ABSTRACT
Hepatitis C virus (HCV) strain JFH-1, which belongs to genotype 2a, replicates autonomously in cultured cells, whereas another genotype 2a strain, J6CF, does not. Previously, we found that replacement of the NS3 helicase and NS5B-to-3′X regions of J6CF with those of JFH-1 confers J6CF replication competence. In this study, we aimed to identify the minimum modifications within these genomic regions needed to establish replication-competent J6CF. We previously identified 4 mutations in the NS5B-to-3′X region that could be used instead of replacement of this region to confer J6CF replication competence. Here, we induced cell culture-adaptive mutations in J6CF by the long-term culture of J6CF/JFH-1 chimeras composed of JFH-1 NS5B-to-3′X or individual parts of this but not the NS3 helicase region. After 2 months of culture, efficient HCV replication and infectious virus production in chimeric RNA-transfected cells were observed, and several amino acid mutations in NS4A were identified in replicating HCV genomes. The introduction of NS4A mutations into the J6CF/JFH-1 chimeras enhanced viral replication and infectious virus production. Immunofluorescence microscopy demonstrated that some of these mutations altered the subcellular localization of the coexpressed NS3 protein and affected the interaction between NS3 and NS4A. Finally, introduction of the most effective NS4A mutation, A1680E, into J6CF contributed to its replication competence in cultured cells when introduced in conjunction with four previously identified adaptive mutations in the NS5B-to-3′X region. In conclusion, we identified an adaptive mutation in NS4A that confers J6CF replication competence when introduced in conjunction with 4 mutations in NS5B-to-3′X and established a replication-competent J6CF strain with minimum essential modifications in cultured cells.
IMPORTANCE The HCV cell culture system using the JFH-1 strain and HuH-7 cells can be used to assess the complete HCV life cycle in cultured cells. This cell culture system has been used to develop direct-acting antivirals against HCV, and the ability to use various HCV strains within this system is important for future studies. In this study, we aimed to establish a novel HCV cell culture system using another HCV genotype 2a strain, J6CF, which replicates in chimpanzees but not in cultured cells. We identified an effective cell culture-adaptive mutation in NS4A and established a replication-competent J6CF strain in cultured cells with minimum essential modifications. The described strategy can be used in establishing a novel HCV cell culture system, and the replication-competent J6CF clone composed of the minimum essential modifications needed for cell culture adaptation will be valuable as another representative of genotype 2a strains.
KEYWORDS: HCV, cell culture, RNA replication, infectious virus production, adaptive mutation
INTRODUCTION
Cell culture of viral pathogens is important for understanding viral characteristics and for developing antiviral strategies. Hepatitis C virus (HCV) is a major cause of chronic liver diseases affecting approximately 150 million people worldwide (1–4). After the discovery of HCV in 1989 (1), HCV research was hindered by the lack of a cell culture system. Several HCV clones were identified to be infectious in chimpanzees, but the clones could not replicate in cultured cells (5–8). In 1999, the HCV subgenomic replicon system was invented (9). This system allows HCV replication in cultured cells, but most of the strains require adaptive mutations for efficient replication. In 2005, we established an HCV cell culture system using the JFH-1 strain (10). This strain was isolated from a fulminant hepatitis patient and belongs to genotype 2a (GT2a) (11). The JFH-1 strain can replicate efficiently in cultured cells without adaptive mutations and is also infectious in chimpanzees (10, 12). This system can be used as a platform to study the entire HCV life cycle, including infection, replication, assembly of infectious viruses, and virus secretion. These systems are useful for the high-throughput screening of antiviral reagents with activity against HCV and have been used to identify novel direct-acting antivirals (DAAs) that efficiently inhibit HCV replication. Since only one amino acid mutation reduces the effect of DAAs, HCV subgenomic replicon or cell culture systems containing fewer or no adaptive mutations are desirable for the accurate characterization of drug-resistant HCV strains. To date, several HCV strains have been reported to be competent for infection and replication in cultured cells. However, all these clones, aside from JFH-1, require many adaptive mutations (13–19). The molecular clone J6CF was constructed from the consensus sequence of HCV isolated from a chimpanzee inoculated with an HCV-infected patient's plasma (20). This strain also belongs to genotype 2a and is similar to the JFH-1 strain at the nucleotide (89% identity) and amino acid (91% identity) levels. Propagation of J6CF in vivo can be observed in chimpanzees after intrahepatic inoculation with in vitro-synthesized full-length HCV RNA (7). However, J6CF does not replicate in cultured cells transfected with J6CF RNA (21).
In a previous study, we described regions of the JFH-1 genome that promote the efficient replication of a J6CF/JFH-1 chimera in cultured cells. We found that replacement of regions of the J6CF NS3 helicase (N3H) and NS5B-to-3′X (N5BX) with those of JFH-1 confers J6CF replication competence (21). Furthermore, we found that three amino acid mutations, A2892S, R2959K, and Y3003F, and one nucleotide mutation, C9458G, in the N5BX region of J6CF confer replication competence without requiring the replacement of the entire N5BX region (22). In this study, we aimed to identify cell culture-adaptive mutations that could be used instead of replacement of N3H of JFH-1 and to establish a replication-competent J6CF clone with minimum essential modifications in cultured cells.
RESULTS
Capacities of viral replication and infectious virus production among J6CF chimeric genomes containing JFH-1 regions.
We previously reported that the N3H and N5BX regions of JFH-1 are essential to confer J6CF replication competence in an otherwise replication-incompetent strain in cultured cells (21). Instead of replacing the entire N5BX region, we clarified that three amino acid mutations (A2892S, R2959K, and Y3003F) in the NS5B region and a nucleotide mutation (C9458G) in the variable region (VR) conferred J6CF replication competence (22). In this study, we aimed to identify the minimum essential modifications that confer J6CF replication competence without having to replace the entire N3H region.
To confirm the replication capacity of J6CF chimeras that did not contain JFH-1 N3H, we constructed a series of J6CF chimeric replicons with JFH-1-derived regions of N5BX, the latter part of the region from NS5B to 3′X (SRX), the region from 5BSL to 3′X (5BSLX), the region from NS5B to VR (5BVR), or NS5B only (N5B) with or without JFH-1 N3H (Fig. 1A). In the reporter replicon assay, efficient replication was observed in all of the J6CF constructs harboring JFH-1 N3H (6.J6/N3H+N5BX-JFH1, 8.J6/N3H+SRX-JFH1, 10.J6/N3H+5BSLX-JFH1, 12.J6/N3H+5BVR-JFH1, and 14.J6/N3H+N5B-JFH1), although replacement of JFH-1 N3H alone (4.J6/N3H-JFH1) did not confer J6CF replication competence (Fig. 1B). Among J6CF chimeric replicon constructs without JFH-1 N3H, the replication capacities of 5.J6/N5BX-JFH1, 7.J6/SRX-JFH1, and 11.J6/5BVR-JFH1 differed. However, other chimeric replicons, 9.J6/5BSLX-JFH1 and 13.J6/N5B-JFH1, did not undergo detectable replication in the reporter replicon assay (Fig. 1B).
FIG 1.
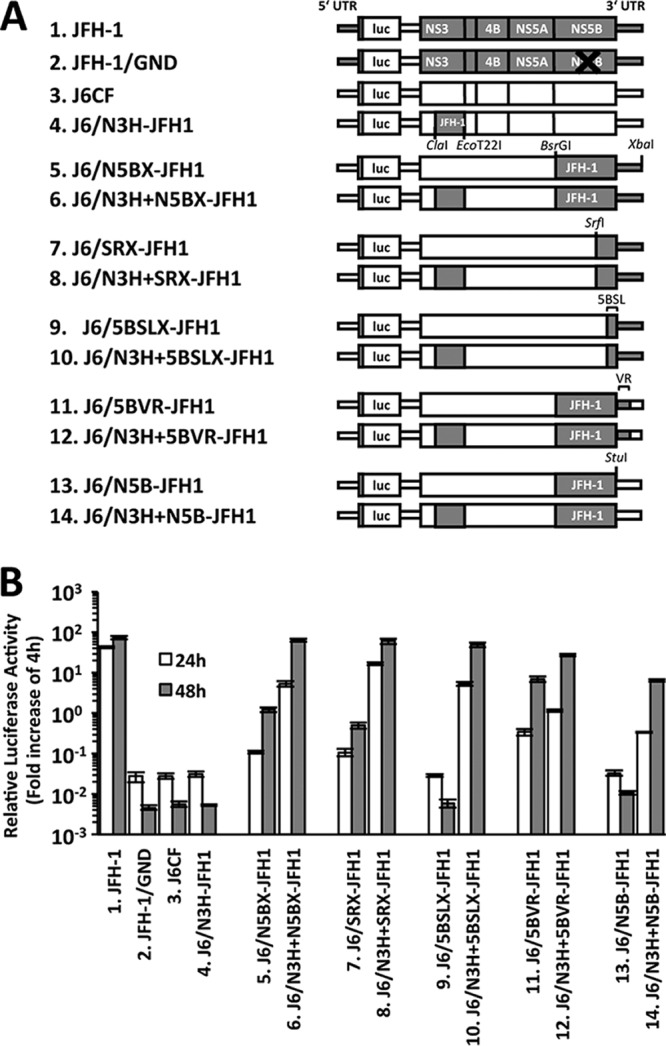
Replication capacity of J6CF chimeric subgenomic replicons containing JFH-1 regions. (A) Schematic structures of chimeric J6CF replicons containing JFH-1 regions. The restriction enzyme recognition sites used for the construction of plasmids are indicated. luc, firefly luciferase gene. (B) Replication capacities of J6CF chimeric subgenomic replicons. In vitro-synthesized RNA was transfected into Huh-7.5.1 cells, and the cells were harvested at 4, 24, and 48 h posttransfection. The luciferase activity in the cell lysates was measured and expressed as the fold increase from the values at 4 h posttransfection to correct for transfection efficiency. Independent assays were performed in triplicate, and the data are presented as the mean number of relative light units (RLU) ± standard deviation at 24 h and 48 h posttransfection.
In assays of full-length HCV RNA transfection, time-dependent increases in the level of the HCV core antigen (Ag) in the cells were observed in all of the J6CF chimeric constructs containing JFH-1 N3H (6.J6/N3H+N5BX-JFH1, 8.J6/N3H+SRX-JFH1, 10.J6/N3H+5BSLX-JFH1, 12.J6/N3H+5BVR-JFH1, and 14.J6/N3H+N5B-JFH1), indicating efficient HCV replication (Fig. 2A and B). Among the J6CF chimeric constructs without JFH-1 N3H, lower levels of replication were observed only in cells transfected with J6/N5BX-JFH1, whereas other transfections did not result in detectable replication. Similar trends in the level of HCV core Ag secreted into the culture media were observed (Fig. 2C). Taken together, among the J6CF chimeric constructs without JFH-1 N3H, we found that 5.J6/N5BX-JFH1 could replicate at low levels when in the form of a subgenomic replicon or full-length construct. The J6CF chimeric constructs 7.J6/SRX-JFH1 and 11.J6/5BVR-JFH1 exhibited detectable replication when they were in the subgenomic replicon form but not in the full-length-construct form. Strains of other chimeric constructs, 9.J6/5BSLX-JFH1 and 13.J6/N5B-JFH1, did not undergo detectable replication when they were in either form.
FIG 2.
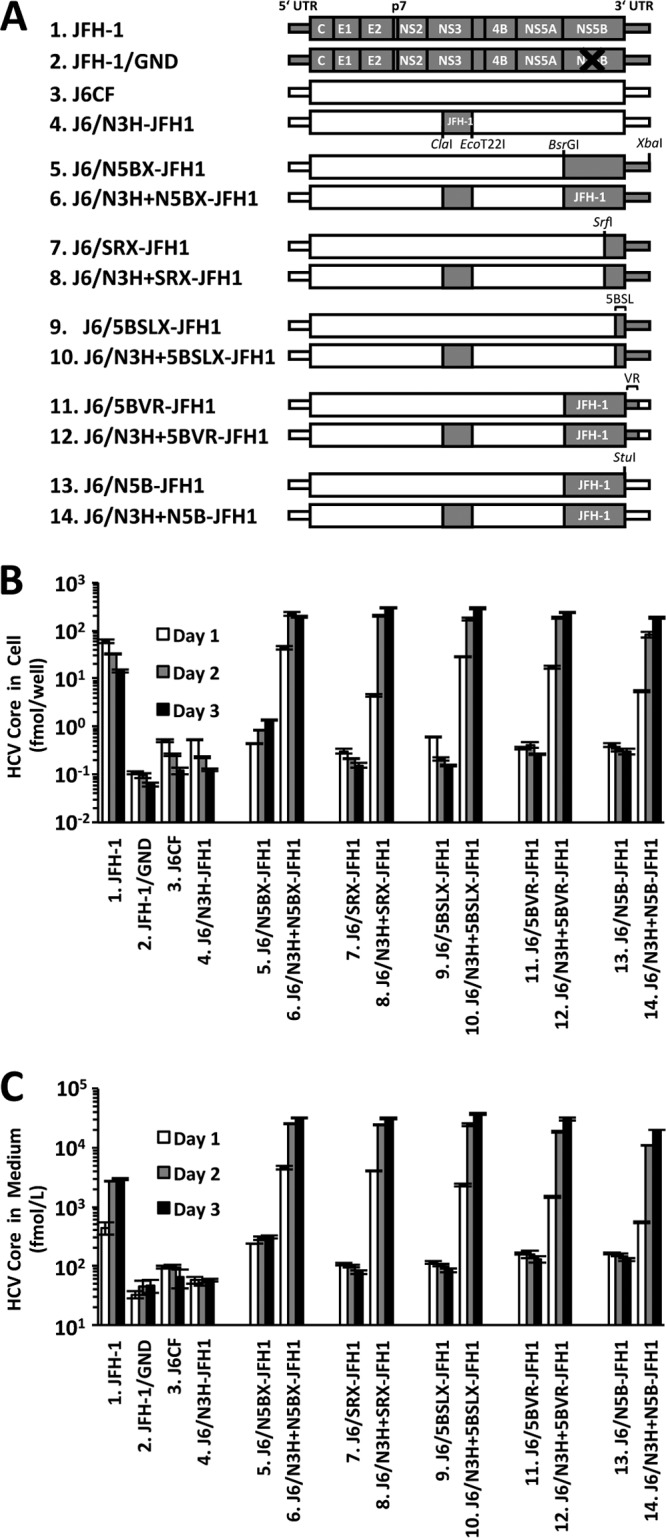
Capacities of viral replication and infectious virus production of full-length J6CF chimeric genomes containing JFH-1 regions. (A) Schematic structures of full-length chimeric J6CF genomes containing JFH-1 regions. The restriction enzyme recognition sites used for the construction of plasmids are indicated. (B, C) HCV core Ag levels in cells (B) and culture media (C) of RNA-transfected cells. In vitro-synthesized RNA was transfected into Huh-7.5.1 cells. Cells and culture media were harvested at 1, 2, and 3 days posttransfection. The amounts of HCV core Ag in cells and culture media were measured. Independent assays were performed in triplicate, and the data are presented as the mean ± standard deviation.
Long-term culture of cells transfected with full-length J6CF chimeric RNA.
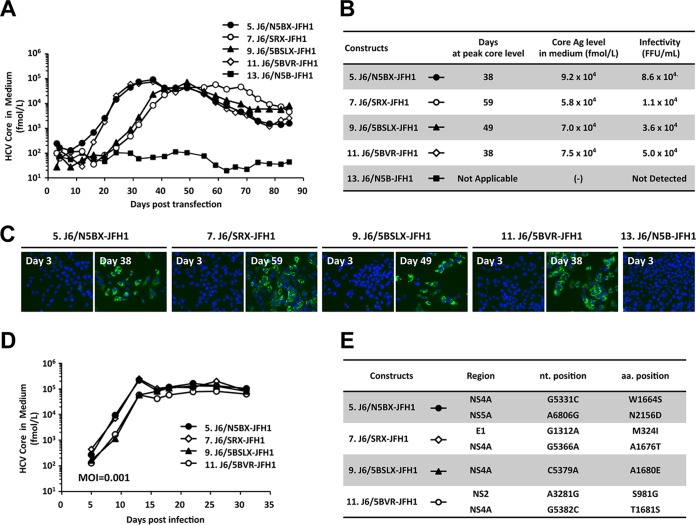
To induce cell culture-adaptive mutations in the J6CF chimeras that undergo low or undetectable levels of replication, we performed the long-term culture of cells transfected with full-length J6CF chimeric RNAs of the constructs 5.J6/N5BX-JFH1, 7.J6/SRX-JFH1, 9.J6/5BSLX-JFH1, 11.J6/5BVR-JFH1, and 13.J6/N5B-JFH1 (Fig. 3A). The HCV core Ag levels in the culture media of cells transfected with 5.J6/N5BX-JFH1 or 11.J6/5BVR-JFH1 RNA started to increase at day 15 and peaked at day 38. The HCV core Ag levels in the culture media of cells transfected with 9.J6/5BSLX-JFH1 or 7.J6/SRX-JFH1 RNA started to increase at day 28 and peaked at days 49 and 59, respectively. The peak levels of HCV core Ag ranged from 5.8 × 104 fmol/liter to 9.2 × 104 fmol/liter, and the infectivity titers ranged from 1.1 × 104 focus-forming units (FFU)/ml to 8.6 × 104 FFU/ml (Fig. 3B). At the times of peak levels of HCV core Ag, most of the cells were HCV positive (Fig. 3C). The increase in the level of HCV core Ag in cells transfected with 13.J6/N5B-JFH1 RNA was not observed until the end of the culture period (day 85).
FIG 3.
Long-term cultures of full-length J6CF chimeric RNA-transfected cells. (A) HCV core Ag levels in the culture media of full-length J6CF chimeric RNA-transfected cells. In vitro-synthesized RNA was transfected into Huh-7.5.1 cells, and the cells were passaged every 2 to 5 days, depending on the conditions of the cells. Culture medium was collected at every passage, and HCV core Ag levels were measured. (B) HCV core Ag levels and infectivity in media at the time of peak HCV core Ag levels (days 38 to 59). (C) Ratios of HCV-positive cells at 3 days posttransfection and at the time of peak HCV core Ag levels. HCV-positive cells were visualized by staining with anti-core Ag antibody (green), and nuclei were visualized with DAPI (4′,6-diamidino-2-phenylindole; blue). (D) Inoculation of culture media at the time of peak HCV core Ag levels into naive Huh-7.5.1 cells at an MOI of 0.001. (E) Amino acid mutations detected in the viral genome after long-term culture.
To assess the replication capacity of the viruses obtained at the times of peak HCV core Ag levels, the culture medium was used to infect naive Huh-7.5.1 cells at a multiplicity of infection (MOI) of 0.001. Immediate increases in HCV core Ag levels were observed after infection at a low MOI compared with the time to the increase in HCV core Ag levels with full-length RNA transfection (compare Fig. 3A and D). The HCV core Ag levels in the media of cells infected with these virus populations peaked at day 13 and ranged from 7.8 × 104 fmol/liter to 2.4 × 105 fmol/liter. These results indicate that the culture media obtained after long-term culture contained cell culture-adapted viruses and these viruses could replicate efficiently even after inoculation at a low MOI. To identify mutations responsible for cell culture adaptation, we determined the sequences of the entire open reading frame of the viral genome and found one or two nonsynonymous mutations in each genome (Fig. 3E). Interestingly, all of these viral genomes contained amino acid mutations in the NS4A region: W1664S in 5.J6/N5BX-JFH1, A1676T in 7.J6/SRX-JFH1, A1680E in 9.J6/5BSLX-JFH1, and T1681S in 11.J6/5BVR-JFH1. These data suggest the possible contribution of the NS4A mutations to the efficient replication of these clones in cultured cells.
Effects of NS4A mutations on the capacity for viral replication and infectious virus production.
To assess the capacity of replication enhancement due to the mutations identified in NS4A, each mutation, W1664S, A1676T, A1680E, or T1681S, was introduced into the subgenomic replicon and the full-length construct of 9.J6/5BSLX-JFH1 that exhibited no detectable replication in either form. In the replicon assay, all four NS4A mutations enhanced the replication of 9.J6/5BSLX-JFH1 but did so with various efficiencies (Fig. 4A). Among the mutations, W1664S and A1680E enhanced the replication of the 9.J6/5BSLX-JFH1 replicon more efficiently than the other mutations. In the full-length-construct assay, the HCV core Ag and infectivity levels of J6/5BSLX-JFH1+A1680E were the highest at 3 days posttransfection at 2.0 × 104 fmol/liter and 1.8 × 104 FFU/ml, respectively. These levels did not equal those obtained with 10.J6/N3H+5BSLX-JFH1, but the levels were comparable (Fig. 4B and C). From these data, we found that the A1680E mutation is the most effective among all identified NS4A mutations.
FIG 4.
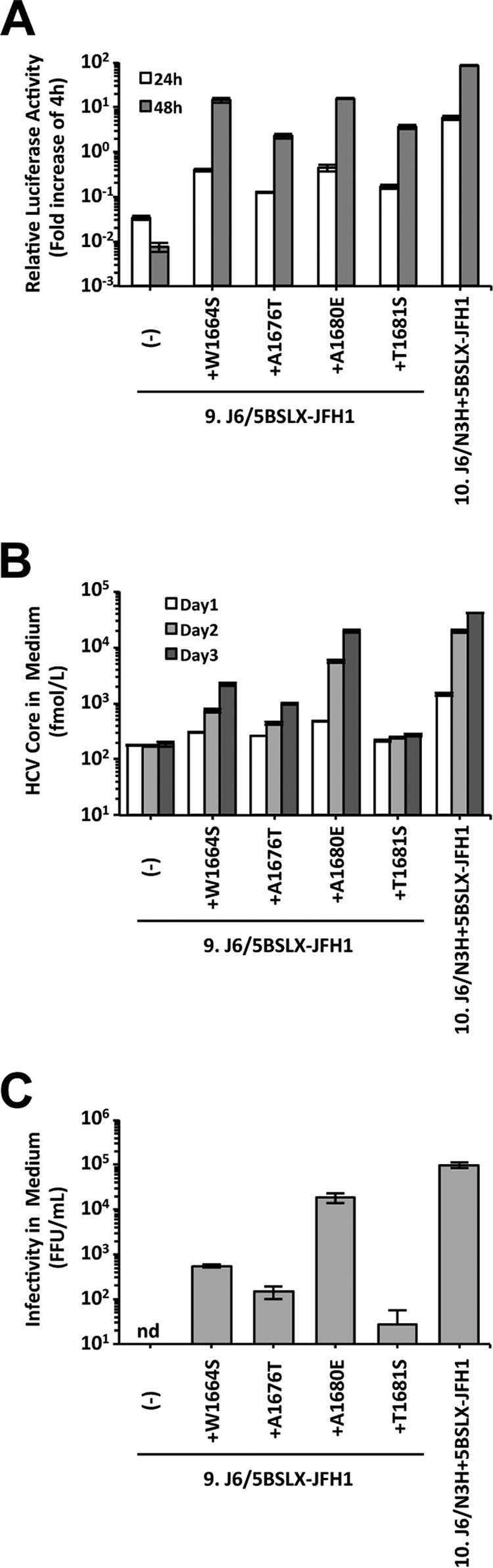
Effects of mutations in NS4A on replication and virus production capacity. (A) Replication capacity of J6/5BSLX-JFH1 subgenomic replicons with NS4A mutations. In vitro-synthesized HCV RNA was transfected into Huh-7.5.1 cells, and the cells were harvested at 4, 24, and 48 h posttransfection. The luciferase activity in the cell lysates was measured and expressed as the fold increase from the values at 4 h posttransfection to correct for transfection efficiency. Independent assays were performed in triplicate, and the data are presented as the mean number of relative light units (RLU) ± standard deviation at 24 h and 48 h posttransfection. (B) HCV core Ag levels in the culture media of full-length HCV RNA-transfected cells. In vitro-synthesized RNA was transfected into Huh-7.5.1 cells. The culture media were harvested at 1, 2, and 3 days posttransfection. The amounts of HCV core Ag in the culture media were measured. Independent assays were performed in triplicate, and the data are presented as the mean ± standard deviation. (C) Infectivity of the culture media of HCV RNA-transfected cells. Culture medium was collected at 72 h posttransfection and serially diluted to infect naive Huh-7.5.1 cells. At 3 days after infection, the cells were fixed, and the infected foci were visualized by staining with anti-core Ag antibody. Infectivity was quantified by counting the infected foci, and the results are expressed as the number of focus-forming units per milliliter. nd, not detected.
Effects of NS4A mutations on subcellular localization of NS3 and NS4A proteins.
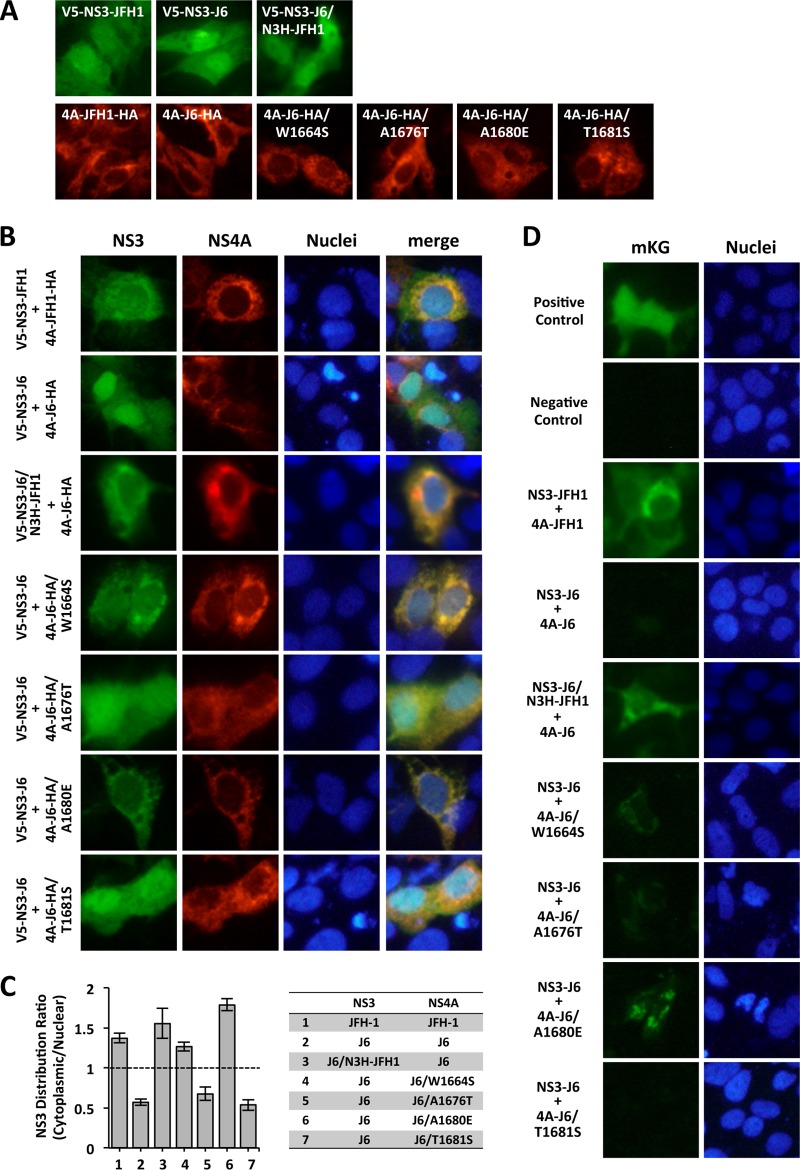
To investigate the mechanisms of replication enhancement from introduction of the identified mutations, we examined the effects of the mutations on the subcellular localization of the NS3 and NS4A proteins in cultured cells. When NS3 was expressed alone, NS3 of JFH-1 (NS3-JFH1), NS3 of J6CF (NS3-J6), and NS3 of J6CF containing JFH-1 N3H (NS3-J6/N3H-JFH1) had similar protein distributions: a diffuse distribution with concentration in the nucleus (Fig. 5A, top). When NS4A was expressed alone, NS4A of JFH-1 (4A-JFH1), NS4A of J6CF (4A-J6), and NS4A with mutations (4A-J6/W1664S, 4A-J6/A1676T, 4A-J6/A1680E, or 4A-J6/T1681S) showed a punctate distribution of protein in the cytoplasm. These observations indicate that the HCV strain (JFH-1 or J6CF) or the NS4A mutations did not affect the distribution of the NS3 or NS4A protein (Fig. 5A, bottom).
FIG 5.
Effects of mutations in NS4A on the subcellular localization of the NS3 and NS4A proteins. (A, B) V5-tagged NS3 and/or HA-tagged NS4A expression plasmids were transfected into Huh-7.5.1 cells. On the following day, the cells were fixed and permeabilized. V5-tagged proteins were visualized by staining with anti-V5 antibody, and HA-tagged proteins were visualized by staining with anti-HA antibody. (C) Ratios of the NS3 distribution (cytoplasmic/nuclear). The average intensities of the NS3 protein in the cytoplasmic and nuclear areas were quantified with ImageJ software, and the ratios were calculated. (D) BiFC assay of NS3 and NS4A. The NS3 expression plasmid with the mKG-N fragment (phmKGN/NS3-JFH1, phmKGN/NS3-J6, or phmKGN/NS3-J6/N3H-JFH1) and the NS4A expression plasmid with the mKG-C fragment (phmKGC/4A-JFH1, phmKGC/4A-J6, phmKGC/4A-J6/W1664S, phmKGC/4A-J6/A1676T, phmKGC/4A-J6/A1680E, or phmKGC/4A-J6/T1681S) were cotransfected into Huh-7.5.1 cells. On the following day, the cells were fixed and examined using a fluorescence microscope. Positive control, mKG-fused p50 and p65 partial domains from NF-κB complex; negative control, empty vectors.
When NS3-JFH1 was coexpressed with 4A-JFH1, the distribution of NS3-JFH1 was in the cytoplasm, although the distribution of 4A-JFH1 was not changed (Fig. 5B). When NS3-J6 was coexpressed with 4A-J6, the distributions of NS3-J6 and 4A-J6 were not altered and were similar to those when these proteins were individually expressed. When NS3-J6/N3H-JFH1 was coexpressed with 4A-J6, the distribution of NS3-J6/N3H-JFH1 was in the cytoplasm, as was the case with the coexpression of NS3-JFH1 and 4A-JFH1. When NS3-J6 was coexpressed with 4A-J6 containing mutations, the distribution of NS3 was altered at different levels. When it was coexpressed with 4A-J6/W1664S or 4A-J6/A1680E, NS3-J6 was mainly distributed in the cytoplasm, as was the case with the coexpression of NS3-JFH1 and 4A-JFH1. However, when it was coexpressed with 4A-J6/A1676T or 4A-J6/T1681S, NS3-J6 exhibited a diffuse distribution with a concentration in the nucleus, as was the case with the coexpression of NS3-J6 and 4A-J6 without mutations. The distributions of NS4A with mutations were similar to those of these molecules expressed individually. Quantitative analysis of the NS3 distribution clearly indicated that NS3-J6 is predominantly distributed in the cytoplasm when it is coexpressed with 4A-J6/W1664S and 4A-J6/A1680E, and these ratios were comparable to or beyond those achieved by coexpression with NS3-JFH1 and 4A-JFH1 or NS3-J6/N3H-JFH1 and 4A-J6 (Fig. 5C). The NS3 distribution ratios of coexpressed NS3-J6 and 4A-J6/A1676T or 4A-J6/T1681S were similar to the ratio of the coexpression of NS3-J6 and 4A-J6 without mutations. These results indicate that the expression of 4A-JFH1 altered the subcellular localization of NS3-JFH1 but the expression of 4A-J6 did not alter the subcellular localization of NS3-J6. Among the adaptive mutations in NS4A, W1664S and A1680E could alter the NS3-J6 distribution from being within the nucleus to being in the cell cytoplasm.
To test if these altered distributions of NS3 were associated with the interaction between NS3 and NS4A, we performed the bimolecular fluorescence complementation (BiFC) assay using NS3 fused to the monomeric Kusabira green (mKG)-N fragment and NS4A fused to the mKG-C fragment. The BiFC assay allows the direct visualization of protein interactions. When NS3-JFH1 was coexpressed with 4A-JFH1, an mKG fluorescent signal was observed (Fig. 5D). When NS3-J6 was coexpressed with 4A-J6, an mKG fluorescent signal was not observed. When NS3-J6/N3H-JFH1 was coexpressed with 4A-J6, an mKG fluorescent signal was also observed. When NS3-J6 was coexpressed with 4A-J6/W1664S and 4A-J6/A1680E, intermediate and strong mKG fluorescent signals were observed, respectively. When NS3-J6 was coexpressed with 4A-J6/A1676T, a faint mKG fluorescent signal was detected. When NS3-J6 was coexpressed with 4A-J6/T1681S, no mKG fluorescent signal was detected. These observations indicate that the A1680E mutation is the most effective for promoting the interaction between NS3-J6 and 4A-J6.
Combined effects of the most effective NS4A mutation and mutations in the N5BX region on J6CF replication.
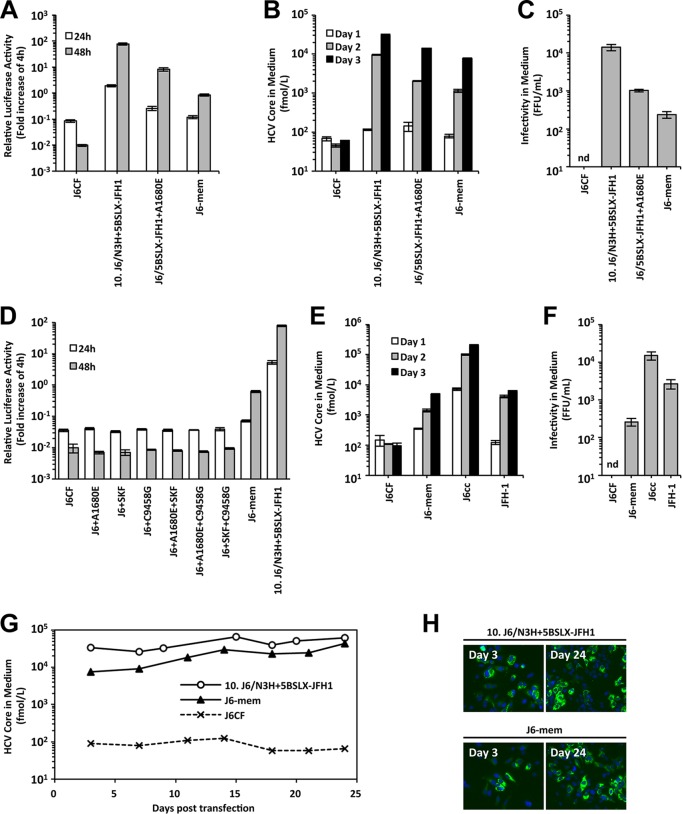
Finally, we tried to establish replication-competent J6CF by introducing the minimum modifications. We previously found that three amino acid mutations, A2892S, R2959K, and Y3003F (SKF), in the NS5B region and a nucleotide mutation, C9458G, in VR are essential modifications for the sufficient replication of J6CF containing N3H of JFH-1 (22). The amino acid mutations of SKF enhance HCV polymerase activity, and the A9348U (in the Y3003F codon) and C9458G mutations are important for optimal RNA structures essential for RNA replication. As a next step, instead of replacing N3H of JFH-1, we introduced the most effective NS4A mutation, A1680E, into the J6CF subgenomic replicon and full-length constructs harboring SKF and C9458G, and we named the construct J6-mem (J6 with minimum essential modifications). In the subgenomic replicon assay, J6-mem replication was observed, but its replication level did not reach the levels of 10.J6/N3H+5BSLX-JFH1 and J6/5BSLX-JFH1+A1680E (Fig. 6A). In the full-length HCV RNA transfection assay, the HCV core Ag levels in the culture media of cells transfected with J6-mem RNA increased in a time-dependent manner (Fig. 6B), and infectivity in the culture medium was also detected (Fig. 6C). However, these titers were lower than those in cells transfected with 10.J6/N3H+5BSLX-JFH1 and J6/5BSLX-JFH1+A1680E RNA. To assess the minimum essential mutations, we tried to detect replication of the J6CF replicon carrying the A1680E, SKF, and C9458G mutations either individually or in combination. However, the J6CF replicon carrying one or two of these mutations failed to replicate (Fig. 6D), indicating that A1680E, SKF, and C9458G were minimum essential modifications for J6CF replication competence in cultured cells. Next, we compared the replication level of J6-mem with that of J6cc, which is a previously reported J6CF variant containing seven modifications, including six amino acid substitutions (F776S, P1100L, F1468L, A1676S, N1931T, and D3001G) and 33 deletions in the poly(U/UC) tract (16). HCV core Ag levels in the culture media of J6-mem-transfected cells were comparable to those in the culture media of JFH-1-transfected cells but lower than those in the culture media of J6cc-transfected cells (Fig. 6E). The infectivity in the culture media of J6-mem-transfected cells was lower than that in the culture media of JFH-1- and J6cc-transfected cells (Fig. 6F). Then, we tested if J6-mem could maintain autonomous replication in cultured cells without a decrease in infectious virus production. J6-mem RNA-transfected cells could continue to secrete HCV core Ag into the culture medium for up to 24 days (Fig. 6G). At 24 days after RNA transfection, most of the cells were HCV positive both for cells transfected with 10.J6/N3H+5BSLX-JFH1 and for cells transfected with J6-mem (Fig. 6H). These results indicate that J6-mem, which contained the J6CF genome containing the minimum essential modifications of A1680E, A2892S, R2959K, Y3003F, and C9458G, could replicate and produce infectious viruses continuously in cultured cells.
FIG 6.
Combined effects of the most effective NS4A mutation and mutations in the N5BX region on J6CF replication. (A) Replication capacity of J6/5BSLX-JFH1 subgenomic replicons with mutations. In vitro-synthesized RNA was transfected into Huh-7.5.1 cells, and the cells were harvested at 4, 24, and 48 h posttransfection. The luciferase activity in the cell lysates was measured and expressed as the fold increase from the values at 4 h posttransfection to correct for transfection efficiency. Independent assays were performed in triplicate, and the data are presented as the mean number of relative light units (RLU) ± standard deviation at 24 h and 48 h after transfection. (B) HCV core Ag levels in the culture media of full-length HCV RNA-transfected cells. In vitro-synthesized RNA was transfected into Huh-7.5.1 cells, and the culture media were harvested at 1, 2, and 3 days posttransfection. The amounts of HCV core Ag in the culture media were measured. Independent assays were performed in triplicate, and the data are presented as the mean ± standard deviation. (C) Infectivity of the culture media of HCV RNA-transfected cells. Culture media of HCV RNA-transfected cells were collected at 72 h posttransfection and were serially diluted to infect naive Huh-7.5.1 cells. Three days after infection, the cells were fixed, and the infected foci were visualized by staining with anti-core Ag antibody. Infectivity was quantified by counting the infected foci, and the results are expressed as the number of focus-forming units per milliliter. nd, not detected. (D) Replication capacity of J6CF subgenomic replicons with modifications introduced individually or in combination. Luciferase activity was measured as described in the legend to panel A. (E, F) Comparisons of HCV core Ag levels (E) and infectivity (F) in the media of J6-mem, J6cc, and JFH-1 RNA-transfected cells. (G) HCV core Ag levels in the culture media of HCV RNA-transfected cells. HCV RNA was transfected into Huh-7.5.1 cells, and the cells were passaged every 2 to 5 days, depending on the conditions of the cells. The culture medium was collected at every passage, and HCV core Ag levels were measured. (H) Ratios of HCV-positive cells at 3 days and 24 days posttransfection. Infected cells were visualized by staining with anti-core Ag antibody (green), and nuclei were visualized with DAPI (4′,6-diamidino-2-phenylindole; blue).
DISCUSSION
The HCV genotype 2a strain JFH-1 is the only HCV strain that replicates efficiently in cultured cells without adaptive mutations. Thus, we aimed to establish replication-competent, genotype 2a J6CF strains by introducing minimum modifications. For this purpose, we constructed a series of J6CF chimeric genomes containing various parts of the JFH-1 genome and obtained J6CF/JFH-1 chimeric constructs with various levels of replication capacity. By using the subgenomic reporter replicon assay, we identified several chimeric constructs that could replicate in cultured cells, but some of the constructs in the full-length-construct form could not replicate following transfection, depending on the region of JFH-1 replaced. We considered that the sensitivity of replication detection might be enhanced in the subgenomic reporter replicon assay compared with the full-length-construct transfection assay because of the length of the constructs (21). Therefore, we used the long-term culture of cells transfected with J6CF/JFH-1 chimeric RNAs and obtained replication-competent cell culture-adapted viruses. This strategy of inducing adaptive mutations has been broadly used, and the established replication-competent HCV strains containing cell culture-adaptive mutations were as follows: TNcc (GT1a) (13), HCV-1cc (GT1a) (14), H77Ccc (GT1a) (14), a NC1 mutant (GT1b) (15), J6cc (GT2a) (16), a JFH-2 mutant (GT2a) (17), J8cc (GT2b) (16), DH8cc (GT2b) (18), DH10cc (GT2b) (18), and an S310 mutant (GT3a) (19).
We previously reported that N3H of JFH-1 is important for the efficient replication of J6CF and that three amino acid mutations, A2892S, R2959K, and Y3003F, and a nucleotide mutation, C9458G, in the N5BX region in lieu of replacement of the entire N5BX region are also important for J6CF replication. The aim of this study was to identify the minimum modifications required for replication in cultured cells. Since the N3H proteins of JFH-1 and J6CF differ by 30 amino acids, we could not narrow down the region or identify the responsible mutations in this area (data not shown). Instead of modifying the N3H region, we successfully identified the adaptive mutations of W1664S, A1676T, A1680E, and T1681S in the NS4A region. Among them, A1680E was the most effective adaptive mutation for efficient replication and infectious virus production of the J6CF/JFH-1 chimera. This mutation emerged individually in the J6/5BSLX-JFH1 genome after long-term culture. The other mutations emerged in combination with mutations in other genomic regions (M324I in E1, S981G in NS2, and N2156D in NS5A). Therefore, A1680E alone may be sufficient to confer replication enhancement, but other mutations, W1664S, A1676T, and T1681S, may require additional adaptive mutations for efficient replication of the J6CF/JFH-1 chimera. We have to note that the W1664S and A1680E mutations enhanced the replication of replicons at similar levels; however, the enhanced levels of infectious virus production obtained by viruses with the W1664S and A1680E mutations were different. Thus, A1680E may be associated with enhancement of both replication and virus production steps. Until now, several cell culture-adapted mutations (W1664C/S and A1676S) in NS4A of genotype 2a strains were reported (16); however, an adaptive mutation at amino acid (aa) 1680 (aa 1676 in the H77 reference strain [GenBank accession number AF009606]) has not yet been described. The residues at aa 1664, aa 1676, aa 1680, and aa 1681 are conserved in J6CF, JFH-1, and the GT2 consensus, and the identified adaptive mutations were not detected in searches of the Hepatitis Virus Database (Table 1). We reasoned that the amino acid change from Ala (hydrophobic) to Glu (acidic) at aa 1680 may affect the characteristics of the NS4A protein or the interaction with other viral proteins.
TABLE 1.
Prevalence of amino acid residues at the positions of mutations identified in NS4A
| J6CF position (aa) | H77 position (aa) | Amino acid |
|||
|---|---|---|---|---|---|
| GT2 consensusa | J6CF | JFH-1 | Adaptive mutation | ||
| 1664 | 1660 | W | W | W | S |
| 1676 | 1672 | A | A | A | T |
| 1680 | 1676 | A | A | A | E |
| 1681 | 1677 | T | T | T | S |
Obtained from a search of the Hepatitis Virus Database (http://s2as02.genes.nig.ac.jp/).
To clarify the molecular mechanisms of the identified adaptive mutations, we assessed the subcellular localization of the NS3 and NS4A proteins. NS4A consists of 54 amino acids, and the hydrophobic N-terminal region anchors the NS3-4A complex to the endoplasmic reticulum and mitochondrial outer membrane (23–26). The central region of NS4A is the viral serine protease cofactor (27), and the C-terminal acidic domain regulates HCV replication (28). A total of 21 amino acids of the NS4A N terminus form a transmembrane α-helix structure that is important for the assembly of the replication complex (29). Several amino acid mutations that deteriorate the replication of subgenomic replicons have been reported in the N-terminal region of NS4A, and some of them were reported to alter the subcellular localization of NS4A itself (29). The NS4A mutations identified in this study are located in the N-terminal region of NS4A; however, the subcellular localization of NS4A itself was not affected by these mutations. Instead, two of these mutations altered the subcellular localization of NS3 protein. NS3-J6 exhibited a diffuse distribution with concentration in the nucleus even when 4A-J6 was coexpressed. However, the expression of 4A-J6/A1680E moved the NS3-J6 distribution to the cytoplasm. This cytoplasmic distribution of NS3-J6 is similar to that of NS3-JFH1 when it is expressed with 4A-JFH1. This altered NS3-J6 distribution was also observed when it was expressed with 4A-J6/W1664S, although not when it was expressed with 4A-J6 containing other mutations. These altered distributions were consistent with the replication enhancement capacity of each adaptive mutation.
Since these identified mutations are located close to the central region of NS4A, which acts as a cofactor, we also assessed the effects of these mutations on the interaction between NS3 and NS4A using a BiFC assay. Although a strong BiFC signal between NS3-JFH1 and NS4A-JFH1 was detected, no BiFC signal between NS3-J6 and NS4A-J6 was detected, indicating that NS4A-J6 could not anchor NS3-J6 to the replication complex. The BiFC signal was detected when NS3-J6 was expressed with NS4A-J6/A1680E, NS4A-J6/A1664S, or NS4A-J6/A1676T. The observed BiFC signal levels were also consistent with the replication enhancement capacity of each adaptive mutation. Thus, the interaction of NS3 and NS4A and the subcellular localization of the NS3 and NS4A complex are essential for replication competence in the cell culture-adapted J6CF strain.
In this study, we constructed a replication-competent J6CF variant, J6-mem, by introducing 5 adaptive mutations, A1680E, A2892S, R2959K, Y3003F, and C9458G (Fig. 7). Among these adaptive mutations, we previously reported that the amino acid mutations A2892S, R2959K, and Y3003F are important for efficient JFH-1 polymerase activity and nucleotide mutations of U9348A in the Y3003F codon and C9458G are responsible for the optimal RNA structure. In this study, we also showed that A1680E contributes to the cytoplasmic localization of NS3. We tested whether each of these three modifications (high polymerase activity, optimal RNA structure, and cytoplasmic localization of NS3) were necessary for J6CF replication in cultured cells and demonstrated that all of them are essential for J6CF replication competence. Using mutational analyses, we could identify the minimum mutations essential for the replication competence of J6CF and uncover molecular mechanisms involved in each mutation. When the replication of J6-mem was compared with that of another replication-competent J6CF variant, J6cc, which has 6 amino acid substitutions (F776S, P1100L, F1468L, A1676S, N1931T and D3001G) and 33 deletions in the poly(U/UC) tract (Fig. 7) and replicates efficiently in cultured cells (16), we found that the replication level of J6-mem was far reduced from that of J6cc. However, the molecular mechanisms involved in each adaptive mutation in J6cc have not yet been determined since J6cc contains 33 deletions in the poly(U/UC) tract, in addition to the other mutations. As we previously reported, a longer (J6CF-type) poly(U/UC) region was advantageous to HCV replication, whereas a shorter J6CF poly(U/UC) region was not (22). Thus, the introduction of a shorter poly(U/UC) region may enhance the replication efficiency of J6-mem. The mutations introduced in J6-mem were minimum and essential for J6CF replication competence in cultured cells because the deletion of any of these mutations deteriorated replication.
FIG 7.
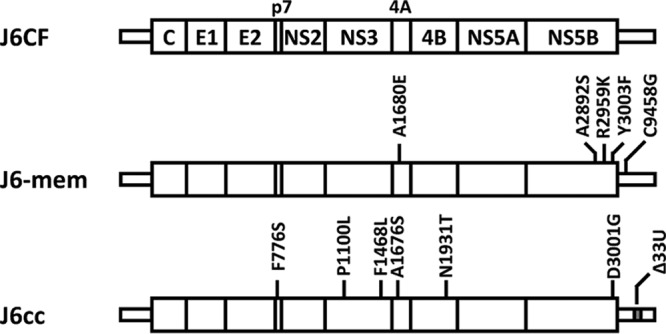
Summary of mutations in J6-mem and J6cc. J6-mem contains A1680E (NS4A), A2892S (NS5B), R2959K (NS5B), Y3003F (NS5B), and C9458G (3′ UTR). J6cc contains F776S (p7), P1100L (NS3), F1468L (NS3), A1676S (NS4A), N1931T (NS4B), and D3001G (NS5B) and 33 U deletions (3′ UTR).
JFH-1 can replicate efficiently and produce infectious viruses in cultured cells. However, in our previous study, the inoculation of a chimpanzee with cell culture-generated JFH-1 virus induced only transient and mild infection, even though JFH-1 was isolated from a patient with fulminant hepatitis (10). In contrast, J6CF and H77C (GT1a) could replicate efficiently in chimpanzees and persistently infected the animals after intrahepatic transfections with each of the viral genomes (5, 7), yet J6CF and H77C could not replicate in cultured cells (21). The inverse relationship between the in vivo and in vitro properties of cell culture-adaptive mutations has also been reported for the Con1 strain (genotype 1b) (8). Because most of the HCV strains infectious in vivo could not replicate in cultured cells without modification, efficient HCV replication in HuH-7 cells may be specific for JFH-1, and the HCV life cycle observed for the JFH-1 strain in HuH-7 cells may be different from that in vivo. HuH-7 cells may lack some factors essential for HCV replication, and JFH-1 or other cell culture-adaptive strains may be able to replicate under such deficient conditions. SEC14L2 has been reported to be an example of such host factors (30). The expression level of SEC14L2 is high in human adult hepatocytes but low in HuH-7 and its derived cells. Ectopic expression of SEC14L2 enhances the replication of HCV strains with a low replication efficiency but does not enhance the replication of JFH-1 in cultured cells. Still, the HCV subgenomic replicon and cell culture system could contribute to developing direct-acting antiviral agents with activity against HCV (31–33). This HCV cell culture system involving various HCV strains of different genotypes can be used to further elucidate the mechanisms of HCV-host interactions or to accurately evaluate the genotype- or strain-specific anti-HCV effects of existing and newly developed drugs. We are confident that our clone, J6-mem, will contribute to these types of studies as another representative of genotype 2a strains.
In conclusion, we identified effective adaptive mutations in the NS4A region that contribute to J6CF replication competence and established a replication-competent J6CF strain with minimum essential modifications in cultured cells.
MATERIALS AND METHODS
Cell culture.
Cells of the Huh-7.5.1 cell line, derived from HuH-7 hepatoma cells and cured of HCV infection, were a kind gift from Francis V. Chisari (Scripps Research Institute, La Jolla, CA) (34). The cells were cultured at 37°C in a 5% CO2 environment using Dulbecco's modified Eagle's medium containing 10% fetal bovine serum.
Plasmid construction.
Schematic structures of the subgenomic replicons and full-length constructs are illustrated in Fig. 1A and 2A, respectively. The HCV full-length constructs of pJ6CF and pJ6cc were kindly provided by Jens Bukh (Copenhagen University Hospital, Hvidovre, Denmark). The reporter subgenomic replicon constructs containing firefly luciferase, pSGR-JFH1, and pSGR-J6CF were previously described (21). The J6CF replicon constructs with JFH-1 N5BX (5.pSGR-J6/N5BX-JFH1), with JFH-1 N3H (4.pSGR-J6/N3H-JFH1), and with both JFH-1 N5BX and N3H (6.pSGR-J6/N3H+N5BX-JFH1), as well as the corresponding full-length constructs (5.pJ6/N5BX-JFH1, 4.pJ6/N3H-JFH1, and 6.pJ6/N3H+N5BX-JFH1), were previously reported (21). Briefly, several sites recognizing the restriction enzymes ClaI (nucleotide [nt] 3929), EcoT22I (nt 5293), and BsrGI (nt 7781) were introduced into the J6CF sequence via nucleotide substitution. The J6CF replicon constructs containing the latter part of JFH-1 N5BX with or without JFH-1 N3H (7.pSGR-J6/SRX-JFH1 and 8.pSGR-J6/N3H+SRX-JFH1) and the corresponding full-length constructs (7.pJ6/SRX-JFH1 and 8.pJ6/N3H+SRX-JFH1) were produced by fragment swapping by digestion with SrfI (nt 8843) and XbaI (nt 9711). This fragment contains previously identified JFH-1-type amino acids that are important for replication (A2892S, R2959K, and Y3003F) and the entire 3′ untranslated region (UTR) of JFH-1. The J6CF replicon constructs containing the JFH-1 region of predicted stem-loop structures in NS5B (5BSL)-to-3′X with or without N3H (9.pSGR-J6/5BSLX-JFH1 and 10.pSGR-J6/N3H+5BSLX-JFH1) and the corresponding full-length constructs (9.pJ6/5BSLX-JFH1 and 10.pJ6/N3H+5BSLX-JFH1) were produced by fusion PCR and fragment swapping (22). This fragment contains the stem-loop structure in the NS5B region and the entire 3′ UTR of JFH-1. The J6CF replicon constructs containing JFH-1 NS5B and the variable region (VR) in the 3′ UTR with or without JFH-1 N3H (11.pSGR-J6/5BVR-JFH1 and 12.pSGR-J6/N3H+5BVR-JFH1) and the corresponding full-length constructs (11.pJ6/5BVR-JFH1 and 12.pJ6/N3H+5BVR-JFH1) were also produced by fusion PCR and fragment swapping. This fragment contains the entire NS5B region and VR of JFH-1. The J6CF replicon constructs containing JFH-1 NS5B with or without JFH-1 N3H (13.pSGR-J6/N5B-JFH1 and 14.pSGR-J6/N3H+N5B-JFH1) and the corresponding full-length constructs (13.pJ6/N5B-JFH1 and 14.pJ6/N3H+N5B-JFH1) were constructed by fragment swapping by digestion with BsrGI (nt 7781) and StuI (nt 9415). This fragment contains the entire NS5B region of JFH-1. The nucleotide positions of the replaced regions in each chimeric construct are listed in Table 2. The W1664S, A1676T, A1680E, and T1681S point mutations were introduced by site-directed mutagenesis PCR.
TABLE 2.
Nucleotide positions of replaced regions in J6CF chimeric constructs
| Construct | Position |
|---|---|
| N3H region | nt 3863–nt 5323 |
| 4.J6/N3H-JFH1 | ClaIa (nt 3929)–EcoT22Ia (nt 5293) |
| N5BX region | nt 7667–nt 9711 |
| 5.J6/N5BX-JFH1 | BsrGIa (nt 7781)–XbaI (3′ end) |
| 7.J6/SRX-JFH1 | SrfI (nt 8843)–XbaI (3′ end) |
| 9.J6/5BSLX-JFH1 | nt 9211–XbaI (3′ end) |
| 11.J6/5BVR-JFH1 | BsrGIa (nt 7781)–nt 9481 |
| 13.J6/N5B-JFH1 | BsrGIa (nt 7781)–StuI (nt 9415) |
Artificially introduced into the J6CF sequence.
To construct expression vectors for the localization assay, we cloned a V5-tagged NS3 fragment and a hemagglutinin (HA)-tagged NS4A fragment of JFH-1 into the pEF1/Myc-His A vector (Invitrogen, Carlsbad, CA) to generate pEF/V5-NS3-JFH1 and pEF/4A-JFH1-HA, respectively. Likewise, we also generated the expression vectors pEF/V5-NS3-J6, pEF/4A-J6-HA, and pEF/4A-J6-HA containing the W1664S, A1676T, A1680E, or T1681S mutation. To construct expression vectors for the BiFC assay, the cDNA fragments of NS3 and NS4A were fused to the N or C terminus of the divided monomeric Kusabira green (mKG) fragments of the phmKGN-MC and phmKGC-MN vectors, respectively (CoralHue Fluo-chase kit; Medical & Biological Laboratories, Nagoya, Japan) (35), and phmKGN/NS3-JFH1, phmKGN/NS3-J6, phmKGN/NS3-J6/N3H-JFH1, phmKGC/4A-JFH1, phmKGC/4A-J6, and phmKGC/4A-J6 with the W1664S, A1676T, A1680E, or T1681S mutation were generated.
RNA synthesis and transfection and determination of infectivity.
RNA synthesis and transfection were performed as previously described (22, 36). The determination of infectivity was also performed as previously described, with infectivity being expressed as the number of focus-forming units (FFU) per milliliter.
Luciferase reporter assay.
Luciferase activity in the lysates of cells transfected with subgenomic reporter replicon RNA was measured as previously described (22, 36).
Quantification of HCV core Ag.
The concentration of HCV core antigen (Ag) in the culture media and in the cell lysate was measured using a chemiluminescent enzyme immunoassay (CLEIA; Lumipulse Ortho HCV antigen; Fujirebio, Tokyo, Japan) in accordance with the manufacturer's instructions (37).
HCV sequencing.
Total RNA in the culture supernatant was extracted using an RNeasy RNA minikit (Qiagen, Valencia, CA). cDNA was synthesized with random primers (TaKaRa Bio, Shiga, Japan) using SuperScript III reverse transcriptase (Invitrogen). cDNA was subsequently amplified with LA Taq DNA polymerase (TaKaRa Bio). Four separate PCR primer sets were used to amplify the fragments from nt 130 to 2445, nt 2285 to 4717, nt 4607 to 7220, and nt 6881 to 9634, covering the entire open reading frame and parts of the 5′ UTR and 3′ UTR of the HCV genome. The sequences of the amplified fragments were determined directly.
Immunostaining.
Immunostaining of infected cells was performed as previously described (38). For the subcellular localization analysis, 1 μg of the V5-tagged NS3 and/or HA-tagged NS4A expression plasmid was transfected into 3 × 105 Huh-7.5.1 cells using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. On the following day, the cells were fixed with 4% paraformaldehyde and then permeabilized. After blocking, the V5-tagged proteins were visualized by staining with anti-V5 antibody (Invitrogen) and Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen), and the HA-tagged proteins were also visualized by staining with anti-HA antibody (Roche, Mannheim, Germany) and Alexa Fluor 555-conjugated goat anti-rat IgG (Invitrogen). We measured the mean fluorescence intensity represented by the mean gray value within randomly selected areas in the nucleus and cytoplasm using ImageJ software (https://imagej.nih.gov), and the NS3 distribution ratio was calculated by dividing the mean intensity of the cytoplasm by that of the nucleus.
BiFC assay.
Huh-7.5.1 cells were cultured on glass coverslips in a 12-well plate at a concentration of 2 × 105 cells/well. One microgram of NS3 expression plasmid (phmKGN/NS3-JFH1, phmKGN/NS3-J6, or phmKGN/NS3-J6/N3H-JFH1) and 1 μg of NS4A expression plasmid (phmKGC/4A-JFH1, phmKGC/4A-J6, or phmKGC/4A-J6 with the W1664S, A1676T, A1680E, or T1681S mutation) were cotransfected into the cells using the Lipofectamine 2000 reagent (Invitrogen). On the following day, the cells were fixed with 4% paraformaldehyde and examined using a fluorescence microscope.
Statistical analysis.
Significant differences were evaluated using Student's t test. A P value of <0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Francis V. Chisari (Scripps Research Institute, La Jolla, CA) for providing the Huh-7.5.1 cell line and Jens Bukh (Copenhagen University Hospital, Hvidovre, Denmark) for providing the pJ6CF and pJ6cc plasmids.
A.M. was supported by grants from JSPS KAKENHI (23790513) and the Ministry of Health, Labor and Welfare of Japan (H22-kanen-wakate-016). T.K. was supported by grants for scientific research from the Ministry of Health, Labor and Welfare of Japan (H23-seisakutansaku-ippan-002) and for research programs on the development of new drugs (15ak0101002h0005) and hepatitis (15fk0310011h0104) from the Japan Agency for Medical Research and Development, AMED.
The funders had no role in study design, data collection and interpretation or the decision to submit the work for publication.
REFERENCES
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, Furuta S, Akahane Y, Nishioka K, Purcell RH, Alter HJ. 1990. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology 12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 3.Feld JJ, Liang TJ. 2006. Hepatitis C—identifying patients with progressive liver injury. Hepatology 43:S194–S206. doi: 10.1002/hep.21065. [DOI] [PubMed] [Google Scholar]
- 4.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med 132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 5.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A 94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanford RE, Lee H, Chavez D, Guerra B, Brasky KM. 2001. Infectious cDNA clone of the hepatitis C virus genotype 1 prototype sequence. J Gen Virol 82:1291–1297. doi: 10.1099/0022-1317-82-6-1291. [DOI] [PubMed] [Google Scholar]
- 7.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 8.Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St Claire M, Bartenschlager R. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci U S A 99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 10.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Kato T, Choi Y, Elmowalid G, Sapp RK, Barth H, Furusaka A, Mishiro S, Wakita T, Krawczynski K, Liang TJ. 2008. Hepatitis C virus JFH-1 strain infection in chimpanzees is associated with low pathogenicity and emergence of an adaptive mutation. Hepatology 48:732–740. doi: 10.1002/hep.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YP, Ramirez S, Jensen SB, Purcell RH, Gottwein JM, Bukh J. 2012. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc Natl Acad Sci U S A 109:19757–19762. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li YP, Ramirez S, Mikkelsen L, Bukh J. 2015. Efficient infectious cell culture systems of the hepatitis C virus (HCV) prototype strains HCV-1 and H77. J Virol 89:811–823. doi: 10.1128/JVI.02877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Date T, Morikawa K, Tanaka Y, Tanaka-Kaneko K, Sata T, Mizokami M, Wakita T. 2012. Replication and infectivity of a novel genotype 1b hepatitis C virus clone. Microbiol Immunol 56:308–317. doi: 10.1111/j.1348-0421.2012.00437.x. [DOI] [PubMed] [Google Scholar]
- 16.Li YP, Ramirez S, Gottwein JM, Scheel TK, Mikkelsen L, Purcell RH, Bukh J. 2012. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc Natl Acad Sci U S A 109:E1101–E1110. doi: 10.1073/pnas.1203829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Date T, Kato T, Kato J, Takahashi H, Morikawa K, Akazawa D, Murayama A, Tanaka-Kaneko K, Sata T, Tanaka Y, Mizokami M, Wakita T. 2012. Novel cell culture-adapted genotype 2a hepatitis C virus infectious clone. J Virol 86:10805–10820. doi: 10.1128/JVI.07235-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez S, Li YP, Jensen SB, Pedersen J, Gottwein JM, Bukh J. 2014. Highly efficient infectious cell culture of three hepatitis C virus genotype 2b strains and sensitivity to lead protease, nonstructural protein 5A, and polymerase inhibitors. Hepatology 59:395–407. doi: 10.1002/hep.26660. [DOI] [PubMed] [Google Scholar]
- 19.Kim S, Date T, Yokokawa H, Kono T, Aizaki H, Maurel P, Gondeau C, Wakita T. 2014. Development of hepatitis C virus genotype 3a cell culture system. Hepatology 60:1838–1850. doi: 10.1002/hep.27197. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto H, Okada S, Sugiyama Y, Kurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M. 1991. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol 72(Pt 11):2697–2704. [DOI] [PubMed] [Google Scholar]
- 21.Murayama A, Date T, Morikawa K, Akazawa D, Miyamoto M, Kaga M, Ishii K, Suzuki T, Kato T, Mizokami M, Wakita T. 2007. The NS3 helicase and NS5B-to-3′X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells. J Virol 81:8030–8040. doi: 10.1128/JVI.02088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murayama A, Weng L, Date T, Akazawa D, Tian X, Suzuki T, Kato T, Tanaka Y, Mizokami M, Wakita T, Toyoda T. 2010. RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells. PLoS Pathog 6:e1000885. doi: 10.1371/journal.ppat.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mottola G, Cardinali G, Ceccacci A, Trozzi C, Bartholomew L, Torrisi MR, Pedrazzini E, Bonatti S, Migliaccio G. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31–43. doi: 10.1006/viro.2001.1229. [DOI] [PubMed] [Google Scholar]
- 24.Nomura-Takigawa Y, Nagano-Fujii M, Deng L, Kitazawa S, Ishido S, Sada K, Hotta H. 2006. Non-structural protein 4A of hepatitis C virus accumulates on mitochondria and renders the cells prone to undergoing mitochondria-mediated apoptosis. J Gen Virol 87:1935–1945. doi: 10.1099/vir.0.81701-0. [DOI] [PubMed] [Google Scholar]
- 25.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol 69:1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolk B, Sansonno D, Krausslich HG, Dammacco F, Rice CM, Blum HE, Moradpour D. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol 74:2293–2304. doi: 10.1128/JVI.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin C, Thomson JA, Rice CM. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J Virol 69:4373–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach BD, Pragai BM, Montserret R, Beran RK, Pyle AM, Penin F, Rice CM. 2007. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J Virol 81:8905–8918. doi: 10.1128/JVI.00937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brass V, Berke JM, Montserret R, Blum HE, Penin F, Moradpour D. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc Natl Acad Sci U S A 105:14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saeed M, Andreo U, Chung HY, Espiritu C, Branch AD, Silva JM, Rice CM. 2015. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature 524:471–475. doi: 10.1038/nature14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemm JA, O'Boyle D II, Liu M, Nower PT, Colonno R, Deshpande MS, Snyder LB, Martin SW, St Laurent DR, Serrano-Wu MH, Romine JL, Meanwell NA, Gao M. 2010. Identification of hepatitis C virus NS5A inhibitors. J Virol 84:482–491. doi: 10.1128/JVI.01360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O'Boyle DR II, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. 2010. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem 53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 34.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki R, Matsuda M, Watashi K, Aizaki H, Matsuura Y, Wakita T, Suzuki T. 2013. Signal peptidase complex subunit 1 participates in the assembly of hepatitis C virus through an interaction with E2 and NS2. PLoS Pathog 9:e1003589. doi: 10.1371/journal.ppat.1003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato T, Date T, Murayama A, Morikawa K, Akazawa D, Wakita T. 2006. Cell culture and infection system for hepatitis C virus. Nat Protoc 1:2334–2339. doi: 10.1038/nprot.2006.395. [DOI] [PubMed] [Google Scholar]
- 37.Murayama A, Sugiyama N, Watashi K, Masaki T, Suzuki R, Aizaki H, Mizuochi T, Wakita T, Kato T. 2012. Japanese reference panel of blood specimens for evaluation of hepatitis C virus RNA and core antigen quantitative assays. J Clin Microbiol 50:1943–1949. doi: 10.1128/JCM.00487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murayama A, Sugiyama N, Wakita T, Kato T. 2016. Completion of the entire hepatitis C virus life cycle in Vero cells derived from monkey kidney. mBio 7:e00273. doi: 10.1128/mBio.00273-16. [DOI] [PMC free article] [PubMed] [Google Scholar]