTF, all available SIVagmSab isolates have been passaged in human cell lines (31, 32), making it difficult to be sure that adaptation had not occurred in vitro and that their coreceptor usage pattern would reflect authentic SIVagmSab phenotypes. Therefore, we cloned envelopes from plasma collected from two sabaeus monkeys (monkeys 89042 and 89044) that were already infected upon their capture in West Africa. SIV sequences of rev, tat, nef, and vpr (but not env) of plasma virus of AGM 89042 were described previously (43), whereas sequences of AGM 89044 had not been previously characterized. We amplified env from plasma viral RNA (vRNA) and compared these two field isolates to each other and to other SIVagmSab sequences. These two env genes share 78% nucleotide sequence homology with each other and represent distinct sequences among reported SIVagmSab env genes, as shown in Fig. 2A.
FIG 2.
Divergent field isolate SIVagmSab Envs efficiently use alternative coreceptors in vitro. SIVagmSab Envs were amplified from the plasma of two wild-infected sabaeus monkeys (animals 89042 and 89044). (A) Maximum likelihood phylogenetic tree showing SIVagmSab89042 and SIVagmSab89044 env genes as well as SIVagmSab92018ivTF and other previously described SIV env nucleotide sequences. Envs used in this study are boxed, and the two env genes cloned here from wild, naturally infected animals are indicated by arrows. Bootstrap values of >70% are indicated by an asterisk. The bar indicates 0.2 nucleotide substitutions per site. (B) 293T cells were transfected with each sabaeus candidate coreceptor in conjunction with sabaeus CD4-31. Cells were then infected with luciferase reporter pseudotypes containing field isolate SIVagmSab Envs or SIVagmSab92018ivTF Env. Cells were lysed and the luciferase content was measured 72 h later. Infections were carried out in triplicate, and data (means ± standard deviations) are representative of results from 3 replicate experiments.
We pseudotyped these SIVagmSab Envs on a luciferase reporter backbone and tested their ability to use the full panel of potential coreceptors in conjunction with sabaeus CD4-31 (Fig. 2B). Both Envs showed robust entry through sabaeus CXCR6, CCR5, and GPR15 in transfected cells. They also showed similar moderate usages of GPR1, but only one of the two Envs used APJ. Additionally, none of the Envs tested used sabaeus CCR5, CXCR6, or GPR15 independently of CD4 (data not shown). Thus, the alternative pattern of coreceptor use of SIVagmSab92018ivTF Env appears to accurately reflect patterns of divergent SIVagmSab isolates from naturally infected animals, although the low-level use of APJ is less consistent.
Efficient use of CXCR6 by SIVagmSab occurs with various CD4 levels.
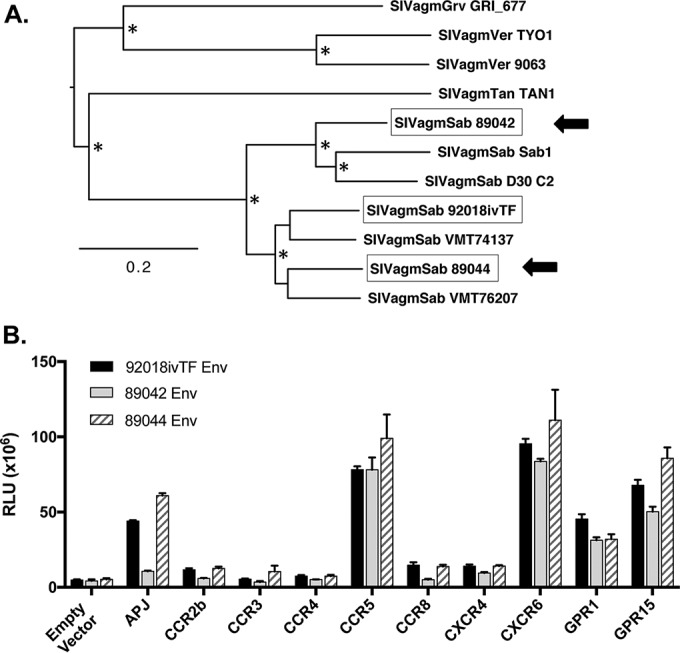
Cells transfected with CD4 and a coreceptor allow the easy identification of potential coreceptor function, but these experimental conditions of overexpression may not reflect entry in the context of primary cells, where coreceptors and CD4 are likely expressed at substantially lower levels. Therefore, we carried out experiments to assess the impact of decreasing amounts of the coreceptor and CD4, in various combinations, on entry supported by each of the coreceptors.
293T cells were first transfected with various amounts of each of the functional sabaeus coreceptors (CCR5, CXCR6, GPR15, GPR1, and APJ), titrating the amount of plasmid in 10-fold serial dilutions from 250 ng to 0.25 ng, in conjunction with our standard amount of the sabaeus CD4 plasmid (250 ng). A parallel set of cells was transfected with human CCR5, CXCR6, and GPR15 plasmids at the same concentrations and stained by fluorescence-activated cell sorter (FACS) analysis, which revealed similar levels of staining (percent positive and mean fluorescence intensity [MFI]) for each coreceptor and decreasing coreceptor expression levels across the titrations that were similar for the three coreceptors (data not shown). Cells transfected with sabaeus CD4 and coreceptor plasmids were then infected with the SIVagmSab92018ivTF reporter virus.
As shown in Fig. 3A, a 10-fold decrease (from 250 to 25 ng) had essentially no impact on entry mediated by CXCR6 and CCR5, whereas entry through GPR15, APJ, and GPR1 was substantially affected. A hundredfold decrease (to 2.5 ng) resulted in an ∼50% loss of entry for CXCR6 and CCR5 but had a substantially greater impact on GPR15 and nearly abrogated entry through APJ and GPR1. Only when it was reduced by 3 orders of magnitude (to 0.25 ng) was entry through CXCR6 and CCR5 markedly attenuated. Thus, high coreceptor levels appear to be less critical for CXCR6 and CCR5 function than for GPR15, GPR1, and APJ.
FIG 3.
Efficient use of sabaeus CXCR6 at low as well as high CD4 levels. (A) 293T cells were transfected with sabaeus CD4-31 (250 ng of the plasmid) and 10-fold serial dilutions (starting at 250 ng) of plasmids encoding sabaeus CCR5, CXCR6, GPR15, GPR1, and GPR15. Empty pcDNA3.1 was used as a filler such that cells under each condition were transfected with an equal quantity of DNA (500 ng). Cells were infected 48 h later with the SIVagmSab92018ivTF Env luciferase reporter pseudotype, and luciferase content was measured 72 h later. (B to D) Experiment performed as described above for panel A but with 10-fold dilutions of CCR5 (B), CXCR6 (C), and GPR15 (D) in conjunction with either large amounts of CD4 (250 ng) or small amounts of CD4 (2.5 ng). Entry levels were normalized to 100% based on RLU expression using maximum CD4 and coreceptor amounts for each coreceptor. Data represent means ± standard deviations of results from three replicate experiments carried out in triplicate.
We then asked whether the amount of CD4 impacts the use of each coreceptor and the relative dependence on coreceptor levels, focusing on CXCR6, CCR5, and GPR15 (Fig. 3B). This question is particularly relevant for SIVagm entry, since AGM CD4+ T cells downregulate CD4 upon entry into the memory pool (36), and CD4 may thus be a limiting factor. For this, we tested a 100-fold difference, utilizing “high-CD4” (250 ng) (comparable to data shown Fig. 3A) and “low-CD4” (2.5 ng) conditions. FACS staining confirmed that titrating the amount of the AGM CD4 plasmid resulted in comparably reduced levels of CD4 expression (data not shown).
As shown in Fig. 3B, the level of entry under low-CD4 conditions was approximately half that under high-CD4 conditions for essentially all coreceptors and coreceptor transfection levels. Furthermore, under low-CD4 conditions, entry through CXCR6 appeared least sensitive to decreasing coreceptor levels, showing relatively preserved entry down to a 100-fold decrease in coreceptor plasmid transfection. These data suggest that low levels of CD4 may be less of an obstacle to SIVagmSab entry through CXCR6 than through other pathways. Collectively, these findings suggest that SIVagmSab entry through sabaeus CXCR6 is at least as robust as entry through sabaeus CCR5, including under conditions of limiting coreceptor and CD4 levels that may be relevant to AGM primary cells. In contrast, GPR15 is used efficiently in standard transfection-overexpression conditions, but entry through this pathway may be more sensitive to lower coreceptor levels, while GPR1 and APJ are less efficient and require higher levels of the coreceptor to support entry.
SIVagmSab uses CXCR6 to infect primary sabaeus PBMC.
We recently demonstrated that replication of SIVsmm in SM primary PBMC was inhibited by blocking CXCR6 with its chemokine ligand CXCL16 and by blocking CCR5 with the small-molecule antagonist maraviroc and was reduced further by using both blocking agents together (28). This indicated that both coreceptor pathways are used for SIVsmm infection of cells of its natural host. Riddick et al. then showed that blocking of CCR5 had no impact on the replication of SIVagmVer in vervet PBMC, implicating the use of non-CCR5 pathways (29). Therefore, having shown that sabaeus CXCR6 functions as an efficient coreceptor in vitro, we asked if CXCR6 was in fact used for SIVagmSab infection of sabaeus primary PBMC. To do this, we tested whether blocking entry through CXCR6 and CCR5, separately and together, would limit the replication of SIVagmSab in sabaeus PBMC.
Since the sequences of these sabaeus coreceptors vary from those of human and sooty mangabey homologs, we first asked if recombinant human CXCL16 and maraviroc would specifically block entry through sabaeus CXCR6 and CCR5, respectively. 293T cells were transfected with plasmids encoding sabaeus CD4 along with CCR5, CXCR6, GPR15, GPR1, and APJ; pretreated for 1 h with CXCL16 (500 ng/ml) or maraviroc (15 μM); and then infected with the SIVagmSab92018ivTF reporter pseudotype (Fig. 4A). Both CXCL16 and maraviroc blocked entry through their cognate coreceptors and were highly specific. However, entry inhibition was incomplete, with 18% entry through CCR5 remaining despite maraviroc treatment and 11% entry through CXCR6 remaining despite CXCL16 treatment. Therefore, the use of these inhibitors is specific, although it could potentially underestimate the amount of SIVagmSab entry through each coreceptor. Unfortunately, no small molecule or other blocker against the orphan receptor GPR15 is available, preventing a similar analysis of this coreceptor.
FIG 4.
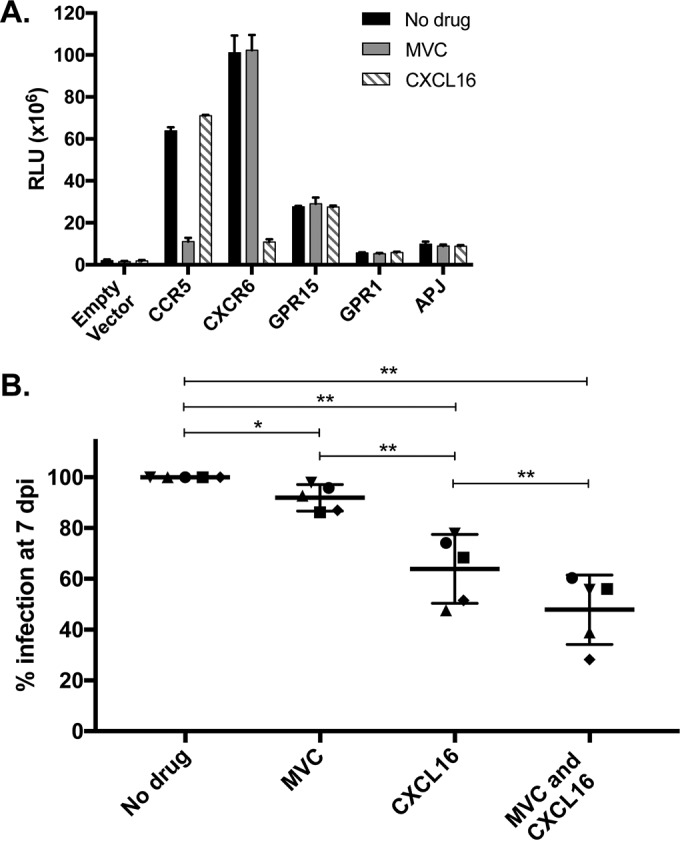
Blocking entry through CXCR6 limits SIVagmSab replication in primary sabaeus lymphocytes. (A) 293T cells were transfected with plasmids encoding sabaeus CCR5, CXCR6, GPR15, GPR1, or APJ or an empty vector (10 ng) in conjunction with sabaeus CD4-31 (1 μg). Cells were treated for 1 h with the CXCR6 ligand CXCL16 (500 ng/ml), the CCR5 blocker maraviroc (MVC) (15 μM), or the vehicle alone and then infected with a luciferase reporter pseudotype containing SIVagmSab92018ivTF Env. Cells were lysed and luciferase content was measured 72 h later. Infections were carried out in triplicate, and data (means ± standard deviations) are representative of results from 3 replicate experiments. (B) Sabaeus PBMC were stimulated for 3 days with PHA/IL-2; treated for 1 h with CXCL16 (500 ng/ml), maraviroc (15 μM), both blocking agents, or the vehicle alone; and then infected in duplicate with SIVagmSab92018ivTF using 5 ng of viral p27 Gag antigen. Infection was measured as p27 production in the supernatant at day 7 and is shown for each treatment as a percentage of the value for the vehicle alone (no drug). Each symbol represents data from a different animal's PBMC, and shown are means and standard deviations under each condition. *, P < 0.05; **, P < 0.01 (two-tailed paired t test). dpi, days postinfection.
We then investigated the use of CXCR6 and CCR5 by SIVagmSab during infection of sabaeus PBMC using cells from five uninfected animals. Sequencing of the CXCR6 and CCR5 genes from the animals revealed identical CXCR6 sequences, while CCR5 revealed previously described polymorphisms at residues 57, 93, and 163 (44). No animals possessed CCR5 sequences predicted to be nonfunctional, but three were heterozygous for R93, which has been reported to support SIVagm entry less efficiently (24), although SIVagmSab replication in PBMC from these animals did not differ from that in other animals (data not shown).
PBMC from the five animals were stimulated with phytohemagglutinin (PHA) and interleukin-2 (IL-2) and then treated with CXCL16, maraviroc, both agents together, or the vehicle alone. Cells were then infected with replication-competent SIVagmSab92018ivTF derived from an infectious molecular clone, and virus replication was quantified by measuring p27 levels in the culture supernatant at day 7. Blockade of CCR5 resulted in a very slight but statistically significant 8% decrease in supernatant p27 levels (range, 2 to 14%; P = 0.026 compared to the vehicle alone). In contrast, blockade of CXCR6 decreased replication by 36% (range, 22 to 52%; P = 0.004 compared to the vehicle alone). The impact of blocking of CXCR6 was significantly greater than that of blocking of CCR5 (P = 0.006). Blocking of both coreceptors together reduced infection further, such that p27 levels were decreased 52% when both agents were used (range, 40 to 72%; P = 0.005 and P = 0.001 compared to CXCL16 alone and CCR5 alone, respectively). In contrast, CXCL16 had no effect on infection of RM PBMC by SIVmac239 (which uses RM CXCR6 poorly and enters RM PBMC entirely through CCR5 [28]) or on infection of human PBMC by HIV-1 BaL, indicating that SIVagmSab/sabaeus PBMC blocking by CXCL16 was not a nonspecific effect of the chemokine (data not shown).
These data confirm that CXCR6 supports SIVagmSab infection of primary lymphocytes in this natural host. These data also suggest that CCR5 does so as well, although the quantitative contribution of CCR5 appears to be less than that of CXCR6 based on these blocking results. Given the incomplete blocking by CXCL16 and maraviroc of their coreceptors, it is unclear whether the remaining infection (∼50%) is due to residual entry through CXCR6 and CCR5 or the use of yet another alternative coreceptor.
DISCUSSION
In contrast to HIV-1- and SIVmac-infected nonprogressors that avoid pathogenesis mainly by controlling viremia, natural hosts are able to tolerate sustained viremia without disease, which is likely the result of prolonged coevolution with their SIVs. Recent evidence suggests that one mechanism for this peaceful coexistence is linked to patterns of host cell targeting in nonpathogenic infections (45). Since cell targeting is determined largely by entry, in this study, we aimed to identify entry pathways used by the natural host virus SIVagmSab. We found that diverse Envs, including those from wild, naturally infected animals, used sabaeus CXCR6 very efficiently, and used GPR15 somewhat less so, in addition to CCR5. Robust CXCR6 use occurred at both high and low CD4 levels. Blocking of CXCR6 inhibited SIVagmSab replication in PBMC, confirming that it not only functions in transfection-based systems but also mediates infection of primary target cells relevant to infection in vivo.
The in vitro use of alternative coreceptors by various SIVs was first observed years ago, although most studies utilized human rather than species-matched molecules (22–24). Nevertheless, CCR5 has long been considered the only coreceptor relevant to SIV infection in vivo. One early-recognized exception was SIVrcm, which infects the natural host red-capped mangabeys (RCM). Most RCM are CCR5 null due to a common CCR5 deletion allele, and SIVrcm was found to use human CCR2 and CXCR6 but not CCR5 (46, 47). More recently, our laboratory identified a common CCR5 deletion allele in SM and showed that homozygous CCR5-null animals were infected at a frequency similar to that of SM with wild-type CCR5 and maintained nearly equivalent viral loads (26). We later identified SM CXCR6 as a robust coreceptor for SIVsmm Envs isolated from both CCR5-null and wild-type animals and showed that CXCR6 mediated entry into SM primary lymphocytes (along with CCR5 if it was present) (27, 28). More recently, it was shown that blocking of entry through CCR5 by SIVagmVer did not hinder replication in vervet PBMC and that species-matched CXCR6 and GPR15 were robust coreceptors in transfected cells (29). Our findings here showing efficient alternative coreceptor usage by SIVagmSab for sabaeus lymphocyte infection identify yet another natural host in which CCR5 is dispensable, bolstering the idea that CCR5-independent alternative coreceptor-mediated infection is a widely shared feature of SIV-natural host relationships. This new paradigm stands in marked contrast to pathogenic SIVmac infection of nonnatural host RM and HIV-1 infection of humans, where CCR5 is indispensable (28, 29, 48–52).
Across natural hosts, CCR5 expression is highly restricted, and the frequency of CCR5+ CD4+ cells is low compared to that in nonnatural hosts (25). This exceedingly low CCR5 expression level has represented a paradox given the sustained high-level viremia seen in natural hosts. Similarly, cell-associated infection in natural hosts exceeds the proportion of cells expressing CCR5 (53). The observation that alternative coreceptors, particularly CXCR6, are used for SIV entry in infections of multiple natural hosts provides an explanation for the robust replication in the face of limited CCR5 expression and suggests that CCR5− CD4+ cells expressing CXCR6 (or other alternative coreceptors) are a major site of virus replication in these species.
The use of alternative coreceptors in CCR5-low natural hosts, in contrast to CCR5-dependent infection of nonnatural hosts, likely also contributes to distinct outcomes of infection. Unique features of natural host infection include the resolution of immune activation after acute infection, maintenance of gut immunity and integrity, an absence of microbial translocation, less lymphoid tissue infection during chronic infection and an absence of inflammation and fibrosis, and a lack of peripheral CD4+ T cell depletion (5, 6). A central mechanism underlying these distinct features is divergent cell subset and anatomical targeting. While infected cells show rapid turnover in both natural and nonnatural hosts, CD4+ Tcm and Tscm cells are infected at a lower frequency in sooty mangabeys than in rhesus macaques (8, 9), which likely allows the maintenance of CD4+ T cell homeostasis. Th17 cells are critical for gut immune barrier function, and although their infection status in natural hosts is not fully defined, they are maintained in infected SM and AGM but lost in SIVmac-infected RM and HIV-1-infected humans (15–17). Associated with this, mucosal barrier integrity is maintained in natural hosts, whereas nonnatural hosts experience a loss of barrier integrity, microbial translocation, and chronic immune activation (18–21). Within lymphoid tissues, Tfh cells are heavily infected throughout the course of HIV-1 and SIVmac infections in humans and RM, with associated inflammation and fibrosis (10). In contrast, chronically infected natural hosts show much less Tfh cell infection and lower levels of lymph node virus, with an absence of inflammation and fibrosis and maintenance of lymph node architecture (11–14). Differential virus targeting to various cell types and anatomical locations indicates that the determinants of cell targeting are important determinants of distinct outcomes of infection.
It has been suggested that on CD4+ T cells, CXCR6 serves as an extralymphoid homing receptor (54), and limited studies in humans suggest that it is expressed mainly by memory T cells, particularly T cells in tissues such as lung and at sites of inflammation and cancer (55–57), although expression on naive cells has been reported as well (58). Studies of CXCR6 expression on nonhuman primate natural host cells have been hindered by a lack of cross-reactivity of anti-human CXCR6 antibodies. In vervet AGM, CXCR6 mRNA was recently described for both naive and memory CD4+ T cell subsets, with divergent patterns of regulation between CCR5 and CXCR6 transcripts, but surface protein expression levels were not determined (29). Thus, while restricted CCR5 expression likely protects certain critical subsets from SIV infection in natural hosts, the CXCR6+ CD4+ cells that enable robust replication without inducing disease remain to be defined. Finally, several studies have investigated SIV transmission and CCR5+ CD4+ target cell availability (59–62). Our data suggest that to fully understand these relationships, it will be important to determine the frequency of CXCR6+ target cells at the sites of transmission in these animals.
Blocking of CXCR6 inhibited infection of sabaeus PBMC to a significantly greater extent (36% reduction) than did blocking of CCR5 (only 8% reduction), suggesting that CXCR6 may be quantitatively dominant in these cells. Alternatively, it is possible that differences in blocking efficiencies for the two agents could be responsible, although we think that this is unlikely to explain the minimal impact of maraviroc on infection (28). Nevertheless, both agents together were not sufficient to completely inhibit SIVagmSab infection. While this result may be due to incomplete blocking of CXCR6 and CCR5, we cannot exclude the possibility that additional coreceptors are used as well. GPR15 is a potential candidate, as it is used in transfected cells (although less efficiently than CXCR6 and CCR5 at limiting amounts of CD4 and the coreceptor) and is expressed on human CD4+ T cells (63). Interestingly, SIVmac can use RM GPR15 to enter transfected cells in vitro, but blocking of CCR5 fully inhibits SIVmac replication in RM PBMC, indicating that GPR15 does not mediate RM PBMC infection (28, 29). Concordant findings were reported for other ex vivo and in vivo studies (64, 65). It is not clear why GPR15 might function in transfected cells but not in primary cells. However, we could not directly test entry through GPR15 since its ligand has not been identified and no small antagonist is available, so a role for GPR15 in addition to CXCR6 is unresolved. Future studies using knockdown approaches could address this point. It is unlikely that GPR1 and APJ serve as coreceptors for SIVagmSab in PBMC, given the low-level function the we found in transfected cells and limited evidence (in humans and rodents at least) for expression on CD4+ T cells (66–70). Finally, while we tested all of the major alternative coreceptors that have been reported for various SIVs, it is possible that there are other, as-yet-unidentified seven-transmembrane receptor (7TMR) molecules that could also mediate entry.
Previous studies of CCR5 and CXCR4 demonstrated that HIV-1 Env engages two coreceptor domains, the N terminus and ECL2 (41, 71). In addition, viruses that use multiple coreceptors may preferentially engage one of the two domains on each coreceptor (72). It will be interesting to determine whether this two-site model applies to CXCR6 use by SIV Envs. SIV use of the CXCR6 N terminus is supported by the observation that a single amino acid change in this region renders RM CXCR6 a poor SIV coreceptor compared to CXCR6 from other primates (30). Further experiments, such as domain swapping and site-directed mutagenesis, will be required to accurately identify the contribution of these domains to SIV Env interactions.
The AGM cells included in this study were polymorphic for CCR5, concordant with previously reported observations (24, 44). Some AGM CCR5 polymorphisms limit SIVagm replication in vitro, which, combined with the low frequency of CCR5+ CD4+ T cells, suggests a long history of coevolution between SIVagm and its host, driven by deleterious consequences of CCR5-mediated infection (24, 44). In contrast, all CXCR6 alleles sequenced here were identical, suggesting distinct evolutionary pressures exerted on these two coreceptors. We speculate that the lack of AGM CXCR6 diversity despite the prolonged AGM-SIVagm coexistence reflects a benign outcome when CXCR6 is used to enter target cells.
With the data reported here, the use of a non-CCR5 pathway in species-matched lymphocyte infection has been shown for three viruses infecting natural hosts, SIVagmSab, SIVsmm, and SIVagmVer, and CXCR6 use specifically has been confirmed for the former two viruses. These viruses are quite diverse: SIVagm and SIVsmm represent two distinct SIV lineages, and studies combining molecular clock analyses with biogeography place their most recent common ancestor >100,000 years ago (73). Furthermore, SIVagmSab and SIVagmVer infect nonsympatric subspecies of African green monkeys in West and East/Southern Africa, respectively, which also diverged over 100,000 years ago (73). Despite these divergences, the common use of non-CCR5 entry pathways suggests that this pattern is the norm among natural hosts that harbor endemic SIV strains and is likely the result of virus-host coevolution. In contrast, CCR5-restricted entry occurs only in more recent zoonotic infections, such as SIVmac and HIV-1 infections, in hosts where CCR5+ CD4+ cells are abundant. While CCR5-only entry is possible because of high CCR5 expression levels in these hosts, why alternative coreceptors no longer support primary lymphocyte infection is unclear. For SIVmac-RM, the main barrier to CXCR6 use is the R31 polymorphism in RM CXCR6 (30), while for HIV-1, it is mainly a function of Env that limits the use of CXCR6 (although exceptions are well described [23, 74]). Thus, in order to understand the emergence of HIV-1, it will be important to define the species-matched coreceptor usage patterns of SIVgsn/SIVmus/SIVmon, which is the natural host virus forerunner for Env of SIVcpz, which gave rise to HIV-1 (75–78).
In summary, our study supports a model of SIV/HIV tropism whereby in nonnatural hosts, an abundance of CCR5+ cells results in infection and disruption of CD4+ T cell subsets and lymphoid tissues, which leads to immunodeficiency. In contrast, in infection of natural hosts, where the frequency of CCR5+ cells is low, the use of CXCR6 (and possibly other coreceptors) directs SIV toward cells that are able to support viremia without causing immunodeficiency. Defining CXCR6+ cells, in both blood and tissues, will be essential for understanding natural host cell targeting.
MATERIALS AND METHODS
Sabaeus African green monkey PBMC.
PBMC from sabaeus AGM (derived from a Caribbean-born colony [79]) were isolated by density gradient centrifugation from whole blood that was collected as previously described (25), cryopreserved, and thawed before use. Animals were housed at the University of Pittsburgh in accordance with the Guide for the Care and Use of Laboratory Animals (83), the Association for Assessment and Accreditation of Laboratory Animal Care, and the Animal Welfare Act. Animal procedures were approved by the IACUC of the University of Pittsburgh.
Cloning of sabaeus CD4 and coreceptor molecules.
Genomic DNA was isolated from resting sabaeus AGM PBMC by using the QIAamp DNA blood minikit (Qiagen), and RNA was isolated from both resting and concanavalin A (ConA)/IL-2-stimulated PBMC by using the RNeasy Plus kit (Qiagen). cDNA was synthesized by using the Superscript III First Strand kit (Invitrogen), using random hexamers (for CD4), oligo(dT) (for CXCR4), or gene-specific primers (for CCR2b and CCR4). Primers for 7TMR cDNA synthesis and PCR amplification were previously described (27), as were the CD4 primers (AGM-CD4-for2 and AGM-CD4-rev) (29). Sabaeus CD4 and full-length 7TMRs were amplified from genomic DNA (APJ, CCR3, CCR5, CCR8, CXCR6, GPR1, and GPR15) or cDNA (CD4, CCR2b, CCR4, and CXCR4) by using Phusion High Fidelity DNA polymerase (New England BioLabs). 7TMRs were amplified from AGM 30, and CD4 was amplified from AGM 30 and AGM 31. Reaction mixtures included 50 ng gDNA or 2 μl cDNA synthesis reaction mix, and cycling conditions were those described in the kit protocol, as previously reported (27), but annealing temperatures were modified as follows: 64°C for APJ, CCR3, CCR4, CCR8, CXCR6, GPR1, GPR15, and CXCR4; 60°C for CCR2b; 61°C for CCR5; or 72°C for CD4. Amplicons were ligated into pcDNA3.1+ by using dual-restriction enzyme digestion (CCR2b, CCR3, CCR4, CCR8, or CXCR6) or into the pcDNA3.1D/V5-His-TOPO vector by TOPO directional cloning (CD4, APJ, GPR1, CCR5, CXCR4, or GPR15) using the pcDNA3.1 Directional TOPO Expression kit (Invitrogen). Clones were screened by restriction digestion and confirmed by Sanger sequencing. Nucleotide and amino acid alignments were performed by using the ClustalW algorithm in MacVector 13.5.2, and 7TMR membrane topology predictions were made by using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html).
Cloning and sequence analysis of SIVagmSab envelopes.
Envs were derived from plasma samples stored at the Institut Pasteur, which were previously collected in Senegal from two sabaeus AGM (animals 89042 and 89044) that were SIV positive (SIV+) upon capture. Viral RNA was isolated from plasma by using the Viral RNA minikit (Qiagen), and cDNA was synthesized by using Superscript III reagents and a specific primer (5′-CTCCWCCCTGGAAAGTCCCKCT-3′) modified from a primer reported previously (33). SIVagmSab Envs were then PCR amplified with primers that were designed by using Primer3 (80) based on previously reported SIVagmSab sequences (forward primer 5′-CACCCCSCTCCAGGCCTGTRNCAATA-3′ and reverse primer 5′-CCARCCATCSACWATDCCCC-3′). Amplicons were cloned into pcDNA3.1D/V5-His-TOPO (Invitrogen) and screened by colony PCR and restriction analysis. Properly sized inserts were screened for function by generating pseudotype virions and testing them on transfected 293T cells expressing sabaeus CD4 and CCR5 or CXCR6.
Functional SIVagmSab env clones were sequenced by using an Illumina MiSeq instrument, and reads were aligned by using Geneious 7.1.7 software (Biomatters Ltd.). A maximum likelihood phylogenetic tree was generated with PhyML v3.0 (81), using the portion of previously reported SIVagm sequences overlapping our two novel env genes (1,882 bp) (59). One thousand bootstrap replicates were performed.
Coreceptor functional analyses using pseudotyped virus.
To generate pseudotype virus, 293T cells were transfected with plasmids encoding SIVagmSab Envs and the pNL43-Luc-E-R+ backbone (82) by using Fugene 6 (Promega), as previously described (27). The following day, cells were washed, and fresh medium was added. The supernatant was collected 2 days later and spun with 5% sucrose to pellet cell debris, and p24 Gag antigen levels were quantified by an enzyme-linked immunosorbent assay (ELISA). Pseudotype virus was treated with DNase (Roche) prior to use (50 U/ml for 15 min) to avoid inadvertent target cell transfection by residual backbone plasmid DNA.
Coreceptor function was tested as previously described (27). Briefly, 2.5 × 105 293T cells were plated per well into 12-well plates and transfected by using Fugene 6 with plasmids encoding CD4 and the coreceptor (or empty pcDNA3.1+), using 250 ng each, except where otherwise noted. Cells were replated 24 h later at 2 × 104 cell/well in 100 μl in 96-well plates and infected the following day with pseudotype viruses using equivalent amounts of virus based on p24 antigen content. Plates were spinoculated for 2 h at 1,200 × g and incubated for 72 h at 37°C. To measure entry, cells were lysed, and luciferase content was quantified as relative light units (RLU), as described previously (27), by using a Luminoskan Ascent instrument (Thermo Labsystems). For all experiments, target cells were infected in parallel with vesicular stomatitis virus G (VSVg) pseudotypes to ensure that differences in entry were SIV/coreceptor specific. To confirm that equivalent amounts of the coreceptor plasmid yielded similar levels of expression and that titration experiments resulted in similar decreases in expression levels for different coreceptors, 293T cells were transfected with human CCR5, CXCR6, and GPR15 and stained by FACS analysis using anti-human coreceptor antibodies 3A9 (CCR5; BD Pharmingen), K041E5 (CXCR6; BioLegend), and 367902 (GPR15; R&D Systems). Validation of AGM CD4 plasmid titration experiments was done similarly, using antibody L200 (BD Pharmingen).
To test the activity and specificity of blocking agents on sabaeus CCR5 and CXCR6, target cells transfected with CD4 and the coreceptor were pretreated for 1 h at 37°C with recombinant human CXCL16 (500 ng/ml; R&D Systems), maraviroc (15 μM; obtained from the NIH AIDS Reagent Program), or the vehicle alone (dimethyl sulfoxide [DMSO]). Cells were infected with pseudotype virus in the continued presence of blocking agents and harvested 72 h later for measurement of luciferase activity.
Sabaeus PBMC infection.
Cryopreserved sabaeus PBMC were thawed and stimulated with PHA-P (5 μg/ml; Sigma) in complete medium (RPMI 1640 with 10% fetal bovine serum [FBS], 1% l-glutamine, and 1% penicillin-streptomycin), and IL-2 (100 U/ml) was added the following day. After 72 h of stimulation, 2 × 105 cells/well were plated into 96-well round-bottom plates and incubated for 1 h at 37°C with recombinant human CXCL16 (500 ng/ml), maraviroc (15 μM), both agents together, or the vehicle alone (DMSO). Cells were then infected by spinoculation (1,200 × g for 90 min) with SIVagmSab92018ivTF derived from an infectious molecular clone (33) using 5 ng viral p27 Gag antigen and then incubated at 37°C overnight. The following morning, cells were washed two times and resuspended in 200 μl complete medium containing IL-2 and appropriate blockers, and twice weekly, half of the supernatant was removed and replaced with fresh medium containing IL-2 and blocking agents. Virus replication was monitored by p27 Gag antigen production in the supernatant by an ELISA (Perkin-Elmer). Infections were carried out in duplicate in 5 independent experiments using cells from different animals, and levels of p27 at day 7 in the absence of blocking agents ranged from 5,000 to 16,000 pg/ml.
Accession number(s).
Sabaeus AGM CD4 and coreceptor sequences cloned here were deposited in GenBank under accession numbers KY225904 to KY225915. SIVagmSab env sequences were deposited in GenBank under accession numbers KY225916 and KY225917.
ACKNOWLEDGMENTS
We thank F. Bibollet-Ruche, R. Warrier, and G. Learn for valuable advice and S. Bryan and F. Shaheen for technical assistance.
This work was funded by NIH grants R56-AI091516 and R01-MH06119 (R.G.C.), 2T32AI007632 (K.S.W.), R37-AI050529 (B.H.H.), and R01HL117715 (C.A. and I.P.). We also acknowledge assistance from multiple cores of the Penn Center for AIDS Research (P30-AI045008).
REFERENCES
- 1.Peeters M, Ma D, Liegeois F, Apetrei C. 2014. Simian immunodeficiency virus infections in the wild, p 37–68. In Ansari A, Silvestri G (ed), Natural hosts of SIV: implication in AIDS, 1st ed Elsevier Inc, New York, NY. [Google Scholar]
- 2.Gordon SN, Dunham RM, Engram JC, Estes J, Wang Z, Klatt NR, Paiardini M, Pandrea IV, Apetrei C, Sodora DL, Lee HY, Haase AT, Miller MD, Kaur A, Staprans SI, Perelson AS, Feinberg MB, Silvestri G. 2008. Short-lived infected cells support virus replication in sooty mangabeys naturally infected with simian immunodeficiency virus: implications for AIDS pathogenesis. J Virol 82:3725–3735. doi: 10.1128/JVI.02408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silvestri G, Sodora DL, Koup RA, Paiardini M, O'Neil SP, McClure HM, Staprans SI, Feinberg MB. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441–452. doi: 10.1016/S1074-7613(03)00060-8. [DOI] [PubMed] [Google Scholar]
- 4.Pandrea I, Ribeiro RM, Gautam R, Gaufin T, Pattison M, Barnes M, Monjure C, Stoulig C, Dufour J, Cyprian W, Silvestri G, Miller MD, Perelson AS, Apetrei C. 2008. Simian immunodeficiency virus SIVagm dynamics in African green monkeys. J Virol 82:3713–3724. doi: 10.1128/JVI.02402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ploquin MJJ, Silvestri G, Müller-Trutwin M. 2016. Immune activation in HIV infection: what can the natural hosts of simian immunodeficiency virus teach us? Curr Opin HIV AIDS 11:201–208. doi: 10.1097/COH.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 6.Raehtz K, Pandrea I, Apetrei C. 6 July 2016. The well-tempered SIV infection: pathogenesis of SIV infection in natural hosts in the wild, with emphasis on virus transmission and early events post-infection that may contribute to protection from disease progression. Infect Genet Evol doi: 10.1016/j.meegid.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lackner AA, Lederman MM, Rodriguez B. 2012. HIV pathogenesis: the host. Cold Spring Harb Perspect Med 2:a007005. doi: 10.1101/cshperspect.a007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, Hallberg PL, Cramer E, Schlub T, Chan ML, Riddick NE, Collman RG, Apetrei C, Pandrea I, Else J, Munch J, Kirchhoff F, Davenport MP, Brenchley JM, Silvestri G. 2011. Low levels of SIV infection in sooty mangabey central memory CD4+ T cells are associated with limited CCR5 expression. Nat Med 17:830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartwright EK, McGary CS, Cervasi B, Micci L, Lawson B, Elliott ST, Collman RG, Bosinger SE, Paiardini M, Vanderford TH, Chahroudi A, Silvestri G. 2014. Divergent CD4+ T memory stem cell dynamics in pathogenic and nonpathogenic simian immunodeficiency virus infections. J Immunol 192:4666–4673. doi: 10.4049/jimmunol.1303193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perreau M, Savoye A-LL, De Crignis E, Corpataux J-MM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenchley JM, Vinton C, Tabb B, Hao XP, Connick E, Paiardini M, Lifson JD, Silvestri G, Estes JD. 2012. Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood 120:4172–4181. doi: 10.1182/blood-2012-06-437608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beer B, Scherer J, zur Megede J, Norley S, Baier M, Kurth R. 1996. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long-term simian immunodeficiency virus infection of African green monkeys. Virology 219:367–375. doi: 10.1006/viro.1996.0262. [DOI] [PubMed] [Google Scholar]
- 13.Diop OM, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Müller-Trutwin MC. 2000. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J Virol 74:7538–7547. doi: 10.1128/JVI.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broussard SR, Staprans SI, White R, Whitehead EM, Feinberg MB, Allan JS. 2001. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J Virol 75:2262–2275. doi: 10.1128/JVI.75.5.2262-2275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, Khoruts A, Frank I, Else J, Schacker T, Silvestri G, Douek DC. 2008. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cecchinato V, Trindade CJ, Laurence A, Heraud JM, Brenchley JM, Ferrari MG, Zaffiri L, Tryniszewska E, Tsai WP, Vaccari M, Parks RW, Venzon D, Douek DC, O'Shea JJ, Franchini G. 2008. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol 1:279–288. doi: 10.1038/mi.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favre D, Lederer S, Kanwar B, Ma ZM, Proll S, Kasakow Z, Mold J, Swainson L, Barbour JD, Baskin CR, Palermo R, Pandrea I, Miller CJ, Katze MG, McCune JM. 2009. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog 5:e1000295. doi: 10.1371/journal.ppat.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12:1365–1371. [DOI] [PubMed] [Google Scholar]
- 19.Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, Hirsch VM, Silvestri G, Douek DC, Miller CJ, Haase AT, Lifson J, Brenchley JM. 2010. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog 6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klatt NR, Harris LD, Vinton CL, Sung H, Briant JA, Tabb B, Morcock D, McGinty JW, Lifson JD, Lafont BA, Martin MA, Levine AD, Estes JD, Brenchley JM. 2010. Compromised gastrointestinal integrity in pigtail macaques is associated with increased microbial translocation, immune activation, and IL-17 production in the absence of SIV infection. Mucosal Immunol 3:387–398. doi: 10.1038/mi.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandrea IV, Gautam R, Ribeiro RM, Brenchley JM, Butler IF, Pattison M, Rasmussen T, Marx PA, Silvestri G, Lackner AA, Perelson AS, Douek DC, Veazey RS, Apetrei C. 2007. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J Immunol 179:3035–3046. doi: 10.4049/jimmunol.179.5.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng HK, Unutmaz D, KewalRamani VN, Littman DR. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 23.Edinger AL, Hoffman TL, Sharron M, Lee B, O'Dowd B, Doms RW. 1998. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- 24.Kuhmann SE, Madani N, Diop OM, Platt EJ, Morvan J, Müller-Trutwin MC, Barré-Sinoussi F, Kabat D. 2001. Frequent substitution polymorphisms in African green monkey CCR5 cluster at critical sites for infections by simian immunodeficiency virus SIVagm, implying ancient virus-host coevolution. J Virol 75:8449–8460. doi: 10.1128/JVI.75.18.8449-8460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandrea I, Apetrei C, Gordon S, Barbercheck J, Dufour J, Bohm R, Sumpter B, Roques P, Marx PA, Hirsch VM, Kaur A, Lackner AA, Veazey RS, Silvestri G. 2007. Paucity of CD4+CCR5+ T cells is a typical feature of natural SIV hosts. Blood 109:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riddick NE, Hermann EA, Loftin LM, Elliott ST, Wey WC, Cervasi B, Taaffe J, Engram JC, Li B, Else JG, Li Y, Hahn BH, Derdeyn CA, Sodora DL, Apetrei C, Paiardini M, Silvestri G, Collman RG. 2010. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog 6:e1001064. doi: 10.1371/journal.ppat.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott ST, Riddick NE, Francella N, Paiardini M, Vanderford TH, Li B, Apetrei C, Sodora DL, Derdeyn CA, Silvestri G, Collman RG. 2012. Cloning and analysis of sooty mangabey alternative coreceptors that support simian immunodeficiency virus SIVsmm entry independently of CCR5. J Virol 86:898–908. doi: 10.1128/JVI.06415-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott ST, Wetzel KS, Francella N, Bryan S, Romero DC, Riddick NE, Shaheen F, Vanderford T, Derdeyn CA, Silvestri G, Paiardini M, Collman RG. 2015. Dualtropic CXCR6/CCR5 simian immunodeficiency virus (SIV) infection of sooty mangabey primary lymphocytes: distinct coreceptor use in natural versus pathogenic hosts of SIV. J Virol 89:9252–9261. doi: 10.1128/JVI.01236-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riddick NE, Wu F, Matsuda K, Whitted S, Ourmanov I, Goldstein S, Goeken RM, Plishka RJ, Buckler-White A, Brenchley JM, Hirsch VM. 2016. Simian immunodeficiency virus SIVagm efficiently utilizes non-CCR5 entry pathways in African green monkey lymphocytes: potential role for GPR15 and CXCR6 as viral coreceptors. J Virol 90:2316–2331. doi: 10.1128/JVI.02529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pöhlmann S, Lee B, Meister S, Krumbiegel M, Leslie G, Doms RW, Kirchhoff F. 2000. Simian immunodeficiency virus utilizes human and sooty mangabey but not rhesus macaque STRL33 for efficient entry. J Virol 74:5075–5082. doi: 10.1128/JVI.74.11.5075-5082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allan JS, Short M, Taylor ME, Su S, Hirsch VM, Johnson PR, Shaw GM, Hahn BH. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol 65:2816–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin MJ, Hui H, Robertson DL, Müller MC, Barré-Sinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH. 1994. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J 13:2935–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gnanadurai CW, Pandrea I, Parrish NF, Kraus MH, Learn GH, Salazar MG, Sauermann U, Töpfer K, Gautam R, Münch J, Stahl-Hennig C, Apetrei C, Hahn BH, Kirchhoff F. 2010. Genetic identity and biological phenotype of a transmitted/founder virus representative of nonpathogenic simian immunodeficiency virus infection in African green monkeys. J Virol 84:12245–12254. doi: 10.1128/JVI.01603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, Choe H, Sodroski J, Newman W, Koup RA, Mackay CR. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med 185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beaumier CM, Harris LD, Goldstein S, Klatt NR, Whitted S, McGinty J, Apetrei C, Pandrea I, Hirsch VM, Brenchley JM. 2009. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat Med 15:879–885. doi: 10.1038/nm.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fomsgaard A, Müller-Trutwin MC, Diop O, Hansen J, Mathiot C, Corbet S, Barré-Sinoussi F, Allan JS. 1997. Relation between phylogeny of African green monkey CD4 genes and their respective simian immunodeficiency virus genes. J Med Primatol 26:120–128. doi: 10.1111/j.1600-0684.1997.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 38.Fomsgaard A, Hirsch VM, Johnson PR. 1992. Cloning and sequences of primate CD4 molecules: diversity of the cellular receptor for simian immunodeficiency virus/human immunodeficiency virus. Eur J Immunol 22:2973–2981. doi: 10.1002/eji.1830221132. [DOI] [PubMed] [Google Scholar]
- 39.Ashkenazi A, Presta LG, Marsters SA, Camerato TR, Rosenthal KA, Fendly BM, Capon DJ. 1990. Mapping the CD4 binding site for human immunodeficiency virus by alanine-scanning mutagenesis. Proc Natl Acad Sci U S A 87:7150–7154. doi: 10.1073/pnas.87.18.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren WC, Jasinska AJ, García-Pérez R, Svardal H, Tomlinson C, Rocchi M, Archidiacono N, Capozzi O, Minx P, Montague MJ, Kyung K, Hillier LW, Kremitzki M, Graves T, Chiang C, Hughes J, Tran N, Huang Y, Ramensky V, Choi O-WW, Jung YJ, Schmitt CA, Juretic N, Wasserscheid J, Turner TR, Wiseman RW, Tuscher JJ, Karl JA, Schmitz JEE, Zahn R, O'Connor DH, Redmond E, Nisbett A, Jacquelin B, Müller-Trutwin MC, Brenchley JM, Dione M, Antonio M, Schroth GP, Kaplan JR, Jorgensen MJ, Thomas GW, Hahn MW, Raney BJ, Aken B, Nag R, Schmitz J, Churakov G, Noll A, Stanyon R, et al. 2015. The genome of the vervet (Chlorocebus aethiops sabaeus). Genome Res 25:1921–1933. doi: 10.1101/gr.192922.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dragic T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J Gen Virol 82:1807–1814. doi: 10.1099/0022-1317-82-8-1807. [DOI] [PubMed] [Google Scholar]
- 42.Pandrea I, Kornfeld C, Ploquin MJ, Apetrei C, Faye A, Rouquet P, Roques P, Simon F, Barré-Sinoussi F, Müller-Trutwin MC, Diop OM. 2005. Impact of viral factors on very early in vivo replication profiles in simian immunodeficiency virus SIVagm-infected African green monkeys. J Virol 79:6249–6259. doi: 10.1128/JVI.79.10.6249-6259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jubier-Maurin V, Sarni-Manchado P, Veas F, Vidal N, Bibollet-Ruche F, Durand JP, Galat-Luong A, Cuny G. 1995. Regulatory genes of simian immunodeficiency viruses from West African green monkeys (Cercopithecus aethiops sabaeus). J Virol 69:7349–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. 1997. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol 71:8642–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanderford T, Paiardini M. 2014. Distinct cellular targets of SIV infection in natural and non-natural hosts of SIV, p 235–256. In Ansari A, Silvestri G (ed), Natural hosts of SIV: implication in AIDS, 1st ed Elsevier Inc, New York, NY. [Google Scholar]
- 46.Chen Z, Kwon D, Jin Z, Monard S, Telfer P, Jones MS, Lu CY, Aguilar RF, Ho DD, Marx PA. 1998. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J Exp Med 188:2057–2065. doi: 10.1084/jem.188.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beer BE, Foley BT, Kuiken CL, Tooze Z, Goeken RM, Brown CR, Hu J, St Claire M, Korber BT, Hirsch VM. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J Virol 75:12014–12027. doi: 10.1128/JVI.75.24.12014-12027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 49.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 50.Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med 2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 51.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien SJ. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856–1862. [DOI] [PubMed] [Google Scholar]
- 52.Veazey RS, Klasse PJ, Ketas TJ, Reeves JD, Piatak M, Kunstman K, Kuhmann SE, Marx PA, Lifson JD, Dufour J, Mefford M, Pandrea I, Wolinsky SM, Doms RW, DeMartino JA, Siciliano SJ, Lyons K, Springer MS, Moore JP. 2003. Use of a small molecule CCR5 inhibitor in macaques to treat simian immunodeficiency virus infection or prevent simian-human immunodeficiency virus infection. J Exp Med 198:1551–1562. doi: 10.1084/jem.20031266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jochems SP, Jacquelin B, Chauveau L, Huot N, Petitjean G, Lepelley A, Liovat A-SS, Ploquin MJJ, Cartwright EK, Bosinger SE, Silvestri G, Barré-Sinoussi F, Lebon P, Schwartz O, Müller-Trutwin MC. 2015. Plasmacytoid dendritic cell infection and sensing capacity during pathogenic and nonpathogenic simian immunodeficiency virus infection. J Virol 89:6918–6927. doi: 10.1128/JVI.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim CH, Kunkel EJ, Boisvert J, Johnston B, Campbell JJ, Genovese MC, Greenberg HB, Butcher EC. 2001. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest 107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unutmaz D, Xiang W, Sunshine MJ, Campbell J, Butcher E, Littman DR. 2000. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J Immunol 165:3284–3292. doi: 10.4049/jimmunol.165.6.3284. [DOI] [PubMed] [Google Scholar]
- 56.Mandai Y, Takahashi D, Hase K, Obata Y, Furusawa Y, Ebisawa M, Nakagawa T, Sato T, Katsuno T, Saito Y, Shimaoka T, Yokosuka O, Yokote K, Ohno H. 2013. Distinct roles for CXCR6(+) and CXCR6(−) CD4(+) T cells in the pathogenesis of chronic colitis. PLoS One 8:e65488. doi: 10.1371/journal.pone.0065488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu W, Liu Y, Zhou W, Si L, Ren L. 2014. CXCL16 and CXCR6 are coexpressed in human lung cancer in vivo and mediate the invasion of lung cancer cell lines in vitro. PLoS One 9:e99056. doi: 10.1371/journal.pone.0099056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharron M, Pöhlmann S, Price K, Lolis E, Tsang M, Kirchhoff F, Doms RW, Lee B. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41–49. [PubMed] [Google Scholar]
- 59.Ma D, Jasinska AJ, Feyertag F, Wijewardana V, Kristoff J, He T, Raehtz K, Schmitt CA, Jung Y, Cramer JD, Dione M, Antonio M, Tracy R, Turner T, Robertson DL, Pandrea I, Freimer N, Apetrei C, International Vervet Research Consortium . 2014. Factors associated with simian immunodeficiency virus transmission in a natural African nonhuman primate host in the wild. J Virol 88:5687–5705. doi: 10.1128/JVI.03606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pandrea I, Onanga R, Souquiere S, Mouinga-Ondéme A, Bourry O, Makuwa M, Rouquet P, Silvestri G, Simon F, Roques P, Apetrei C. 2008. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J Virol 82:5501–5509. doi: 10.1128/JVI.02555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chahroudi A, Cartwright E, Lee ST, Mavigner M, Carnathan DG, Lawson B, Carnathan PM, Hashempoor T, Murphy MK, Meeker T, Ehnert S, Souder C, Else JG, Cohen J, Collman RG, Vanderford TH, Permar SR, Derdeyn CA, Villinger F, Silvestri G. 2014. Target cell availability, rather than breast milk factors, dictates mother-to-infant transmission of SIV in sooty mangabeys and rhesus macaques. PLoS Pathog 10:e1003958. doi: 10.1371/journal.ppat.1003958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandrea I, Parrish NF, Raehtz K, Gaufin T, Barbian HJ, Ma D, Kristoff J, Gautam R, Zhong F, Haret-Richter GS, Trichel A, Shaw GM, Hahn BH, Apetrei C. 2012. Mucosal simian immunodeficiency virus transmission in African green monkeys: susceptibility to infection is proportional to target cell availability at mucosal sites. J Virol 86:4158–4168. doi: 10.1128/JVI.07141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen LP, Pan J, Dinh TT, Hadeiba H, O'Hara E, Ebtikar A, Hertweck A, Gökmen MR, Lord GM, Jenner RG, Butcher EC, Habtezion A. 2015. Role and species-specific expression of colon T cell homing receptor GPR15 in colitis. Nat Immunol 16:207–213. doi: 10.1038/nrm3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiene M, Marzi A, Urbanczyk A, Bertram S, Fisch T, Nehlmeier I, Gnirss K, Karsten CB, Palesch D, Münch J, Chiodi F, Pöhlmann S, Steffen I. 2012. The role of the alternative coreceptor GPR15 in SIV tropism for human cells. Virology 433:73–84. doi: 10.1016/j.virol.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 65.Pöhlmann S, Stolte N, Munch J, Ten Haaft P, Heeney JL, Stahl-Hennig C, Kirchhoff F. 1999. Co-receptor usage of BOB/GPR15 in addition to CCR5 has no significant effect on replication of simian immunodeficiency virus in vivo. J Infect Dis 180:1494–1502. doi: 10.1086/315097. [DOI] [PubMed] [Google Scholar]
- 66.Puffer BA, Sharron M, Coughlan CM, Baribaud F, McManus CM, Lee B, David J, Price K, Horuk R, Tsang M, Doms RW. 2000. Expression and coreceptor function of APJ for primate immunodeficiency viruses. Virology 276:435–444. doi: 10.1006/viro.2000.0557. [DOI] [PubMed] [Google Scholar]
- 67.Rourke JL, Muruganandan S, Dranse HJ, McMullen NM, Sinal CJ. 2014. Gpr1 is an active chemerin receptor influencing glucose homeostasis in obese mice. J Endocrinol 222:201–215. doi: 10.1530/JOE-14-0069. [DOI] [PubMed] [Google Scholar]
- 68.Banas M, Zegar A, Kwitniewski M, Zabieglo K, Marczynska J, Kapinska-Mrowiecka M, LaJevic M, Zabel BA, Cichy J. 2015. The expression and regulation of chemerin in the epidermis. PLoS One 10:e0117830. doi: 10.1371/journal.pone.0117830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang C, Parrish NF, Wilen CB, Li H, Chen Y, Pavlicek JW, Berg A, Lu X, Song H, Tilton JC, Pfaff JM, Henning EA, Decker JM, Moody MA, Drinker MS, Schutte R, Freel S, Tomaras GD, Nedellec R, Mosier DE, Haynes BF, Shaw GM, Hahn BH, Doms RW, Gao F. 2011. Primary infection by a human immunodeficiency virus with atypical coreceptor tropism. J Virol 85:10669–10681. doi: 10.1128/JVI.05249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med 186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Z, Berson JF, Chen Y, Turner JD, Zhang T, Sharron M, Jenks MH, Wang Z, Kim J, Rucker J, Hoxie JA, Peiper SC, Doms RW. 1997. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci U S A 94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma D, Jasinska A, Kristoff J, Grobler JP, Turner T, Jung Y, Schmitt C, Raehtz K, Feyertag F, Martinez Sosa N, Wijewardana V, Burke DS, Robertson DL, Tracy R, Pandrea I, Freimer N, Apetrei C, International Vervet Research Consortium . 2013. SIVagm infection in wild African green monkeys from South Africa: epidemiology, natural history, and evolutionary considerations. PLoS Pathog 9:e1003011. doi: 10.1371/journal.ppat.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tscherning-Casper C, Vodros D, Menu E, Aperia K, Fredriksson R, Dolcini G, Chaouat G, Barre-Sinoussi F, Albert J, Fenyo EM. 2000. Coreceptor usage of HIV-1 isolates representing different genetic subtypes obtained from pregnant Cameroonian women. European Network for In Utero Transmission of HIV-1. J Acquir Immune Defic Syndr 24:1–9. doi: 10.1097/00042560-200005010-00001. [DOI] [PubMed] [Google Scholar]
- 75.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 76.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 78.Sharp PM, Shaw GM, Hahn BH. 2005. Simian immunodeficiency virus infection of chimpanzees. J Virol 79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, Jacquelin B, Bohm R, Marx PA, Barre-Sinoussi F, Hirsch VM, Müller-Trutwin MC, Lackner AA, Veazey RS. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J Virol 80:4858–4867. doi: 10.1128/JVI.80.10.4858-4867.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 81.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 82.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 83.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC. [Google Scholar]