ABSTRACT
The envelope glycoproteins (Envs) from human immunodeficiency virus type 1 (HIV-1) mediate viral entry. The binding of the HIV-1 gp120 glycoprotein to CD4 triggers conformational changes in gp120 that allow high-affinity binding to its coreceptors. In contrast to all other Envs from the same phylogenetic group, M, which possess a serine (S) at position 375, those from CRF01_AE strains possess a histidine (H) at this location. This residue is part of the Phe43 cavity, where residue 43 of CD4 (a phenylalanine) engages with gp120. Here we evaluated the functional consequences of replacing this residue in two CRF01_AE Envs (CM244 and 92TH023) by a serine. We observed that reversion of amino acid 375 to a serine (H375S) resulted in a loss of functionality of both CRF01_AE Envs as measured by a dramatic loss in infectivity and ability to mediate cell-to-cell fusion. While no effects on processing or trimer stability of these variants were observed, decreased functionality could be linked to a major defect in CD4 binding induced by the replacement of H375 by a serine. Importantly, mutations of residues 61 (layer 1), 105 and 108 (layer 2), and 474 to 476 (layer 3) of the CRF01_AE gp120 inner domain layers to the consensus residues present in group M restored CD4 binding and wild-type levels of infectivity and cell-to-cell fusion. These results suggest a functional coevolution between the Phe43 cavity and the gp120 inner domain layers. Altogether, our observations describe the functional importance of amino acid 375H in CRF01_AE envelopes.
IMPORTANCE A highly conserved serine located at position 375 in group M is replaced by a histidine in CRF01_AE Envs. Here we show that H375 is required for efficient CRF01_AE Env binding to CD4. Moreover, this work suggests that specific residues of the gp120 inner domain layers have coevolved with H375 in order to maintain its ability to mediate viral entry.
KEYWORDS: HIV-1, CRF01_AE, Env, gp120, Phe43 cavity, CD4, inner domain layers
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) has a very high genetic variability owing to the error-prone reverse transcriptase (lacking a proofreading function), the many rounds of replication allowing a high viral turnover rate, and significant immune pressure from the host during the chronic infection (1–6). The vast genetic variability of HIV-1 resulted into its classification into 4 major groups, M, N, O, and P. The HIV-1 group M is responsible for the majority of cases in the global pandemic. It comprises 9 major subtypes (A1, A2, B, C, D, F1, F2, G, H, J, and K) and circulating recombinant forms (CRFs) (7–11). These recombinant forms initially result from multiple infections within the same individual, which favors recombination between two different genomic RNA copies upon coinfection of the same cell and production of heterozygous virions (12); CRFs are recombinants that are transmitted and repeatedly sampled (13–15). The utilization of the two distinct alternating templates by the reverse transcriptase allows the generation of a chimeric DNA molecule and in major instances results in the creation of a new HIV-1 intersubtype recombinant virus (13, 16, 17).
Southern Asia is particularly affected by the CRFs. CRF01 was originally established in central Africa, where it had undergone diversification by the time the Asian CRF01 epidemic began in the late 1980s in Thailand (14, 15). During the past decade, an increasing prevalence of the CRF01_AE subtype was observed, and it accounted for 42.5% of all national infections in China in 2007 (18) and 95% of those in Thailand in 2004 (19). CRF01_AE has a mosaic genome composed of the gag-pol region, most of the accessory genes from clade A, and the env gene from clade E (20). No parental clade E strain matching the CRF01_AE Env has been identified (20). Altering or exchanging fragments of genes encoding crucial proteins such as the envelope can have a sizeable impact on viral fitness (21, 22), virulence, and ability to evade the host immune system, thus providing evolutionary advantages to the virus (8).
Host cell entry is the first step of the HIV-1 replication cycle and requires the mature viral envelope glycoproteins, which result from the proteolytic cleavage of gp160 viral protein into exterior gp120 and transmembrane gp41 subunits. These subunits are linked by noncovalent links allowing conformational changes of the trimer during the entry process (23–25). The gp120 exterior subunit is important for the initial interaction with the CD4 receptor. The Phe43 cavity, located at the interface of the inner and outer domains of gp120, allows the engagement to CD4 via its Phe43 residue (26). Then, rearrangement of the variable regions V1 and V2 exposes the binding site for the coreceptors (CCR5 and CXCR4) (27). A major conformational change occurs in gp41, where a six-helix bundle structure, composed of three N-terminal heptad repeat (HR1) and three C-terminal heptad repeat (HR2) regions, brings together the viral envelope and the target cell membrane (28, 29).
It has been shown that some of these conformational rearrangements could be impacted by large alterations in the Phe43 cavity. Replacement of the well-conserved group M serine at position 375 by a large hydrophobic residue such as tryptophan alters Env conformation by predisposing gp120 to spontaneously assume a state closer to the CD4-bound conformation (30). In agreement with an important role played by Env conformation on CD4 interaction (23, 30–32), a recent study using the simian-human immunodeficiency virus (SHIV) model in rhesus macaques showed that replacing residue 375 by larger hydrophobic or basic amino acids enhanced Env affinity to rhesus macaque CD4 (rhCD4) and allowed a better entry into rhCD4 lymphocyte T cells, in vivo (33), highlighting the importance of this residue and its context for viral replication.
In addition to the Phe43 cavity, other gp120 elements play a role in CD4 interaction and Env conformation. For example, it has been described that three topological layers located in the inner domain of gp120 affect CD4 interaction and subsequent conformational changes. These three layers (layer 1, layer 2, and layer 3) of the inner domain secure CD4 binding by helping to expose the CD4-binding site (layer 3) (31, 34) and then by keeping CD4 in place by decreasing the off-rate (layers 1 and 2) (23). It has been suggested that the gp120 inner domain layers have evolved to accommodate for changes occurring in the Phe43 cavity (32, 34).
Despite high genetic diversity, some functional glycoprotein domains in HIV-1 envelope have been preserved over time, such as the Phe43 cavity. However, CRF01_AE Envs appear to have evolved differently from other subtypes. Their envelope glycoproteins possess a histidine at position 375. Here, we investigate the functional relevance of 375H residue in the Phe43 cavity of CRF01_AE Envs and its interplay with the inner domain layers.
RESULTS
Comparison of residues at position 375 in the Phe43 cavity among HIV-1 strains.
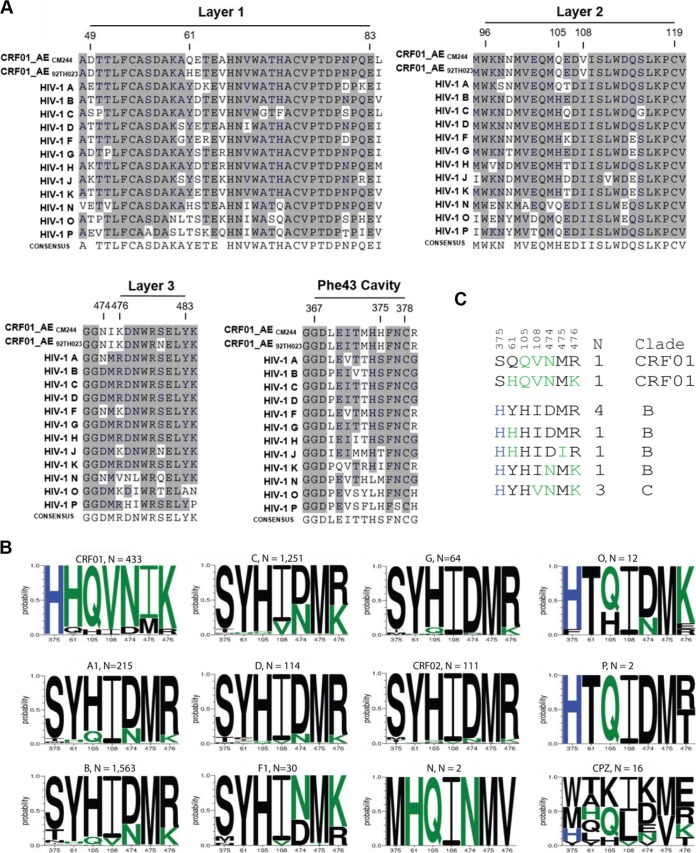
The sequence comparison of CRF01_AE strains 92TH023 and CM244 to all the other HIV-1 types highlights the differences in the amino acids at position 375 in the Phe43 cavity (Fig. 1A). CRF01_AE sequences have a conserved histidine at this position, whereas the other clades have a serine (Fig. 1B). Since it was previously suggested that the inner domain layers have coevolved with the Phe43 cavity to accommodate the residue located at position 375 (32), we investigated whether residues located in the gp120 inner domain layers were enriched in sequences with residue H375 and by extension in CRF01_AE relative to other HIV-1 clades. Visual inspection of an initial alignment of Env protein sequences suggested differences between CRF01_AE and the rest of group M strains at positions 61 (layer 1), 105 and 108 (layer 2), and 474 to 476 (layer 3) (Fig. 1A). This was further supported by a more extensive exploration of distinctive subtype variation in these sites, by determining the frequency of amino acid variants at each of these positions independently (Fig. 1B), based on the Los Alamos database curated Env alignment, which contained one sequence per person; the 2016 database alignment contained 4,632 sequences, 433 of which were from CRF01_AE. It is interesting that HIV groups O and P share this histidine at position (H375 (Fig. 1) and group N has a unique form with a methionine in this position (M375). Each of these groups arose from separate zoonosis events, from chimpanzee or gorilla into humans (35), and position 375 is highly variable among SIVcpz viruses isolated from chimpanzees (Fig. 1B). These groups share some of the amino acids that are enriched in CRF01 along with H375, in particular Q105, highlighting its likely importance (Fig. 1C).
FIG 1.
Phe43 cavity and inner-domain layer HIV-1 Env sequences. (A) Primary sequence alignment of gp120 residues form the Phe43 cavity, layer 1, layer 2, andlayer 3 from a single sequence from each clade: HIV-1 A (accession number ABB29387.1), HIV-1 B (accession number K03455), HIV-1 C (AAB36507.1), HIV-1 D (P04581.1), HIV-1 F (ACR27173.1), HIV-1 G (ACO91925.1), HIV-1 H (AAF18394.1), HIV-1 J (ABR20452.1), HIV-1 K (CAB59009.1), HIV-1 N (AAT08775.1), HIV-1 O (AAA99883.1), HIV-1 P (GQ328744), and two studied viruses, CRF01_AE CM244 (AY713425) and CRF01_AE 92TH023. Residue numbering is based on that of the HXBc2 strain of HIV-1. The shading highlights residues that are similar to those of CRF01_AE from the other subtype from group M. (B) A Logo depiction of the frequency of each amino acid in the positions that we originally noted by eye, based on the data shown in panel A, to be correlated with 375 H and 375 S/T. The height of the letter indicates its frequency. CRF01_AE has >99% 375 H, and the amino acids that are shown to be enriched among CRF01_AE sequences, therefore also associated with 375 H, are shown in green. Black indicates amino acids that are favored in other clades or are rare variants. Logos for sequences from the following subtypes and groups are shown: HIV-1 M group major subtypes A1, B, C, D, F1, and G; M group CRFs 01 and 02; HIV-1 groups N, O, and P; and Envs isolated from chimpanzees (CPZ). Subtype and groups are noted above each Logo, followed by their count. The 2016 Los Alamos database curated Env alignment was used as the basis for this figure, which contains 4,632 HIV-1 and related SIVcpz sequences. (C) Alignment of the positions of interest in the inner domain, ordered as described for panel B. The two instances of an H375S being found in CRF01_AE are indicated, and the 10 naturally occurring sequences from other M group major subtypes with the S375H mutation are also shown. As in panel B, CRF01_AE common amino acids are shown in green, the positions are indicated above the alignment, and the number of times the pattern was found (N) and the clade in which the variant arose are noted on the right.
The histidine at position 375 (H375) was found in greater than 99% of the 433 CRF01 sequences. The other inner domain amino acids were more than 75% conserved within CRF01_AE. We found two H375S mutations in the context of the CRF01_AE lineage in the HIV-1 database, as well as a small number of S375H substitutions in other M groups, clades, and subtypes. When H375S naturally arose in CRF01, it tended to be accompanied by some of the inner domain substitutions commonly found in the M group, and when S375H arose naturally in other clades, it was generally accompanied by some of the inner domain amino acids commonly found in CRF01, indicative of coevolution. These occurrences suggest that distinct mutations in the inner domain preferentially accompany the shifts at position 375 but that not all changes are simultaneously required in all contexts.
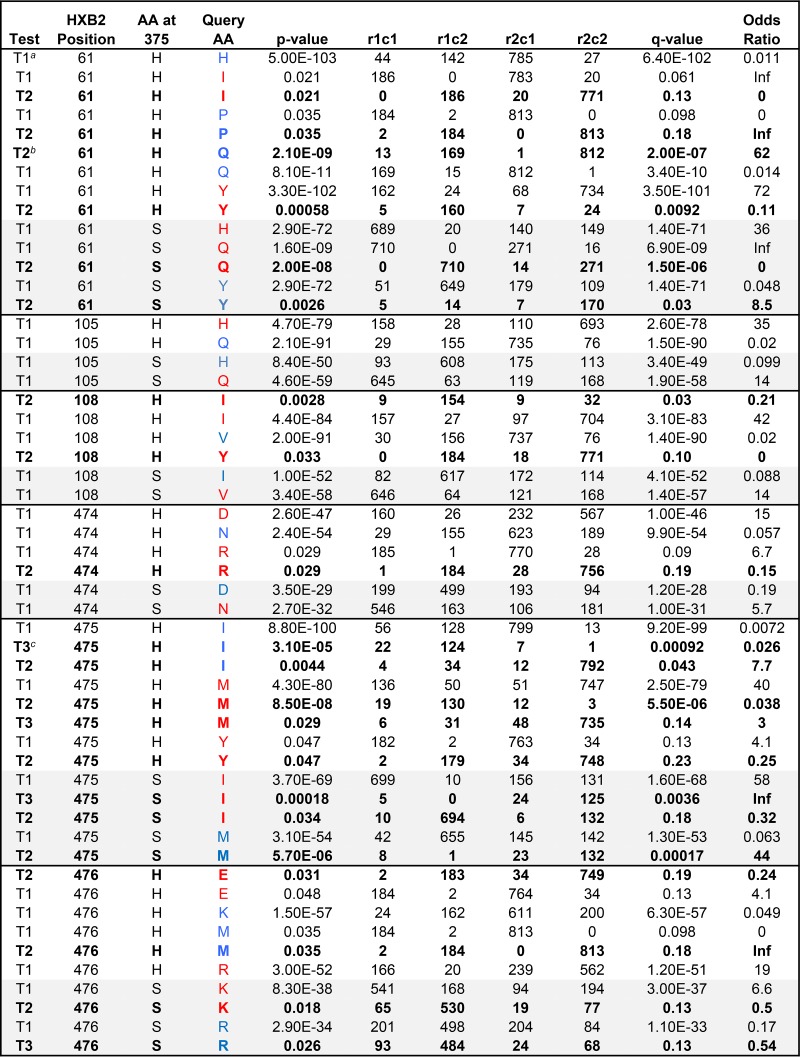
To more explicitly explore the associations between position 375 and the other sites that we identified to be of interest within the inner domain, we statistically characterized the frequency of association between amino acid variants in position 375 and the other positions. A simple Fisher's test yields very highly significant associations with the sites of interest that we identified in the inner domain (Table 1), as expected given the profound associations with well-sampled clades illustrated in Fig. 1B. In such cases, founder effects and functional constraints are difficult to disentangle, and hence, it is not possible to resolve whether or not the associations observed are due to direct functional constraints or whether both amino acids are simply associated with the CRF01_AE lineage. Thus, we also used phylogenetic strategies to explore whether particular mutational events were more like to occur in the context of either H or S at position 375 (Table 1). All but one (residue 105) of the sites of interest had significant phylogenetically supported associations, and some of the less common amino acids at each position were implicated as well as the consensus forms of the M group and CRF01 (Table 1). Of note, residue 105 was not associated toward the polymorphism more commonly found with residue 375 (H or S) in CRF01_AE, thus highlighting the limitation of visual inspection in identifying phylogenetic associations.
TABLE 1.
Summary of amino acids in positions within the inner domain layers that covary with amino acids found in Env position 375d
The test “T1” is just a simple Fisher's exact test using a 2 × 2 contingency table with no phylogenetic correction (see footnote d below). It cannot resolve whether correlations between amino acids in position 375 and other sites are due to functional dependence or founder effects.
To determine whether or not a given association could have resulted from a phylogenetic artifact due to a founder effect, we also did a phylogenetically corrected association analysis. For the H375-H61 T1 association, the H61 and H375 are both very common in CRF01_AE, and so this could be just a founder, or lineage-based, association. In this case, the association was not supported by a phylogenetically corrected analysis, thus leaving the possibility that there may be a functional constraint or that the association may simply result from CRF01_AE being enriched for both H375 and H61. On the other hand, several other amino acids at position 61 were statistically robust even after a phylogenetic correction, strongly supporting a covariation pattern between the two sites. To do the phylogenetic correction, maximum likelihood was used to estimate the probability of a given amino acid at each ancestral node in a phylogenetic tree. The most probable amino acid in a given position in the last node prior to a leaf is considered to be the most likely ancestral amino acid in that position, given the tree and the evolutionary model. This approach is described in detail in reference 59. Changes to and from a given ancestral state in the query position, given the amino acid at 375, were tallied and then used as a basis for Fisher's exact test. An example of a “T2” corrected table can be found in row 6; T2 tests when the ancestral state is not the query amino acid. In this case, position 61 is much more likely to change to Q if there is an H at 375 than if there is not (P = 2.1−09).
The third kind of comparison we evaluated is test T3; it determines, when the query amino acid is present in the ancestral state, the frequency with which it mutates away from that state associated with the amino at 375. The first example of a T3 test is I475 (raw 32), in association with H375. In this case, I475 remains I 124/125 times when H is present at 375, while I475 mutates away from I much more frequently when an amino acid other than H is found at 375.
For position 375, starting with either H or not-H (or S or not-S), we tallied the number of times the position 375 amino acid is present in association with each amino acid found in each other position in the alignment; thousands of tests were done. The signature analysis identified amino acids that covaried with amino acids at position 375: either H375, the common amino acid in CRF01, or S375, the most common amino acid at position 375 in HIV-1 sequences from all other subtypes. We prioritized Env positions within the inner domain. T1 is a simple Fisher's test that tests for correlations between amino acids in position 375 and amino acids in other positions. It cannot resolve whether correlations are due to functional dependence or founder effects. Therefore, a phylogenetically corrected signature analysis (59, 60) was also used for mutational events that were enriched in the sites of interest in the context of either H375 or S375 (tests T2 and T3, shown in boldface). These tests evaluate whether amino acids tend either to mutate or to stay the same in the context of either H375 or S375, and these tests correct for overt clade effects. Query amino acids (AA) are blue if they are enriched in the context of the given AA at 375 and red if they are negatively associated. r1c1, r1c2, r2c1, and r2c2 are the rows (r) and columns (c), i.e., the elements in a 2 × 2 contingency table; for example, in the first row, an H in position 375 is found 142 times in conjunction with an H at position 61 (r1c2) and 44 times with other amino acids at position 61 (r1c1), and H375 is significantly enriched for an H in position 61. P values are 2-sided Fisher's exact test based on this table. q values, or false-positive rates, were calculated as described in references 59 and 60) to address multiple-test issues, and only associations with q values of <0.20 are included here. Comparisons with H375 are white, and those with S375 are lightly shaded. All amino acids in all positions in the Env alignment were evaluated for associations with the amino acids at position 375, H375 and S375. The q values to address multiple tests were calculated based on comparisons of each amino acid in all Env positions, and associations with a q value of <0.20 were deemed of interest.
Effects of gp120 Phe43 cavity changes on processing and subunit association of CRF01_AE 92TH023 and CRF01_AE CM244 envelope glycoproteins.
To evaluate the role of H375 on CRF01_AE Env function, we replaced histidine 375 by a serine (H375S), which is the most common form in the remaining group M. These mutations were introduced into CRF01_AE 92TH023 and CM244 Envs. Site-directed mutagenesis was also used to modify the CRF01_AE Env residues 61, 105, 108, and 474 to 476; these mutations (H61Y, Q105H, V108I, NIK474-476DMR) correspond to the most prevalent amino acids that are found in non-CRF01_AE subtypes and are further referred to as the layer mutations (LM).
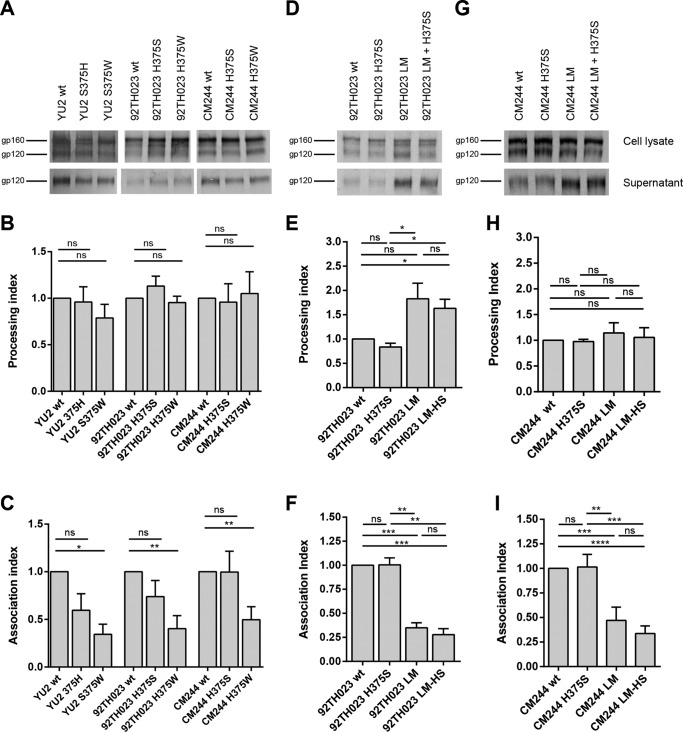
We then evaluated the effects of these substitutions on processing and stability of the Env trimer. All mutants were efficiently expressed (Fig. 2A to D), and the introduction of a larger amino acid (histidine [H] or tryptophan [W]) in the Phe43 cavity did not affect the processing of Yu2 Env (clade B) or 92TH023 and CM244 (clade CRF01_AE) Envs relative to the corresponding wild-type Env (Fig. 2B). Similar levels of gp120 in the cell supernatant and the cell lysate were also observed in mutants harboring H or S at position 375 (Fig. 2A), suggesting that these mutations do not impact the association between the two subunits (gp41 and gp120). Furthermore, a loss of association of the Yu2, 92TH023, and CM244 Env subunits was observed for 375W mutants (Fig. 2C). We also evaluated the effects of the LM in the presence of a serine or histidine at position 375. Introduction of the LM described above increased the processing of Envs independently of the residue present at position 375 (Fig. 2E) and decreased by almost 75% the association between the gp120 and the gp41 subunits (Fig. 2F).
FIG 2.
Effects of position 375 changes on processing and subunit association of CRF01_AE 92TH023 and CM244 Envs. Cell lysates and supernatants (SN) of 35S-labeled cells transiently expressing the HIV-1Yu2, HIV-1CRF01_AE 92TH023, or HIV-1CM244 wild-type and the indicated mutant envelope glycoproteins were precipitated with HIV+ sera. The precipitated proteins were loaded onto polyacrylamide gels (A, D, and G) and analyzed by autoradiography and densitometry (B, C, E, F, H, I). Processing index (B, E and H) and association index (C, F, and I) values of each envelope, calculated as described in Materials and Methods. Data represent the averages ± standard errors of the means (SEM) from 3 independent experiments. Statistical significance was tested using unpaired t test (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Effects of gp120 Phe43 cavity changes on CRF01_AE 92TH023 and CRF01_AE CM244 envelope glycoprotein function.
We then evaluated the role of H375 and LM on Env function. The entry phenotype was assessed by measuring the relative infectivity and cell-to-cell fusion ability of the variants described above. Briefly, relative infectivity was assessed by incubating Cf2Th-CD4/CCR5 cells with normalized amounts of recombinant HIV-1 virions pseudotyped with wild-type and mutant 92TH023 or CM244 Env variants. Cell-to-cell fusion was assessed by coincubation of 293T cells expressing the envelope glycoprotein variants with the reporter TZM-bl cells for 6 h at 37°C as described in Materials and Methods.
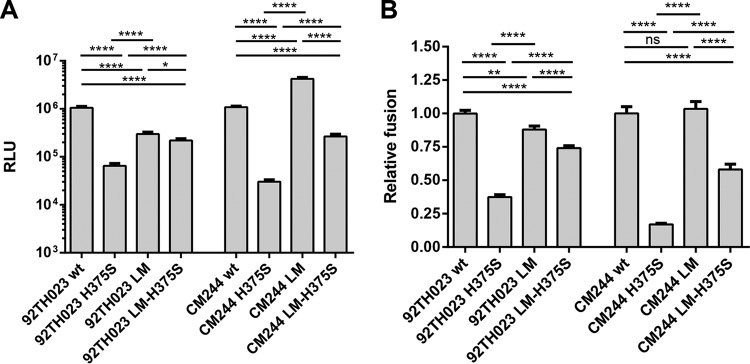
The introduction of a serine at position 375 in CR01_AE Envs significantly decreased the infectivity of HIV-1 viral particles (Fig. 3A) and presented lower ability to mediate cell-to-cell fusion (Fig. 3B). Interestingly, introduction of the LM variants partially restored the ability of H375S to mediate viral entry and cell-to-cell fusion (Fig. 3 and Table 2), suggesting that the LM compensate for the H375S phenotype. Of note, we noticed that LM variants, on their own, enhanced viral entry and cell-to-cell fusion for CM244 but not 92TH023 Env, suggesting that some structural differences between these two strains exist.
FIG 3.
Infectivity and cell-to cell fusion of CRF01_AE variants. (A) Relative infectivity was assessed by incubating Cf2Th-CD4/CCR5 cells with recombinant HIV-1 pseudotyped with wild-type and mutant CRF01_AE envelope glycoproteins, normalized according to reverse transcriptase. Data shown represent the means ± SEM obtained from three independent experiments. (B) Cell-cell fusion ability was assessed by coincubation of 293T cells expressing the envelope glycoprotein variants with the reporter TZM-bl cells for 6 h at 37°C. The activities of the mutant envelope glycoproteins were normalized to that of the wild-type (wt) envelope glycoproteins. The results shown represent the means ± SEM obtained from four independent experiments. Statistical significance was tested using unpaired t test (ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001).
TABLE 2.
Phenotypes of CRF01_AE gp120 variantsa
| CRF01_AE gp 120 variant | Amino acid or mutation at position 375b | Processing indexc | Association indexd | Relative infectivitye | Cell-to-cell fusion abilityf | CD4-Igg |
|---|---|---|---|---|---|---|
| CRF01_AE 92TH023 | 375H (wt) | 1h | 1h | 1 ± 0.06590 | 1 ± 0.02257 | 1h |
| H375S | 0.8369 ± 0.07743 | 1.004 ± 0.07331 | 0.0671 ± 0.007230 | 0.3739 ± 0.0181 | 0.1478 ± 0.06188 | |
| H375W | 0.9534 ± 0.06886 | 0.4035 ± 0.1356 | 0.8202 ± 0.1556 | 1.514 ± 0.06388 | ND | |
| LM | 1.828 ± 0.3201 | 0.3493 ± 0.05255 | 0.2824 ± 0.02690 | 0.8793 ±0.02617 | 1.039 ± 0.2101 | |
| LM-375S | 1.632 ± 0.1865 | 0.2786 ± 0.06254 | 0.2090 ± 0.01721 | 0.7412 ± 0.01638 | 0.6248 ± 0.1039 | |
| CRF01_AE CM244 | 375H (wt) | 1h | 1h | 1 ± 0.05814 | 1 ± 0.05007 | |
| H375S | 0.9751 ± 0.04242 | 1.014 ± 0.06434 | 0.02800 ± 0.002716 | 0.1619 ± 0.00874 | ||
| H375W | 1.051 ± 0.2340 | 0.4974 ± 0.1355 | 0.8026 ± 0.2523 | 0.8326 ± 0.05030 | ||
| LM | 1.144 ± 0.1963 | 0.4709 ± 0.06737 | 3.876 ± 0.2968 | 1.1534 ± 0.05501 | ||
| LM-375S | 1.057 ± 0.1890 | 0.3363 ± 0.03885 | 0.2464 ± 0.02595 | 0.6263 ± 0.03909 | ||
| YU2 | 375S (wt) | 1h | 1h | 1 ± 0.0558 | 1 ± 0.01773 | |
| S375H | 0.9599 ± 0.1636 | 0.5956 ± 0.1735 | 1.205 ± 0.3382 | 0.9529 ± 0.06498 | ||
| S375W | 0.7885 ± 0.1457 | 0.3427 ± 0.1062 | 0.9868 ± 0.3191 | 0.9605 ± 0.02366 |
Values represent the means ± SEM of results from at least three independent experiments.
The numbering of the CRF01_AE and Yu2 gp120 envelope glycoprotein amino acid residues is based on that of the prototypic HXBc2 strain of HIV-1, where 1 is the initial methionine (54). The mutations result in the replacement of the amino acid residue shown on the left by the amino acid residue shown on the right of the number; for example, H375S indicates a substitution of a serine residue for the histidine residue at position 375. LM represents the combination of the following mutations: H61Y, Q105H, V108I, and NIK474-476DMR.
The processing index is a measure of the conversion of the mutant gp160 envelope glycoprotein precursor to mature gp120 relative to that of the wild-type envelope glycoproteins. The processing index was calculated by the following formula: processing index = ([total gp120]mt × [gp160]wt)/([gp160]mt × [total gp120]wt), where mt is mutant and wt is wild type.
The association index is a measure of the ability of the mutant gp120 molecule to remain associated with the envelope glycoprotein complex on the expressing cell relative to that of the wild-type envelope glycoproteins. The association index is calculated as follows: association index = ([mt gp120]cell × [wt gp120]supernatant)/([mt gp120]supernatant × [wt gp120]cell), where mt is mutant and wt is wild type.
Relative infectivity was assessed by infecting Cf2Th-CD4/CCR5 cells with similar amounts of recombinant HIV-1 pseudotyped with wild-type and mutant CRF01_AE envelope glycoproteins, normalized according to reverse transcriptase (see Materials and Methods). The ratio of mutant/wild-type virus infectivity is reported.
Cell-cell fusion ability was assessed by coincubation for 6 h at 37°C of 293T cells expressing the 92TH023 or CM244 envelope glycoprotein variants with the reporter TZM-bl cells, as described in Materials and Methods.
Normalized amounts of radiolabeled wild-type and mutant gp120 glycoproteins were incubated with 13 nM CD4-Ig for 1 h at 37°C. The immunoprecipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. ND, not determined.
For association/processing indexes and CD4-Ig immunoprecipitation, no SEM are reported for wt, since values were normalized to 1 for the wt.
Ability of mutant envelope glycoproteins to interact with CD4 and susceptibility to soluble CD4 (sCD4) neutralization.
Since residue 375 is known to modulate Env affinity for CD4 (23, 30, 33), we first evaluated the ability of 92TH023 LM and 375 mutants to interact with CD4 both by immunoprecipitation and by surface plasmon resonance (SPR). For immunoprecipitation analysis, normalized amounts of radiolabeled wild-type and mutant gp120 glycoproteins were incubated with human CD4-Ig. The precipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. For SPR analysis, purified gp120 variants were evaluated using a sensor chip where CD4-Ig was immobilized.
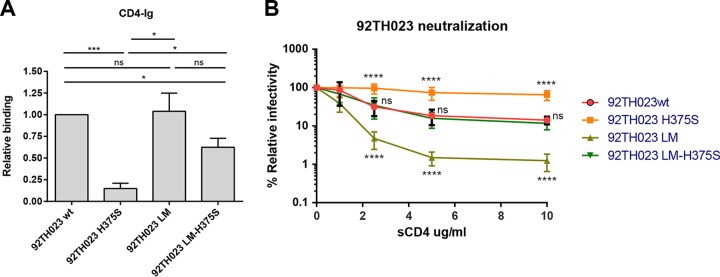
Interestingly, replacing H375 by a serine dramatically decreased the affinity for CD4 (Fig. 4A). Introduction of the LM alone did not significantly affect CD4 binding compared to the wild-type Env but partially rescued CD4 binding for the H375S variant. A detailed analysis of the interaction by SPR indicated that the decreased recognition by the H375S variant was due to an enhanced off-rate compared to that of the wild-type protein (Table 3). Interestingly, introduction of the LM variants restored CD4 binding of the H375S Env by decreasing the off-rate (Table 3). This is reminiscent of the role played by gp120 inner domain layer 1 residues (such as H66 and W69), which were previously shown to maintain CD4 binding by decreasing the off-rate, despite being located far from the CD4-binding site (23).
FIG 4.
CD4 interaction and sCD4 neutralization of CRF01_AE variants. (A) Normalized amounts of radiolabeled wild-type and mutant gp120 glycoproteins were immunoprecipitated with CD4-Ig as described in Materials and Methods for 1 h at 37°C. The precipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. Fold increase in the binding of gp120 variants to ligands was normalized to wt. Data shown represent the means ± SEM from four independent experiments. Statistical significance was tested using the unpaired t test (ns, not significant; *, P < 0.05; ***, P < 0.001). (B) Normalized amounts of recombinant HIV-1 virions expressing luciferase and bearing different Env mutations were incubated with serial dilutions of sCD4 for 1 h at 37°C before infecting Cf2Th-CD4/CCR5 cells. Luciferase activity in cell lysates was measured to determine the infectivity. Infectivity was normalized to 100% in the absence of the ligand. Data shown are representative of results from at least three independent experiments, performed in quadruplicate. ns, not significant; ****, P < 0.0001.
TABLE 3.
Ability of HIV-1 CRF01_AE 92TH023 gp120 variants to interact with CD4-Ig by surface plasmon resonancea
| CRF01_AE 92TH023 gp120 variant | On-rate constant (M−1 s−1) | Off-rate constant (s−1) | KD (M) (fold change) |
|---|---|---|---|
| wt | 3.34E+03 | 9.76E−04 | 2.92E−07 (1.00) |
| H375S | 2.71E+03 | 1.89E−03 | 6.95E−07 (2.38) |
| LM | 1.10E+04 | 1.49E−04 | 1.35−08 (0.05) |
| LM + H375S | 8.59E+03 | 3.22E−04 | 3.74E−08 (0.13) |
CD4-Ig was immobilized directly onto a CM5 sensor chip, and the binding of the indicated gp120 protein was evaluated as described in Materials and Methods. KD, equilibrium dissociation constant; wt, wild type; LM, inner-domain mutants H61Y, Q105H, V108I, and NIK474-476DMR.
As expected from decreased affinity for CD4 (Fig. 4A), viruses bearing the H375S variant were resistant to sCD4 neutralization (Fig. 4B and C). While introduction of the LM alone increased sCD4 sensitivity, introduction in the context of the H375S variant restored wild-type levels of sCD4 neutralization. Thus, these results indicate that in the context of CRF01_AE Envs, sCD4 neutralization correlates with Env affinity for CD4.
DISCUSSION
Since the first emergence of CRF01_AE in Africa and its subsequent spread to Asia, where it established a major epidemic, these viruses have become the dominant strains in many Asian countries, including Southern China and Thailand, where they have the highest prevalence (18, 36, 37). Why this lineage of viruses was able to spread so rapidly is unclear at the moment; it did not spread particularly rapidly in Africa (14). Higher viral loads and shorter survival times than in people infected in western countries have been associated with CRF01_AE infections (38–40).
Interestingly, we noticed that from all group M subtypes present in the NIH Los Alamos HIV database, only CRF01_AE has a highly conserved histidine at position 375. Since it has been previously shown that replacement of serine 375 by a histidine modified the Env conformation toward a state closer to the CD4-bound conformation (30), we evaluated whether histidine 375 affected Env function in the CRF01_AE Env context.
Besides the Phe43 cavity, additional gp120 elements have been shown to play a role in the transition to the CD4-bound conformation (23, 41–45). Interestingly, potential coevolution between certain gp120 inner domain layer residues and the Phe43 cavity has been suggested (32, 34). Therefore, we evaluated whether additional residues of the gp120 inner domain might be associated with H375. Comparisons of the CRF01_AE Env sequences to sequences from other subtypes allowed the identification of six residues of the gp120 inner domain layers (residues 61, 105, 108, and 474 to 476) that were conserved in CRF01_AE but not in other subtypes; these were likely candidates to compensate for the presence of a large residue at position 375 (H375) in CRF01_AE. Replacement of residue 375 by a tryptophan but not a histidine decreased the association between gp120 and gp41 both in the clade B Yu2 and in CRF01_AE Envs; however, no effect on Env processing was observed for all the variants tested. Replacement of H375 by a serine did not result in a significant decrease of Env stability or processing.
Mutating the six residues that were initially identified by eye but then confirmed to be important by phylogenetically corrected signature analysis in 5/6 cases in the inner domain layers had a significant impact on the association and processing of the Env, and this was independent of the amino acid (His or Ser) at position 375. Inner domain layer variants enhanced the processing of the Env and decreased the association of gp120 and gp41. CRF01_AE Envs had a better overall stability without the layer mutations. Our results indicate that the Phe43 cavity and the inner domain layers modulate the stability of CRF01_AE Envs. Of note, none of the individual inner domain layer mutations present in the LM variant restored the phenotype of H375S on their own, indicating that the combination of all mutations is required to compensate for the Phe43 cavity change (data not shown).
It is well known that HIV-1 Env is a metastable and highly flexible protein (46). Therefore, functional Env trimers need to maintain a balance between the envelope stability, the magnitude of the conformational changes, and binding affinity to CD4. In fact, premature triggering of the Env complex to downstream conformations results in functional inactivation (47), the exposure of nonneutralizing epitopes (48), and rapidly decaying intermediate states (47, 48). We observed that the capacity of CRF01_AE Envs to interact with CD4 depends on the presence of a histidine at position 375. Interestingly, the poor recognition of CD4 by CRF01_AE Envs with the H375S mutation could be compensated by restoring the nature of 6 layer residues conserved in the majority of non-CRF01_AE M group strains. Decreased CD4 interaction correlated with a lack of Env function as measured by infectivity and the ability to mediate cell-to-cell fusion. Moreover, decreased CD4 interaction explained a dramatic loss in sCD4 sensitivity of the H375S mutant Envs, which was restored upon introduction of these inner domain mutations. Our results suggest that CRF01_AE strains have evolved to secure CD4 binding with the presence of a bigger amino acid in the Phe43 cavity and that additional changes in the inner domain layers coevolved to compensate for the presence of H375.
Unlike the majority of HIV-1 Envs, some SIV groups have larger residues than a serine at Env position 375 (methionine, histidine, tyrosine, tryptophan). In several cases, the strains with these larger amino acids successfully made the transition from primates into humans (the P, N, and O groups). Since the presence of larger amino acids at 375 is known to enhance the affinity of Env for CD4 (23, 30, 33), it is possible that their presence in simian immunodeficiency virus (SIV) strains is required to infect cells that express lower levels of CD4 on their surface, as has been shown for rhesus macaques macrophages (49–51). However, why a histidine was selected for in subtype CRF01_AE at position 375 for a virus infecting human cells is not clear. Additional work is needed to assess the evolutionary pressures that resulted in the fixation of a histidine at position 375 in this rapidly spreading HIV-1 subtype.
MATERIALS AND METHODS
Cell lines.
293T human embryonic kidney, Cf2Th canine thymocytes (American Type Culture Collection), and TZM-bl cell lines (NIH AIDS Research and Reference Reagent Program) were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Sigma) and 100 μg/ml of penicillin-streptomycin (Mediatech). Cf2Th cells stably expressing human CD4 and CCR5 (Cf2Th-CD4-CCR5) (52) were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen) and 0.15 mg/ml of hygromycin B (Roche Diagnostics). The TZM-bl cell line is a HeLa cell line stably expressing high levels of CD4 and CCR5 and possessing an integrated copy of the luciferase gene under the control of the HIV-1 long terminal repeat (53).
Site-directed mutagenesis.
Mutations were introduced individually or in combination into the pcDNA3.1 vector (Invitrogen) expressing the CRF01_AE 92TH023 and CRF01_AE CM244 envelope glycoproteins. Site-directed mutagenesis was performed using the QuikChange II XL site-directed mutagenesis protocol (Stratagene). The presence of the desired mutations was determined by automated DNA sequencing. The numbering of the CRF01_AE 92TH023 and CRF01_AE CM244 envelope glycoprotein amino acid residues is based on that of the prototypic HXBc2 strain of HIV-1, where 1 is the initial methionine (54).
Immunoprecipitation of envelope glycoproteins.
For pulse-labeling experiments, 3 × 105 293T cells were transfected with pcDNA3.1-HIV-1 Yu2, CRF01-AE 92TH023, or CM244 gp160 variants using a standard calcium phosphate method. One day after transfection, cells were metabolically labeled for 16 h with 100 μCi/ml [35S]methionine-cysteine (35S protein labeling mix; Perkin-Elmer) in Dulbecco's modified Eagle's medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. Cells were subsequently lysed in radioimmunoprecipitation assay (RIPA) buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP-40, and 0.05% sodium dodecyl sulfate [SDS]). Precipitation of radiolabeled Yu2, 92TH023, and CM244 envelope glycoproteins from cell lysates or medium was performed with a mixture of sera from HIV-infected individuals.
The association index is a measure of the ability of the mutant gp120 molecule to remain associated with the Env trimer complex on the expressing cell relative to that of the wild-type Env trimer. The association index is calculated as follows: association index = ([mt gp120]cell × [wt gp120]supernatant)/([mt gp120]supernatant × [wt gp120]cell), where mt is mutant and wt is wild type. The processing index is a measure of the conversion of the mutant gp160 Env precursor to mature gp120 relative to that of the wild-type Env trimer. The processing index was calculated by the following formula: processing index = ([total gp120]mt × [gp160]wt)/([gp160]mt × [total gp120]wt), where mt is mutant and wt is wild type.
Alternatively, medium containing radiolabeled gp120 variants was immunoprecipitated with CD4-Ig for 1 h at 37°C in the presence of 50 μl of 10% protein A-Sepharose (General Electric). All samples were loaded onto polyacrylamide gels and analyzed by autoradiography using PhosphorImager (Molecular Dynamics).
Recombinant luciferase viruses.
Recombinant viruses containing the firefly luciferase gene were produced by calcium phosphate transfection of 293T cells with the HIV-1 proviral vector pNL4.3 Env-Luc and the plasmid expressing the wild-type or mutant Yu2, 92TH023, or CM244 envelope glycoproteins at a ratio of 2:1. Two days after transfection, the cell supernatants were harvested and centrifuged for 5 min at 3,000 rpm to pellet cell debris. The reverse transcriptase activity was measured for all viral preparations as described previously (55). The virus-containing supernatants were stored in aliquots at −80°C.
Cell-to-cell fusion.
To assess cell-to-cell fusion, 3 × 105 293T cells were cotransfected by the calcium phosphate method with an HIV-1 Tat-expressing plasmid, pLTR-Tat, and the Yu2, 92TH023, or CM244 envelope glycoproteins in an expression vector. Two days after transfection, 3 × 104 293T cells were added to TZM-bl target cells that were seeded at a density of 3 × 104 cells/well in 96-well luminometer-compatible tissue culture plates 24 h before the assay. Cells were coincubated for 6 h at 37°C, after which they were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. An LB 941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM 170 dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (Prolume).
Purification of recombinant 92TH023 and CM244 gp120 glycoproteins.
To produce soluble gp120, 10 × 106 293T cells were transfected with a plasmid expressing wild-type or mutant 92TH023 or CM244 gp120 using PEI MAX 40000 as directed by the manufacturer (Polysciences). Three days after transfection, supernatant was collected and cells were pelleted and discarded. The supernatants were filtered (0.22-μm filter; Corning), and soluble gp120 was purified by lectin affinity columns. Briefly, agarose-bound Galanthus Nivalis Lectin (Vector Laboratories) was added to the filtered supernatant and the mixture was incubated overnight on a shaking platform. The mixture (supernatant-containing beads) was then passed through columns (for washing with cold phosphate-buffered saline [PBS]), and proteins were eluted with methyl-α-d-pyranoside (0.5 M), which was followed by fast protein liquid chromatography (FPLC) purification of monomeric gp120, as described previously (56). gp120 preparations were dialyzed against PBS and stored in aliquots at −80°C. To assess purity, recombinant proteins were loaded on SDS-PAGE polyacrylamide gels and stained with Coomassie blue.
SPR biosensor analysis.
Surface plasmon resonance biosensor data were collected on a Biacore 3000 optical biosensor (General Electric). CD4-Ig was immobilized onto separate flow cells within the same sensor chip (CM5; GE) to a surface density of around 500 response units (RU) using standard amine coupling chemistry (57). The binding capacities of CD4 surfaces were kept low to avoid mass transport effects and steric hindrance. Flow cell 1 or 3 was left blank as a control for nonspecific binding and refractive index changes. With the instrument operating in a parallel sensing mode, soluble gp120 was injected over flow cells 1 and 2 or 3 and 4 at different concentrations ranging from 100 to 750 nM at a flow rate of 30 μl/min for 3 min. This was followed by a 10-min dissociation phase to allow an estimation of off-rates and binding affinities. Sensor data were prepared for kinetic analysis by subtracting binding responses collected from the blank reference surface. The association and dissociation phase data were fitted simultaneously with BIAevaluation, version 3.2, RC1 software using a 1:1 Langmuir model of binding.
Infection by single-round luciferase viruses.
Cf2Th-CD4-CCR5 target cells were seeded at a density of 2 × 104 cells/well in 96-well luminometer-compatible tissue culture plates (PerkinElmer) 24 h before infection. Recombinant viruses in a final volume of 100 μl were then added to the target cells containing different dilutions of antibodies followed by incubation for 48 h at 37°C. The medium was then removed from each well, and the cells were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. An LB 941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM 170 dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (Prolume).
Sequence analyses.
The Logo plots (58) for HIV were made using the Analyze Align tool at the HIV database and are based on the LOGO program (https://www.hiv.lanl.gov/content/sequence/ANALYZEALIGN/analyze_align.html) and the HIV-1 database global curated and filtered alignment circa October 2016, including 1 HIV-1 protein Env sequence per person Env from 4,632 individuals.
Signature analyses (59, 60) were used to track amino acids that covaried with either H375 or S375. A subset of 1,000 M group sequences from the more than 4,500 M group sequences were sampled from the full Los Alamos database alignment to use as input, reducing the data set size to facilitate computationally intensive phylogenetic analyses and ancestral state reconstruction. To make the analysis more informative, the 1,000 sequences were picked after first narrowing the field to include only subtypes and recombinant forms that had at least some sequences with H375, to enable identification of covariation patterns across the M group. Because of their structural proximity, we were most interested in evaluating signature associations within the inner domain layer, and so we present these fully in Table 1. Other phylogenetically corrected associations were found in other regions of Env; however, that may be of interest to explore in future studies. In particular, H375 was positively associated with A101, K231, N289, V341, A430, I519, and H629 and negatively associated with I307 and V430 (all with very low P values in phylogenetically corrected tests, ranging from 10−10 to 10−15), raising the possibility that these sites may also somehow functionally interact with position 375.
ACKNOWLEDGMENTS
This work was supported by CIHR foundation grant 352417 to A.F. A.F. is the recipient of a Canada Research Chair on Retroviral Entry. D.Z. is the recipient of FRQS master award 30888. M.V. is the recipient of CIHR Doctoral Research Award 291485. K.W. and B.K. were supported by grant AI100645 from the Duke Center for HIV/AIDS Vaccine Immunology and Immunogen Design (CHAVI-ID). Our funding sources had no role in data collection, analysis, or interpretation and were not involved in the writing of the manuscript. We have no conflicts of interest to report.
The opinions expressed herein are those of the authors and should not be construed as official or representing the views of the U.S. Department of Defense or Department of the Army. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
REFERENCES
- 1.Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol 69:5087–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Neil PK, Sun G, Yu H, Ron Y, Dougherty JP, Preston BD. 2002. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J Biol Chem 277:38053–38061. doi: 10.1074/jbc.M204774200. [DOI] [PubMed] [Google Scholar]
- 3.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. 2010. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol 84:9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin JM. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 5.Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A 102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 7.McCutchan FE. 2006. Global epidemiology of HIV. J Med Virol 78(Suppl 1):S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 8.Burke DS. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg Infect Dis 3:253–259. doi: 10.3201/eid0303.970301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor BS, Hammer SM. 2008. The challenge of HIV-1 subtype diversity. N Engl J Med 359:1965–1966. doi: 10.1056/NEJMc086373. [DOI] [PubMed] [Google Scholar]
- 10.Hemelaar J. 2012. The origin and diversity of the HIV-1 pandemic. Trends Mol Med 18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Hemelaar J. 2013. Implications of HIV diversity for the HIV-1 pandemic. J Infect 66:391–400. doi: 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Saksena NK, Lau KA, Dwyer DE, Wang B. 2011. HIV recombination and pathogenesis—biological and epidemiological implications, HIV and AIDS—updates on biology, immunology, epidemiology and treatment strategies. InTech doi: 10.5772/23819. [DOI] [Google Scholar]
- 13.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. 2000. HIV-1 nomenclature proposal. Science 288:55–56. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, Stanke M, Morgenstern B, Korber B, Leitner T. 2010. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 16.Hu WS, Temin HM. 1990. Retroviral recombination and reverse transcription. Science 250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 17.Temin HM. 1991. Sex and recombination in retroviruses. Trends Genet 7:71–74. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Li W, Zhong P, Fang K, Zhu K, Musa TH, Song Y, Du G, Gao R, Guo Y, Yan W, Xuan Y, Wei P. 2016. Nationwide trends in molecular epidemiology of HIV-1 in China. AIDS Res Hum Retroviruses 32:851–859. doi: 10.1089/AID.2016.0029. [DOI] [PubMed] [Google Scholar]
- 19.Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 20.Carr JK, Salminen MO, Koch C, Gotte D, Artenstein AW, Hegerich PA, St Louis D, Burke DS, McCutchan FE. 1996. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol 70:5935–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon-Loriere E, Galetto R, Hamoudi M, Archer J, Lefeuvre P, Martin DP, Robertson DL, Negroni M. 2009. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog 5:e1000418. doi: 10.1371/journal.ppat.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archer J, Pinney JW, Fan J, Simon-Loriere E, Arts EJ, Negroni M, Robertson DL. 2008. Identifying the important HIV-1 recombination breakpoints. PLoS Comput Biol 4:e1000178. doi: 10.1371/journal.pcbi.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helseth E, Olshevsky U, Furman C, Sodroski J. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol 65:2119–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. 2003. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313:117–125. doi: 10.1016/S0042-6822(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 26.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol 69:5723–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol 2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 29.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 30.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol 76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desormeaux A, Coutu M, Medjahed H, Pacheco B, Herschhorn A, Gu C, Xiang SH, Mao Y, Sodroski J, Finzi A. 2013. The highly conserved layer-3 component of the HIV-1 gp120 inner domain is critical for CD4-required conformational transitions. J Virol 87:2549–2562. doi: 10.1128/JVI.03104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finzi A, Pacheco B, Xiang SH, Pancera M, Herschhorn A, Wang L, Zeng X, Desormeaux A, Kwong PD, Sodroski J. 2012. Lineage-specific differences between human and simian immunodeficiency virus regulation of gp120 trimer association and CD4 binding. J Virol 86:8974–8986. doi: 10.1128/JVI.01076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, Brocca-Cofano E, Deleage C, Hao X, Chuang GY, Gorman J, Gardner M, Lewis MG, Hatziioannou T, Santra S, Apetrei C, Pandrea I, Alam SM, Liao HX, Shen X, Tomaras GD, Farzan M, Chertova E, Keele BF, Estes JD, Lifson JD, Doms RW, Montefiori DC, Haynes BF, Sodroski JG, Kwong PD, Hahn BH, Shaw GM. 2016. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113:E3413–E3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding S, Medjahed H, Prevost J, Coutu M, Xiang SH, Finzi A. 2016. Lineage-specific differences in the gp120 inner domain layer 3 of human and simian immunodeficiency viruses. J Virol 90:10065–10073. 10.1128/JVI.01215-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong P, Pan Q, Ning Z, Xue Y, Gong J, Zhen X, Zhou L, Sheng F, Zhang W, Gai J, Cheng H, Yue Q, Xing H, Zhuang M, Lu W, Shao Y, Kang L. 2007. Genetic diversity and drug resistance of human immunodeficiency virus type 1 (HIV-1) strains circulating in Shanghai. AIDS Res Hum Retroviruses 23:847–856. doi: 10.1089/aid.2006.0196. [DOI] [PubMed] [Google Scholar]
- 37.He X, Xing H, Ruan Y, Hong K, Cheng C, Hu Y, Xin R, Wei J, Feng Y, Hsi JH, Takebe Y, Shao Y, Group for HIVMES . 2012. A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One 7:e47289. doi: 10.1371/journal.pone.0047289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu DJ, Vanichseni S, Mastro TD, Raktham S, Young NL, Mock PA, Subbarao S, Parekh BS, Srisuwanvilai L, Sutthent R, Wasi C, Heneine W, Choopanya K. 2001. Viral load differences in early infection with two HIV-1 subtypes. AIDS 15:683–691. doi: 10.1097/00002030-200104130-00003. [DOI] [PubMed] [Google Scholar]
- 39.Rangsin R, Piyaraj P, Sirisanthana T, Sirisopana N, Short O, Nelson KE. 2007. The natural history of HIV-1 subtype E infection in young men in Thailand with up to 14 years of follow-up. AIDS 21(Suppl 6):S39–S46. [DOI] [PubMed] [Google Scholar]
- 40.Rangsin R, Chiu J, Khamboonruang C, Sirisopana N, Eiumtrakul S, Brown AE, Robb M, Beyrer C, Ruangyuttikarn C, Markowitz LE, Nelson KE. 2004. The natural history of HIV-1 infection in young Thai men after seroconversion. J Acquir Immune Defic Syndr 36:622–629. doi: 10.1097/00126334-200405010-00011. [DOI] [PubMed] [Google Scholar]
- 41.Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol 19:893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang CY, Hariharan K, Nara PL, Sodroski J, Moore JP. 1994. Immunization with a soluble CD4-gp120 complex preferentially induces neutralizing anti-human immunodeficiency virus type 1 antibodies directed to conformation-dependent epitopes of gp120. J Virol 68:5854–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A 97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veillette M, Desormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 47.Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB III, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog 5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mori K, Rosenzweig M, Desrosiers RC. 2000. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J Virol 74:10852–10859. doi: 10.1128/JVI.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcon L, Choe H, Martin KA, Farzan M, Ponath PD, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. 1997. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol 71:2522–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcon L, Sodroski J. 1997. High degree of sensitivity of the simian immunodeficiency virus (SIVmac) envelope glycoprotein subunit association to amino acid changes in the glycoprotein 41 ectodomain. AIDS Res Hum Retroviruses 13:441–447. doi: 10.1089/aid.1997.13.441. [DOI] [PubMed] [Google Scholar]
- 52.LaBonte JA, Patel T, Hofmann W, Sodroski J. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J Virol 74:10690–10698. doi: 10.1128/JVI.74.22.10690-10698.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HXB2CG. https://hfv.lanl.gov/content/sequence/HIV/COMPENDIUM/1998/III/HXB2.pdf.
- 55.Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360. doi: 10.1016/0042-6822(81)90642-5. [DOI] [PubMed] [Google Scholar]
- 56.Coutu M, Finzi A. 2015. HIV-1 gp120 dimers decrease the overall affinity of gp120 preparations for CD4-induced ligands. J Virol Methods 215-216:37–44. doi: 10.1016/j.jviromet.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 57.Johnsson B, Lofas S, Lindquist G. 1991. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem 198:268–277. doi: 10.1016/0003-2697(91)90424-R. [DOI] [PubMed] [Google Scholar]
- 58.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 60.Gnanakaran S, Daniels MG, Bhattacharya T, Lapedes AS, Sethi A, Li M, Tang H, Greene K, Gao H, Haynes BF, Cohen MS, Shaw GM, Seaman MS, Kumar A, Gao F, Montefiori DC, Korber B. 2010. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput Biol 6:e1000955. doi: 10.1371/journal.pcbi.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]