ABSTRACT
Myxomatosis is a recurrent problem on rabbit farms throughout Europe despite the success of vaccines. To identify gene variations of field and vaccine strains that may be responsible for changes in virulence, immunomodulation, and immunoprotection, the genomes of 6 myxoma virus (MYXV) strains were sequenced: German field isolates Munich-1, FLI-H, 2604, and 3207; vaccine strain MAV; and challenge strain ZA. The analyzed genomes ranged from 147.6 kb (strain MAV) to 161.8 kb (strain 3207). All sequences were affected by several mutations, covering 24 to 93 open reading frames (ORFs) and resulted in amino acid substitutions, insertions, or deletions. Only strains Munich-1 and MAV revealed the deletion of 10 ORFs (M007L to M015L) and 11 ORFs (M007L to M008.1L and M149R to M008.1R), respectively. Major differences were observed in the 27 immunomodulatory proteins encoded by MYXV. Compared to the reference strain Lausanne, strains FLI-H, 2604, 3207, and ZA showed the highest amino acid identity (>98.4%). In strains Munich-1 and MAV, deletion of 5 and 10 ORFs, respectively, was observed, encoding immunomodulatory proteins with ankyrin repeats or members of the family of serine protease inhibitors. Furthermore, putative immunodominant surface proteins with homology to vaccinia virus (VACV) were investigated in the sequenced strains. Only strain MAV revealed above-average frequencies of amino acid substitutions and frameshift mutations. Finally, we performed recombination analysis and found signs of recombination in vaccine strain MAV. Phylogenetic analysis showed a close relationship of strain MAV and the MSW strain of Californian MYXV. However, in a challenge model, strain MAV provided full protection against lethal challenges with strain ZA.
IMPORTANCE Myxoma virus (MYXV) is pathogenic for European rabbits and two North American species. Due to sophisticated strategies in immune evasion and oncolysis, MYXV is an important model virus for immunological and pathological research. In its natural hosts, MYXV causes a benign infection, whereas in European rabbits, it causes the lethal disease myxomatosis. Since the introduction of MYXV into Australia and Europe for the biological control of European rabbits in the 1950s, a coevolution of host and pathogen has started, selecting for attenuated virus strains and increased resistance in rabbits. Evolution of viruses is a continuous process and influences the protective potential of vaccines. In our analyses, we sequenced 6 MYXV field, challenge, and vaccine strains. We focused on genes encoding proteins involved in virulence, host range, immunomodulation, and envelope composition. Genes affected most by mutations play a role in immunomodulation. However, attenuation cannot be linked to individual mutations or gene disruptions.
KEYWORDS: DNA sequencing, genome analysis, myxoma virus, poxvirus
INTRODUCTION
The release of myxoma virus (MYXV) in the 1950s for the biological control of European rabbits (Oryctolagus cuniculus) in Australia and France is a well-documented example of unprecedented virus spread and unexpected host-pathogen coevolution leading to attenuated virus strains and the natural selection of rabbits with genetic resistance to MYXV in the environment (1–3). Despite the success of vaccines, MYXVs remain a recurrent problem on rabbit farms throughout Europe. There is not much validated information about vaccine breaks, and many problems after vaccination seem to be due to vaccine and application failures (4). However, a surveillance of the efficacy of established vaccine strains against new field viruses is highly recommended. Today, modern sequencing techniques enable the rapid genetic characterization of large numbers of MYXV field isolates in comparison to reference strains.
MYXV is a member of the Poxviridae family. It is the type species of the genus Leporipoxvirus of the subfamily of Chordopoxvirinae, which additionally comprises Hare fibroma virus, Rabbit fibroma virus (RFV), and Squirrel fibroma virus (5). In its evolutionary host, the North American brush rabbit (Sylvilagus bachmani) or the South American tapeti (Sylvilagus brasiliensis), MYXV causes a mild, benign infection accompanied by cutaneous fibromas restricted to the site of inoculation. However, European rabbits (Oryctolagus cuniculus) infected with MYXV develop the rapidly systemic and highly lethal disease myxomatosis. This virus is passively transmitted by mosquitoes or other biting arthropods but does not replicate within the vector.
European settlers first introduced European rabbits into Australia to satisfy their need for meat and fur in 1788. The introduction of several rabbits in 1859 resulted in the continent-wide spread of rabbits within 50 years and became responsible for major ecological damages. In order to control the rabbit population, the MYXV standard laboratory strain (SLS), isolated in Brazil, was introduced into Australia in 1950 and spread within 5 years across the entire, highly susceptible rabbit population (6). Within 2 years after the introduction of the SLS, slightly attenuated MYXV strains emerged in the field. At the same time, the wild-rabbit population was selected for rabbits that were resistant to myxomatosis. This host-pathogen coevolution resulted in a reduction of the virulence of field strains of MYXV in wild rabbits compared to laboratory rabbits. Within a few years, less virulent strains became predominant in the field (7). This evolutionary experiment was repeated in Europe in 1952. Like all poxviruses, MYXV possess a large, linear, double-stranded DNA genome of ∼160 kb encapsidated within brick-shaped virions of about 200 to 300 nm. In contrast to other DNA viruses, poxvirus replication takes place exclusively in the cytoplasm of infected cells. To date, the complete genomes of 33 members of the Leporipoxvirus genus, 32 MYXV strains and 1 RFV strain (Table 1), are available.
TABLE 1.
Origins of MYXV sequences obtained from GenBank
| Virus | Geographic origin | Source | Reference | Genome size (bp) | Region sequenced (bp)a | GenBank accession no. |
|---|---|---|---|---|---|---|
| Lausanne | Campina, Brazil | Unknown | 8 | 161,777 | 1–161777 | AF170726 |
| SG33 | France | Vaccine strain | 18 | 148,244 | 1–161777 | GQ409969 |
| 6918 | Girona, Spain | Wild rabbit | 17 | 161,766 | 1–161777 | EU552530 |
| MSW | California, USA | Rabbit tissue | 19 | 164,600 | 1–161777 | KF148065 |
| SLS (Moses strain/strain B) | Brazil | Rabbit tissue stock (F. Fenner) | 2 | 161,763 | 1–161777 | JX565574 |
| Glenfield | Central NSW, Australia | CV-1 cell stock | 2 | 161,742 | 15–161763 | JX565567 |
| KM13 | Southern NSW, Australia | Rabbit tissue stock (F. Fenner) | 2 | 161,771 | 1–161777 | JX565569 |
| Uriarra | Canberra District, Australia | CV-1 cell stock | 2 | 161,768 | 1–161777 | JX565577 |
| SWH | Canberra District, Australia | Wild rabbit | 2 | 161,797 | 1–161777 | JX565576 |
| BRK | Canberra District, Australia | Wild rabbit | 2 | 161,701 | 1–161777 | JX565562 |
| Bendigo | Central Victoria, Australia | Wild rabbit | 2 | 161,738 | 1–161777 | JX565565 |
| Meby | Tasmania, Australia | Wild rabbit | 2 | 161,542 | 87–161691 | JX565571 |
| Cornwall | Cornwall, UK | Rabbit tissue stock (F. Fenner) | 2 | 161,775 | 1–161777 | JX565566 |
| Sussex | Sussex, UK | Rabbit tissue stock (F. Fenner) | 2 | 161,778 | 1–161777 | KC660084 |
| Nottingham attenuated | Nottingham, UK | Rabbit tissue stock (F. Fenner) | 2 | 161,777 | 1–161777 | JX565572 |
| Gung/91 | Canberra District, Australia | Wild rabbit | 2 | 161,443 | 151–161627 | JX565568 |
| Wellington | Central NSW, Australia | Wild rabbit | 2 | 161,688 | 29–161749 | JX565582 |
| BRK/12-2-93 | Canberra District, Australia | Wild rabbit | 2 | 161,496 | 140–161638 | JX565563 |
| BD23 | Southwest Queensland, Australia | Wild rabbit | 2 | 161,971 | 285–161555 | JX565584 |
| BD44 | Southwest Queensland, Australia | Wild rabbit | 2 | 162,847 | 1–161777 | KC660079 |
| BRK/897 | Canberra District, Australia | Wild rabbit | 2 | 161,545 | 103–161675 | JX565564 |
| OB1/406 | Canberra District, Australia | Wild rabbit | 2 | 161,612 | 87–161691 | JX565573 |
| OB2/W60 | Canberra District, Australia | Wild rabbit | 2 | 162,483 | 1–161777 | KC660081 |
| OB3/Y317 | Canberra District, Australia | Wild rabbit | 2 | 161,748 | 1–161777 | KC660083 |
| OB3/1120 | Canberra District, Australia | Wild rabbit | 2 | 161,722 | 1–161777 | KC660082 |
| WS1/234 | Canberra District, Australia | Wild rabbit | 2 | 161,754 | 1–161777 | JX565578 |
| WS6/1071 | Canberra District, Australia | Wild rabbit | 2 | 161,752 | 41–161737 | JX565580 |
| WS1/328 | Canberra District, Australia | Wild rabbit | 2 | 161,483 | 156–161622 | JX565579 |
| WS6/346 | Canberra District, Australia | Wild rabbit | 2 | 161,430 | 140–161638 | JX565581 |
| SWH/8-2-93 | Canberra District, Australia | Wild rabbit | 2 | 161,740 | 1–161777 | JX565575 |
| SWH/805 | Canberra District, Australia | Wild rabbit | 2 | 161,780 | 1–161777 | KC660085 |
| SWH/1209 | Canberra District, Australia | Wild rabbit | 2 | 162,413 | 33–161745 | JX565583 |
| Rabbit fibroma virus (Kasza strain) | Ohio, USA | Wild rabbit | 63 | 159,857 | 11–161788 | AF170722 |
The first MYXV, which has been fully sequenced, was the Lausanne strain. It is considered the reference genome (8). The genome consists of 161,777 bp with terminal inverted repeats (TIRs) of 11.5 kb. MYXV harbors 170 nonoverlapping open reading frames (ORFs). Much of the understanding of poxvirus replication was obtained from research on vaccinia virus (VACV). As with all members of the Poxviridae, 2 distinct forms of infectious virions, mature virions (MV) and enveloped virions (EV), are produced (9, 10). In VACV, both membranes contain surface proteins providing epitopes for neutralizing antibodies. MV membrane-associated proteins also have orthologs in MYXV: A17 (MYXV M107), A27 (MYXV M115), A28 (MYXV M116), D8 (MYXV M083), H3 (MYXV M071), and L1 (MYXV M055). The same counts for the EV-specific proteins B5 (MYXV M144) and the F13L protein, referred to as p37 (MYXV M022) (11–13).
Poxviruses reveal a high mutation rate that enables them to evolve at rates nearly as high as those of RNA viruses (14). Therefore, it is important to investigate MYXV vaccine strains, as well as MYXV field strains, for mutations in genes encoding envelope proteins and proteins involved in immunomodulation or virulence. An effective vaccination strategy is required for successful protection from myxomatosis. Only live vaccines of attenuated MYXV provide full protection from myxomatosis. They can be divided into 2 groups. The first group of live vaccines consists of orthologous attenuated MYXV strains. Vaccine strain MAV has an unclear history, but initially, it was derived from a cell culture-attenuated Californian MSD strain (15). The second group of vaccines consists of the closely related, nonpathogenic leporipoxvirus RFV. In RFV, 7 ORFs are missing or fragmented compared to MYXV. However, RFV is genetically and antigenically sufficiently closely related to MYXV to provide cross-protection from MYXV (16).
The aim of this study was the examination of the genetic stability of MYXV vaccine and field strains. We generated complete genome sequences of six MYXV vaccine, challenge, and field strains (Table 2) and compared them in detail to those of the reference strain Lausanne, the vaccine strains SG33 and 6918, as well as RFV. The genome analysis concerned primarily virulence, host range, immunomodulating, and envelope protein-encoding genes. The phylogenetic relationship among the 6 sequenced MYXV strains, the reported RFV sequence, and all 32 available MYXV sequences was analyzed by a maximum likelihood phylogenetic analysis. The identification of nonessential gene regions that may serve for the integration of exogenous DNA for the development of vector-based vaccines may improve existing MYXV vaccines.
TABLE 2.
Origins of strains of MYXV sequenced for this study
| MYXV strain | Geographic origin | Source | Yr of isolation | Genome size (bp) | AT content (%) | Sequence coverage (n-fold) | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| MAV | California, USA | Commercial vaccine strain | 1964 | 147,574 | 56.15 | 47.7 | KP723391 |
| ZA | Pulawy, Poland | Commercial challenge strain for MAV | 1985 | 161,609 | 56.46 | 67.4 | KP723386 |
| Munich-1 | Munich, Germany | Wild rabbit with myxomas | 1985 | 150,884 | 56.72 | 183.1 | KP723387 |
| FLI-H | Greifswald, Germany | Rabbit with myxomatosis | 2004 | 161,790 | 56.47 | 10.3 | KP723390 |
| 2604 | Oldenburg, Germany | Rabbit with myxomatosis | 2004 | 161,715 | 56.47 | 22.5 | KP723389 |
| 3207 | Greifswald, Germany | Pygmy rabbit with myxomatosis | 2007 | 161,799 | 56.47 | 6.7 | KP723388 |
RESULTS AND DISCUSSION
We sequenced the complete genomes of 6 MYXV isolates: the commercial vaccine strain MAV, the challenge strain ZA, 3 field strains (Munich-1, 2604, and 3207) isolated between 1985 and 2007 from rabbits that died of myxomatosis, and field strain FLI-H, isolated from a vaccinated rabbit that also died of myxomatosis (Table 2). Challenge strain ZA was used to prove the vaccine efficacy of strain MAV in animal experiments for drug approval. These six whole-genome sequences and the reported sequences of the Spanish isolate 6918 (17) and the French vaccine strain SG33 (18) were compared to the those of the Lausanne strain and RFV as a reference. First, we aligned the nucleotide sequences ORF by ORF to that of the Lausanne strain as a reference. We then checked for homopolymers and possible sequencing errors. At suspicious positions, we took a closer look at the sequencing raw data. After excluding any error, we translated the nucleotide sequences into amino acid sequences and performed an alignment. These ORF-by-ORF analyses at the amino acid level revealed several differences compared to the Lausanne strain.
The genomes of vaccine strain MAV and field strain Munich-1 revealed the most differences among the 6 sequenced strains compared to the genome of the Lausanne strain. Only these strains revealed severe truncations and deletions of several ORFs. The genome of strain MAV is 147,574 bp with an AT content of 56.15% (compared to 56.42% for the Lausanne strain and 60.46% for RFV). The genome is 14.2 kb shorter than that of strain Lausanne (161,777 bp) due to deletions in the TIR. In strain MAV, 160 of the 170 ORFs previously assigned to the Lausanne strain are present (Table 3); 6 are severely disrupted (Table 4). At the amino acid level, there are 2,783 differences involving 94 predicted proteins compared to the Lausanne strain. The differences consist of 1,307 amino acid (aa) substitutions as well as 1,455 deletions and 21 insertions that were the result of frameshift mutations. Of 160 predicted proteins, 66 (41.25%) are identical to those of the Lausanne strain, and 17 (10.63%) showed a substitution of 1 aa only (Tables 3 and 4).
TABLE 3.
Distribution of ORF deletions and amino acid substitutions of the sequenced strains compared to the Lausanne strain
| MYXV strain | Genome size (bp) | Full-length genome |
Terminal genomic regionsa |
Central genomic regionb |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of mutated ORFs | No. of deleted ORFs | No. of substitutions (aa) | No. of deletions (aa) | No. of insertions (aa) | Total no. of changes (aa) | No. of mutated ORFs | No. of deleted ORFs | No. of substitutions (aa) | No. of deletions (aa) | No. of insertions (aa) | Total no. of changes (aa) | No. of mutated ORFs | No. of deleted ORFs | No. of substitutions (aa) | No. of deletions (aa) | No. of insertions (aa) | Total no. of changes (aa) | ||
| Lausanne | 161,777 | ||||||||||||||||||
| MAV | 147,574 | 94 | 11 | 1,307 | 1,455 | 21 | 2,783 | 5 | 4 | 41 | 84 | 125 | 89 | 7 | 1,266 | 1,371 | 21 | 2,658 | |
| ZA | 161,609 | 25 | 50 | 281 | 331 | 4 | 4 | 4 | 21 | 46 | 281 | 327 | |||||||
| Munich-1 | 150,884 | 24 | 10 | 34 | 429 | 1 | 464 | 7 | 3 | 6 | 400 | 406 | 17 | 7 | 28 | 29 | 1 | 58 | |
| FLI-H | 161,790 | 33 | 58 | 324 | 382 | 10 | 12 | 12 | 23 | 46 | 324 | 370 | |||||||
| 2604 | 161,715 | 37 | 71 | 193 | 12 | 276 | 12 | 16 | 16 | 25 | 55 | 193 | 12 | 260 | |||||
| 3207 | 161,799 | 36 | 62 | 965 | 11 | 1,038 | 6 | 7 | 7 | 30 | 55 | 965 | 11 | 1,031 | |||||
Terminal genomic regions of ORFs M000.5L to M008.1L and M008.1R to M000.5R.
Central genomic regions of ORFs M009L to M156R.
TABLE 4.
Summary of genes of the sequenced strains and comparison of amino acid sequences to those of orthologs in MYXV Lausanne
| MYXV gene | MAV |
Munich-1 |
FLI-H |
2604 |
3207 |
ZA |
SG33 |
6918 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Difference in length (aa) | No. of aa substitutions | Difference in length (aa) | No. of aa substitutions | Difference in length (aa) | No. of aa substitutions | Difference in length (aa) | No. of aa substitutions | Difference in length (aa) | No. of aa substitutions | Difference in length (aa) | No. of aa substitutions | Difference in length (aa) | No. of aa substitutions | Difference in length (aa) | No. of aa substitutions | |
| M000.5L | ||||||||||||||||
| M000.5R | ||||||||||||||||
| M001L | 1 | 1 | ||||||||||||||
| M001R | 1 | —b | ||||||||||||||
| M002L | 1 | 3 | ||||||||||||||
| M002R | 1 | Deleted | 3 | |||||||||||||
| M003.1L | 1 | 2 | 2 | 2 | 1 | |||||||||||
| M003.1R | 1 | 2 | 2 | Deleted | 1 | |||||||||||
| M003.2L | ||||||||||||||||
| M003.2R | ||||||||||||||||
| M004L | 1 | 1 | 1 | 1 | 1 | |||||||||||
| M004R | 1 | 1 | 1 | Deleted | 1 | |||||||||||
| M004.1L | ||||||||||||||||
| M004.1R | Deleted | |||||||||||||||
| M005L | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| M005R | 1 | 1 | 1 | 1 | Deleted | 1 | ||||||||||
| M006L | −83a | 36 | −400 | |||||||||||||
| M006R | 2 | Deleted | ||||||||||||||
| M007L | Deleted | Deleted | 1 | 1 | ||||||||||||
| M007R | 1 | Deleted | 1 | |||||||||||||
| M008L | Deleted | Deleted | 1 | 1 | 1 | 1 | ||||||||||
| M008R | −1 | 1 | 1 | 1 | Deleted | 1 | ||||||||||
| M008.1L | Deleted | Deleted | 1 | 2 | 1 | 2 | ||||||||||
| M008.1R | Deleted | 1 | 2 | Deleted | 2 | |||||||||||
| M009L | −408a | Deleted | −162 | 1 | −162 | 1 | −162 | 1 | 1 | −93 | 23 | |||||
| M010L | Deleted | |||||||||||||||
| M011L | −133 | 2 | Deleted | −133 | 2 | |||||||||||
| M012L | 1 | 10 | Deleted | 1 | 1 | |||||||||||
| M013L | −14 | 37 | Deleted | 1 | 1 | 2 | ||||||||||
| M014L | 73 | Deleted | 1 | 1 | 1 | 1 | ||||||||||
| M015L | 12 | Deleted | 10 | 1 | 1 | 1 | ||||||||||
| M016L | 20 | 8 | ||||||||||||||
| M017L | −43 | 5 | 1 | |||||||||||||
| M018L | 2 | −54 | 10 | |||||||||||||
| M019L | 7 | |||||||||||||||
| M020L | ||||||||||||||||
| M021L | 3 | 1 | 1 | 1 | 1 | |||||||||||
| M022L | 14 | 1 | ||||||||||||||
| M023R | −29 | 17 | ||||||||||||||
| M024L | 7 | |||||||||||||||
| M025L | 5 | 1 | ||||||||||||||
| M026L | 1 | |||||||||||||||
| M027L | 1 | 2 | 2 | 2 | ||||||||||||
| M028L | ||||||||||||||||
| M029L | ||||||||||||||||
| M030L | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| M031R | 1 | 1 | 1 | 1 | ||||||||||||
| M032R | 18 | 1 | −107 | 8 | 1 | 1 | 1 | |||||||||
| M033R | 3 | 1 | ||||||||||||||
| M034L | 1 | 32 | 1 | 1 | 1 | 1 | 1 | |||||||||
| M035R | 9 | 1 | 1 | |||||||||||||
| M036L | 58 | 1 | 1 | 1 | 2 | −145 | 15 | |||||||||
| M037L | ||||||||||||||||
| M038L | 14 | |||||||||||||||
| M039L | ||||||||||||||||
| M040L | 1 | |||||||||||||||
| M041L | ||||||||||||||||
| M042L | 8 | |||||||||||||||
| M043L | 5 | 3 | ||||||||||||||
| M044R | 22 | 1 | 1 | 1 | 1 | 3 | 1 | |||||||||
| M045L | 21 | 1 | 1 | |||||||||||||
| M046L | 6 | |||||||||||||||
| M047R | 8 | −8 | 2 | 1 | ||||||||||||
| M048L | 1 | 4 | −1 | 2 | −1 | 2 | ||||||||||
| M049R | 13 | 2 | 1 | 1 | ||||||||||||
| M050R | ||||||||||||||||
| M051R | ||||||||||||||||
| M052L | 1 | |||||||||||||||
| M053R | 1 | 1 | ||||||||||||||
| M054R | 1 | 7 | 1 | |||||||||||||
| M055R | ||||||||||||||||
| M056R | ||||||||||||||||
| M057L | 1 | |||||||||||||||
| M058R | 2 | 1 | 1 | |||||||||||||
| M059R | 1 | |||||||||||||||
| M060R | 3 | 1 | 2 | |||||||||||||
| M061R | 2 | |||||||||||||||
| M062R | 1 | 1 | ||||||||||||||
| M063R | −5 | 5 | 1 | 1 | ||||||||||||
| M064R | −1 | 1 | 1 | 1 | ||||||||||||
| M065R | ||||||||||||||||
| M066R | 1 | |||||||||||||||
| M067L | 1 | 1 | 1 | |||||||||||||
| M068R | −374 | 3 | 1 | |||||||||||||
| M069L | ||||||||||||||||
| M070R | ||||||||||||||||
| M071L | 1 | 1 | ||||||||||||||
| M072L | 2 | −107 | 2 | 2 | ||||||||||||
| M073R | 1 | 1 | 1 | 1 | ||||||||||||
| M074R | 1 | 1 | 1 | |||||||||||||
| M075R | ||||||||||||||||
| M076R | 35 | 1 | 1 | 1 | 1 | 11 | ||||||||||
| M077L | 8 | 23 | 9 | |||||||||||||
| M078R | 21 | 12 | ||||||||||||||
| M079R | 7 | 4 | ||||||||||||||
| M080R | 10 | 6 | ||||||||||||||
| M081R | 3 | |||||||||||||||
| M082R | ||||||||||||||||
| M083L | −128 | 7 | 5 | |||||||||||||
| M084R | −5 | |||||||||||||||
| M085R | 1 | 1 | ||||||||||||||
| M086L | 7 | 1 | ||||||||||||||
| M087L | 6 | |||||||||||||||
| M088L | −1 | 16 | 1 | |||||||||||||
| M089L | 10 | |||||||||||||||
| M090L | 2 | 1 | 5 | |||||||||||||
| M091L | ||||||||||||||||
| M092L | 1 | 1 | ||||||||||||||
| M093L | ||||||||||||||||
| M094R | ||||||||||||||||
| M095L | 11 | 1 | ||||||||||||||
| M096L | 18 | 1 | 1 | 1 | 1 | |||||||||||
| M097R | 7 | |||||||||||||||
| M098L | ||||||||||||||||
| M099L | 40 | 2 | 1 | |||||||||||||
| M100R | ||||||||||||||||
| M101L | 1 | |||||||||||||||
| M102L | ||||||||||||||||
| M103L | ||||||||||||||||
| M104L | ||||||||||||||||
| M105L | 1 | |||||||||||||||
| M106L | ||||||||||||||||
| M107L | 1 | |||||||||||||||
| M108R | 13 | |||||||||||||||
| M109L | 3 | |||||||||||||||
| M110L | 6 | |||||||||||||||
| M111R | 20 | 4 | 1 | |||||||||||||
| M112R | ||||||||||||||||
| M113R | 3 | 3 | ||||||||||||||
| M114R | 10 | 1 | ||||||||||||||
| M115L | 5 | 39 | ||||||||||||||
| M116L | ||||||||||||||||
| M117L | 6 | |||||||||||||||
| M118L | 2 | |||||||||||||||
| M119L | ||||||||||||||||
| M120L | ||||||||||||||||
| M121R | 1 | |||||||||||||||
| M122R | −1 | 8 | ||||||||||||||
| M123R | 1 | 1 | 1 | |||||||||||||
| M124R | ||||||||||||||||
| M125R | ||||||||||||||||
| M126R | 14 | |||||||||||||||
| M127L | 39 | −279 | 18 | |||||||||||||
| M128L | 1 | 9 | 1 | 1 | ||||||||||||
| M129R | 15 | |||||||||||||||
| M130R | 5 | 24 | ||||||||||||||
| M131R | −123 | 10 | 10 | 1 | ||||||||||||
| M132L | 2 | 25 | ||||||||||||||
| M133R | 40 | 2 | ||||||||||||||
| M134R | 216 | 2 | 1 | −1 | 2 | 1 | −2 | 4 | 1 | |||||||
| M135R | 12 | 16 | 8 | −138 | 21 | |||||||||||
| M136R | 2 | 28 | ||||||||||||||
| M137R | 48 | |||||||||||||||
| M138L | 1 | 53 | ||||||||||||||
| M139R | 2 | 13 | ||||||||||||||
| M140R | 29 | 1 | 3 | 2 | −322 | 10 | 1 | 51 | ||||||||
| M141R | 5 | 53 | 1 | 2 | 1 | 2 | 1 | 5 | 53 | |||||||
| M142R | 3 | 4 | 26 | |||||||||||||
| M143R | 11 | 13 | ||||||||||||||
| M144R | 5 | 1 | −2 | 70 | ||||||||||||
| M146R | 17 | |||||||||||||||
| M147R | 1 | −1 | 32 | |||||||||||||
| M148R | −476 | 11 | 3 | 1 | 2 | −1 | 217 | −211 | 19 | |||||||
| M149R | Deleted | 63 | ||||||||||||||
| M150R | Deleted | 1 | −3 | 104 | 2 | |||||||||||
| M151R | Deleted | —b | 2 | |||||||||||||
| M152R | Deleted | Deleted | ||||||||||||||
| M153R | Deleted | Deleted | 1 | |||||||||||||
| M154L | Deleted | Deleted | ||||||||||||||
| M156R | Deleted | Deleted | ||||||||||||||
In-frame fusion of truncated ORFs M006L and M009L.
In-frame fusion of truncated ORFs M151R and M001R.
The genome of the ZA challenge strain is 161,609 bp long with an AT content of 56.46% (Table 2). The genome is 168 bp shorter than that of strain Lausanne. There are 331 amino acid differences in 25 predicted MYXV proteins (Table 3). All of the 170 ORFs assigned to the Lausanne strain are present, and 146 of the predicted viral proteins (85.38%) are identical to those of the Lausanne strain (Table 3). Compared to the Lausanne strain, we detected a substitution of 50 aa as well as a loss of 281 aa in coding regions due to frameshift mutations. The genome of the field isolate Munich-1 is 150,884 bp long with an AT content of 56.72% (Table 2). The genome is 10.9 kb shorter than that of the Lausanne strain. In strain Munich-1, 160 of the 170 ORFs assigned to the Lausanne strain are present, whereas 10 ORFs are deleted and 2 are severely truncated inside or in close proximity to the left TIR. Six of the deleted ORFs encode proteins with immunomodulatory functions. The deletion spans from the end of M006L to the beginning of M016L. Gene M006L encodes a putative E3 ubiquitin ligase of 509 aa (19), whereas the function of the 77-aa M016 protein is unknown. As a consequence, the truncated ORFs M006L and M016L are fused in frame, whereas M007L, M008L, M008.1L, M009L, M010L, M011L, M012L, M013L, M014L, and M015L are missing. At the amino acid level, there are 464 differences in 24 predicted proteins consisting of 34 amino acid substitutions, 1 insertion, and 429 deletions compared to the Lausanne strain. The majority of these amino acids were lost because of the in-frame fusion of ORFs M006L and M016L. The remaining 9 amino acids were lost by premature stop codons in the corresponding ORFs. Thus, 137 predicted viral proteins (85.09%) are identical to those of the Lausanne strain, 18 (11.18%) revealed a 1-aa substitution, and 4 (2.48%) revealed a substitution of 2 aa (Tables 3 and 4). The genome of strain FLI-H consists of 161,790 bp with an AT content of 56.47% (Table 2). This genome is 13 bp longer than that of the Lausanne strain. All 170 ORFs assigned to the Lausanne strain are present. At the amino acid level, there are 382 differences in 33 predicted proteins involving 324 amino acid deletions, caused by frameshift mutations, and 58 amino acid substitutions compared to the Lausanne strain (Table 3). Therefore, 80.7% of these predicted viral proteins are identical to those of the Lausanne strain. The genome of field strain 2604 is 161,715 bp, which is 62 bp smaller than that of the Lausanne strain. The AT content is 56.47% (Table 2). All 170 ORFs assigned to the Lausanne strain are present. At the amino acid level, there are 276 differences in 37 predicted viral proteins. Thus, 134 (78.36%) of these predicted viral proteins are identical to those of the Lausanne strain (Table 3). Finally, the genome of strain 3207 is 161,799 bp, which is 22 bp longer than that of the Lausanne strain, and has an AT content of 56.47% (Table 2). All 170 ORFs assigned to the Lausanne strain are present. At the amino acid level, there are 1,038 differences in 36 predicted viral proteins compared to the Lausanne strain. Therefore, 135 (78.95%) of these predicted viral proteins are identical to those of the Lausanne strain (Table 3).
TIR.
The genome of MYXV can be divided into 3 parts: the central part and the 2 flanking TIRs. The length of the TIR regions varies from 11,577 bp in strain Lausanne to 15,464 bp in strain MSW (19). The left and right TIRs of strain Lausanne consist of 11,577 bp each (Table 3) and include 12 ORFs duplicated in each TIR. The mechanism determining the length of the TIRs is not known; however, changes in the size of the TIRs can occur readily (19). The TIRs of field strains FLI-H, 2604, and 3207 and challenge strain ZA are slightly shorter than those of the Lausanne strain (∼11,450 bp each) (Table 3). Each TIR incorporates all homologs of the genes present in strain Lausanne. No deletions or disruptions are observed. In strain Munich-1, the boundary of the TIRs has shifted due to deletions. The M006L gene is truncated, whereas the homologs of M007L (M-T7), M008L, and M008.1L (SERP-1) are deleted compared to the Lausanne strain. Vaccine strain MAV reveals the deletion of 11 ORFs and the truncation of 3 ORFs inside or in close proximity to the left and right TIRs. Ten of the deleted ORFs encode proteins with immunomodulatory function. The TIRs consist of 7,681 bp. In the left TIR of strain MAV, the deletion spans from the beginning of M006L to the end of M009L. The consequences of this deletion are the absence of ORFs M007L, M008L, and M008.1L as well as an in-frame fusion of the truncated ORFs M006L and M009L flanking the deletion. The M006L and M009L genes encode putative E3 ubiquitin ligases of 509 aa each. The strain MAV fusion gene aligns with the last 1,276 nucleotides (nt) of the strain Lausanne M006L sequence; after this, the next 302 nt align with the beginning of the strain Lausanne M009L sequence. The ORF, however, encodes the first 101 aa of M009 and the last 425 aa of the M006 protein. The impact of the M009L mutation on MYXV virulence is unknown since the virulent field isolates analyzed in this study and recently reported Australian isolates also reveal a disruption of the M009L ORF (6). The M008L gene is predicted to encode an E3 ubiquitin ligase and is missing in strain MAV. Finally, ORFs M007L and M008.1L encode proteins with immunomodulatory functions. M007L encodes the interferon gamma (IFN-γ) receptor homolog M-T7, which downregulates inflammation (20). M008.1L encodes the secreted serine protease inhibitor SERP1 and has a major role in virulence. It belongs to the serpin family of serine protease inhibitors, which downregulate the inflammatory response to virus infection (21). In the right TIR of strain MAV, the deletion spans from the first one-third of M148R to M008.1R. Because of this deletion, M148R is severely truncated, and M149R, M150R, M151R, M152R, M153R, M154L, M156R, and M008.1R are deleted completely. All encoded proteins play a role in immunomodulation (21–28).
Immunomodulatory proteins.
Poxviruses encode a vast variety of immunomodulatory proteins aimed at circumventing the host's immune defense and ensuring successful viral replication. Understanding the mechanisms by which poxviruses evade and disrupt the immune system is essential for the design and production of vaccines and therapeutics. Some of these secreted immunomodulatory proteins are already expressed in heterologous systems and used for the treatment of inflammatory conditions (29). Immunomodulatory proteins aim at various targets in the cell and recognize immune targets in rabbits as well as in mice or humans. The predicted proteins can be divided into 6 major groups (Table 5): MYXV proteins with antiapoptotic functions, MYXV serpins that inhibit cellular proinflammatory or proapoptotic proteases, MYXV proteins that interfere with leukocyte chemotaxis, MYXV proteins that interfere with leukocyte activation, MYXV proteins with sequence similarity to human immunodeficiency virus (HIV) proteins, and MYXV proteins with other immune functions. Compared to MYXV Lausanne, the following proteins show major differences between the sequenced strains and MYXV Lausanne.
TABLE 5.
Genes of the sequenced MYXV strains and RFV encoding immunomodulatory proteins with homologs in MYXV Lausannea
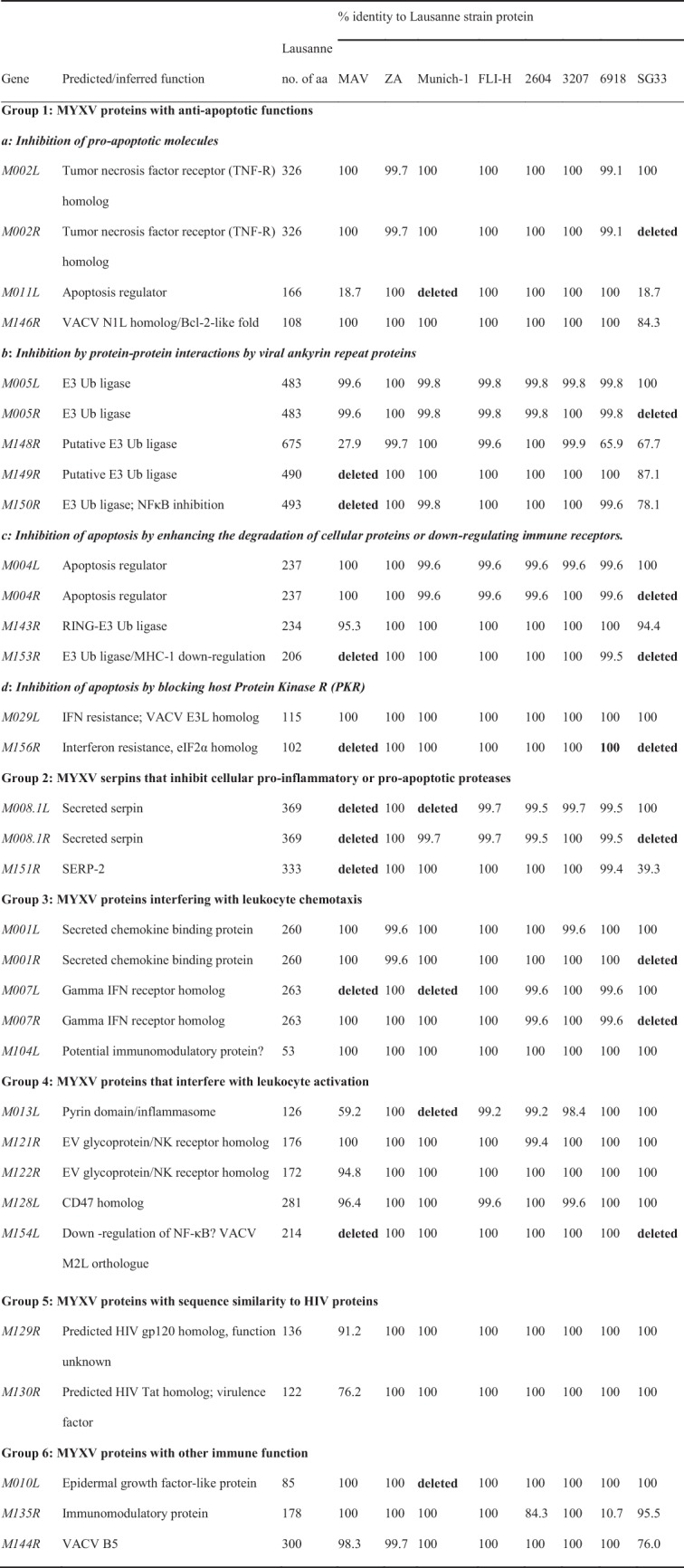 |
Ub, ubiquitin.
Proteins with antiapoptotic functions. (i) Inhibition of proapoptotic molecules.
Three proteins inhibit proapoptotic molecules: M-T2 (M002L/R), M011, and M146. M-T2 is a tumor necrosis factor receptor homolog (30). In strain SG33, M002R is deleted completely, but due to the presence of M002L, the protein is still expressed. The M011L gene product prevents the loss of mitochondrial membrane potential (31). In strains MAV and SG33, the predicted protein is severely truncated due to a premature stop codon and is 33 aa.
(ii) Inhibition of protein-protein interactions by viral ankyrin repeat proteins.
M148R and M149R encode members of the ankyrin repeat (ANK) family of poxviruses involved in protein-protein interactions and encode a putative E3 ubiquitin ligase (22). The M148 protein of strain MAV is severely truncated due to a premature stop codon and the resulting frameshift mutation. We also observed a frameshift mutation in strain 6918. The predicted protein is also severely truncated. In strain MAV, the M149 protein is missing completely, whereas in strain SG33, 63 aa are replaced. M150R encodes myxoma nuclear factor (MNF), which is critical for productive viral infection in rabbits, since its deletion generates an almost apathogenic virus that still replicates in cells (23). In strain MAV, the ORF is deleted completely. In strain SG33, 104 amino acid substitutions are present in the resulting protein.
(iii) Inhibition of apoptosis by enhancing the degradation of cellular proteins or downregulating immune receptors.
M004L/R (i.e., M004L and M004R) encodes the intracellular virulence factor M-T4. This protein modulates the inflammatory response to virus infection and inhibits the apoptosis of infected lymphocytes (32–34) in a way that is still poorly understood. In strain SG33, M004R is deleted completely. M153R encodes the endoplasmic reticulum (ER)-resident protein MV-LAP (myxoma virus leukemia-associated protein). MV-LAP acts as a ubiquitin ligase and promotes the downregulation of the surface-bound molecules major histocompatibility complex class I (MHC-I) and CD4 by targeting them for degradation within the lysosome (35). In both vaccine strains MAV and SG33, the whole ORF is missing.
(iv) Inhibition of apoptosis by blocking host protein kinase R.
The M156R protein is a structural mimic of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α) and inhibits protein kinase R (PKR) (36). This ORF is missing in vaccine strains MAV and SG33.
(v) MYXV serpins that inhibit cellular proinflammatory or proapoptotic proteases.
M008.1L/R codes for the protein SERP-1, which belongs to the serpin family of serine protease inhibitors. Serpins are widely used in eukaryotic systems to regulate proteinase-dependent processes. SERP-1 is secreted from infected cells and downregulates the inflammatory response to virus infections by inhibiting serine proteases irreversibly. Rabbits infected with MYXV deficient in SERP-1 show inflammation, less severe secondary bacterial infections, and significant recovery at 14 days postinfection. When rechallenged with the parental MYXV, rabbits are resistant to myxomatosis (21). M008.1L/R is missing in vaccine strain MAV (due to large deletions at the left and right ends of the genome). In strains SG33 and Munich-1, either M008.1L or M008.1R is present. The ORF encoding SERP-2 is deleted in vaccine strain MAV and severely truncated in vaccine strain SG33 due to a deletion in the right end of the genome.
(vi) MYXV proteins interfering with leukocyte chemotaxis.
M007L/R encodes the IFN-γ receptor homolog M-T7, which downregulates inflammation (20, 37). In strains MAV, Munich-1, and SG33, one copy is deleted.
(vii) MYXV proteins that interfere with leukocyte activation.
M013L encodes a pyrin domain-containing MYXV protein that binds to the inflammasome and inhibits the activation of interleukin-1β (IL-1β) and IL-18 (38). The M013 protein also inhibits cellular NF-κB signaling, which regulates the secretion of proinflammatory cytokines (39). The whole ORF is missing in strain Munich-1 and is severely truncated in strain MAV due to a premature stop codon. M154R encodes a protein with homology to the VACV M2 protein. There is little information about the VACV M2 protein since it is unique to poxviruses. The VACV M2 protein is produced early during infection and is sequestered in the ER (40). During viral replication, high levels of viral proteins cause ER stress. To circumvent this problem, ER overload response (EOR) and unfolded-protein response (UPR) pathways become activated (41). The M2 protein of VACV blocks an early event in the EOR pathway to ensure successful viral infection. The coding ORF M154R is deleted in both vaccine strains MAV and SG33.
(viii) MYXV proteins with sequence similarity to HIV proteins.
M129R and M130R share partial sequence similarity with key proteins of HIV, which play a role in virus entry and decreased apoptosis (42, 43). In vaccine strain MAV, 15 aa are replaced in the M129 protein (91.2% amino acid identity to the Lausanne strain), and 24 aa are replaced in the M130 protein.
(ix) MYXV proteins with other immune functions.
M010L codes for a secreted epidermal growth factor, which is necessary for successful infection in vivo. M010L is missing in Munich-1. M135R is an important virulence factor of MYXV with unknown functions (44). In strain 2604, a frameshift mutation results in the reduced amino acid identity of the M135 protein (82%) to the Lausanne strain. In strain 6918, the M135 protein is severely truncated due to a frameshift mutation. M144R shows similarity to VACV C3L and B5R, whose encoded proteins have complement-binding and structural functions, respectively (43). We observed amino acid substitutions only in strain SG33.
Both vaccine strains MAV and SG33 reveal the deletion of 9 ORFs, whereas strain Munich-1 shows the deletion of 5 ORFs encoding proteins with immunomodulatory functions. Some deletions are shared (Table 5). Taking a more detailed look by considering the duplication of ORFs at both ends of the genome, vaccine strain MAV reveals the absence of the immunomodulatory proteins M008.1, M149, M150, M151, M153, M154, and M156. The IFN-γ receptor homolog is still expressed by M007R. In vaccine strain SG33, only proteins M153, M154, and M156 are missing. In strain Munich-1, proteins M10, M11, and M13 are missing.
Overall, it seems likely that the M148 protein plays an important role in the attenuation of vaccine strains MAV and SG33 as well as in Spanish field strain 6918. In strains MAV and 6918, the ORF is severely truncated due to frameshift mutations and shows 27.9% amino acid identity (strain MAV) or 65.9% a amino acid identity (strain 6918) to the Lausanne strain. In vaccine strain SG33, 32.3% of the ORF is mutated, resulting in a protein with 67.7% amino acid identity to the Lausanne strain. Previously, it was shown that the deletion of M148R results in a faster resolution of inflammation and that the M148 protein is partially localized in the nucleus (22).
Envelope proteins.
The antibodies that provide protection from MYXV are not known, whereas VACV surface proteins that provide targets for neutralizing antibodies are well known (12). Since leporipoxviruses encode orthologs of the immunodominant envelope proteins of VACV, it can be assumed that neutralizing antibodies are also directed against these envelope proteins. Considering this, we analyzed these orthologous genes from the whole genomes of 9 leporipoxvirus strains, the 6 MYXV strains sequenced for this work (Table 2), SG33, 6918, and RFV and compared them to that of the Lausanne strain (Table 6; see also Fig. S1 in the supplemental material). Only RFV and vaccine strain MAV showed significant amounts of amino acid substitutions and deletions in the envelope proteins affecting potentially antigenic sites and transmembrane regions (Fig. S1a to S1g). One frameshift mutation was detected in MYXV MAV M083L (Fig. S1d). This ORF codes for the M083 protein (VACV D8) and reveals a frameshift mutation due to the insertion of 2 nucleotides. The resulting protein is truncated and is only 158 aa. Due to this truncation, the transmembrane region (aa 242 to 269 in MYXV Lausanne) is lost. Considering this, it seems to be impossible for the M083 protein to be located in the envelope of strain MAV.
TABLE 6.
Immunodominant MYXV, VACV, and RFV envelope proteinsc
| MYXV gene | VACV ortholog | RFV ortholog | Function of gene product | No. of aa of Lausanne strain protein | % identity to Lausanne strain protein |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAV | ZA | Munich | FLI-H | 2604 | 3207 | 6918 | SG33 | RFV | |||||
| M022L | F13L | S13L | EV protein | 371 | 96.2 | 100 | 100 | 100 | 100 | 100 | 99.7 | 100 | 93.0a |
| M055R | L1R | S055R | Structural protein | 242 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 96.3 |
| M071L | H3L | S071L | Structural protein | 324 | 100 | 100 | 100 | 100 | 100 | 100 | 99.7 | 100 | 90.4 |
| M083L | D8L | S083L | Carbonic anhydrase homolog/structural protein? | 286 | 52.8a | 100 | 100 | 100 | 100 | 100 | 100 | 98.6a | 84.3 |
| M107L | A17L | S107L | MV membrane protein | 200 | 100 | 100 | 100 | 100 | 100 | 100 | 99.5 | 100 | 83.0a |
| M115L | A27L | S115L | Fusion protein/EV formation/MV surface protein | 188 | 76.6b | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 72.9a |
| M144R | B5R | Missing | EV spread | 300 | 98.3 | 99.7 | 100 | 100 | 100 | 100 | 100 | 76.7a | 76.0a |
Truncated compared to the MYXV Lausanne protein.
Lengthened compared to the MYXV Lausanne protein.
Shown is a comparison of amino acid sequences with those of orthologs in MYXV Lausanne.
Obviously, these mutations do not affect vaccination efficacy since vaccination with each virus protects rabbits from myxomatosis successfully. From our in silico analyses, we cannot conclude how these mutations affect the formation of transmembrane regions and the production of neutralizing antibodies.
Vaccine efficacy.
For drug approval, MYXV vaccines need to pass challenge tests to ensure both efficacy and safety. For drug approval in Germany, naive New Zealand White rabbits and those vaccinated with strain MAV (103.1 to 104.68 50% tissue culture infective doses [TCID50]; n = 50) were infected with the ZA challenge strain (106.0 TCID50) at 17 to 24 days postvaccination (Table 7). The vaccine was considered effective if 90% of the vaccinated animals showed a protective effect and at least 90% of the naive animals revealed symptoms of myxomatosis. Clinical signs of myxomatosis were observed in nonvaccinated animals only, starting with conjunctival inflammation and resulting in swollen head, closed eyelids, and impaired respiration. These rabbits developed the first symptoms of myxomatosis at 8 to 11 days postinfection, which resulted in full myxomatosis, with a mortality rate of 100%. The vaccinated animals did not show any signs of myxomatosis except for localized small nodules at the inoculation site of challenge strain ZA. However, despite the numerous deletions, insertions, and mutations in vaccine strain MAV, the vaccinated rabbits showed full protection (100% survival) against lethal challenges with strain ZA, which is closely related to the Lausanne strain (see below).
TABLE 7.
Vaccine efficacy of vaccine strain MYXV MAVd
| Virus batch | TCID50 for immunizationa | Day of challenge postinoculationb | Mortality (no. of dead animals/total no. of animals) |
|
|---|---|---|---|---|
| Vaccinated animals | Control animals | |||
| 249 06 92 | 103.1 | 21 | 0/4 | 2/2 |
| VM 07/92c | 103.11 | 26 | 0/4 | 2/2 |
| 147 01 91 | 103.21 | 21 | 0/4 | 2/2 |
| 08 04 90 | 103.46 | 24 | 0/2 | 2/2 |
| 268 08 92 | 103.75 | 17 | 0/4 | 2/2 |
| 177 05 91 | 104.20 | 21 | 0/2 | 2/2 |
| 368/369 08 93 | 104.25 | No data | 0/4 | 2/2 |
| 212 02 92 | 104.68 | 21 | 0/4 | 2/2 |
| 257 07 92 | 104.75 | 21 | 0/4 | 2/2 |
MYXV MAV TCID50 subcutaneously.
A total of 106.0 TCID50 of MYXV ZA subcutaneously.
Lyophilized virus.
Data for drug approval in Germany.
Recombination analysis and phylogenetic reconstruction.
The complete genome sequences of the 6 MYXV isolates (Table 2) were combined with all reported genome sequences of leporipoxviruses (32 MYXV isolates and 1 RFV isolate) (Table 1) isolated in Australia, Europe, and the United States. Multiple-sequence alignments of these 39 complete genomes were conducted with Clustal Omega (45, 46). We checked the sequences of strains 2604, 3207, FLI-H, MAV, Munich-1, and ZA for signals of recombination. To this end, we determined the similarity of these sequences to the other 33 sequences by using a customized R script. The sequences reported under GenBank accession numbers JX565562 to JX565569, JX565571 to JX565584, and KC660079 to KC660085 and the sequence of strain Lausanne exhibit a pairwise distance of at least 99.91%. Therefore, we chose strain Lausanne as a representative of this group, and for this group, the 6 query sequences were compared to that of the Lausanne strain only. The obtained similarity plots are shown in Fig. S2 in the supplemental material. The query sequences can be divided into three groups by the strength of the evidence for recombination that we found.
(i) Group 1.
The sequences of strains 2604 and Munich-1 show uniformly high sequence similarity to the Lausanne strain or one of the other query sequences throughout the whole genome; that is, they show no signs of recombination.
(ii) Group 2.
The sequences of strains 3207, FLI-H, and ZA exhibit the same uniform, high similarity to the Lausanne strain or the other query sequences for most parts of the genome but display several drops in similarity to the other query sequences by about 0.5 to 1.5% over a length of about 1 to 1.5 kbp each. The most pronounced example of this kind of feature can be observed at around position 73000 for the sequence of strain 3207. These kinds of drops can be explained by either convergence or recombination; that is, they could be due to the virus developing a distinct biological feature, manifesting in its genome differing in the parts of the genome encoding this feature. Alternatively, these drops could be the result of recombination between a sequence similar to that of the Lausanne strain and a sequence that is as yet unsampled and slightly different from that of MYXV Lausanne. This region of lower similarity harbors genes M071L and M072L, which encode a structural protein and an RNA polymerase I (Pol I)-associated transcription factor. Interestingly, a sliding Bayes analysis performed previously by Kerr et al. (19) revealed recombination events between M076R and M080R of strains MSW and SG33. It is currently unknown why these two regions of recombination are located at nearly the same position. However, the fact that the locations of these regions do not coincide for these three sequences indicates that we are probably facing recombination.
(iii) Group 3.
The sequence of strain MAV shows unequivocal signs of recombination between two sequences similar to those of strains Lausanne and MSW. No recombination with RFV was detected.
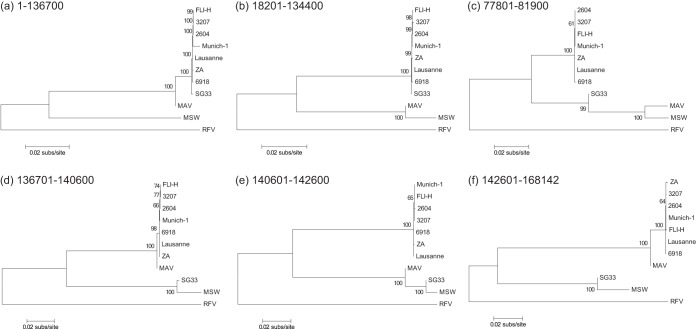
To conduct a phylogenetic analysis of the 11 sequences considered, we partitioned the genomes into six sets of coordinate intervals (denoted regions A to F), which share the same phylogenetic history for all sequences. These sets are 90,200 bp (region A), 46,400 bp (region B), 4,100 bp (region C), 3,900 bp (region D), 2,000 bp (region E), and 25,542 bp (region F), and Fig. S3 in the supplemental material shows how the 11 sequences analyzed are segmented by this partition. This is in line with the recombination pattern determined previously for strain SG33 (19). Since the implied variance in sequence similarity is small, we neglect the alleged recombination from which strains ZA, FLI-H, and 3207 emerged. For each of these putatively recombination-free regions, we built a phylogenetic tree (Fig. 1).
FIG 1.
Recombination in the investigated MYXV strains. Shown is the maximum likelihood phylogeny of 11 complete genome sequences of strains of MYXV and RFV. Separate maximum likelihood trees were estimated for the six sets of coordinate intervals that share the same phylogenetic history for all sequences (see Fig. S2 in the supplemental material). The trees were constructed with the putatively recombination-free regions of the multiple-sequence alignment and midpoint rooted. Bootstrap support values are shown for key nodes, and horizontal branch lengths are drawn at the scale of nucleotide substitutions per site.
For regions A, D, and F, which comprise the majority of the genome, strain MAV appears to be a close sister group of strain Lausanne. In contrast, strain MAV is more closely related to MSW in regions B, C, and E. Since strain MAV has an unclear history, our findings of the relatedness of strain MAV and the Californian strain are in line with a statement made previously by Müller et al. (15), who said that strain MAV was derived from a Californian MSD strain. Consequently, strain MAV is a recombinant of European and Californian strains. RFV was the most differing lineage and therefore represents an outgroup.
In this study, we sequenced and analyzed the genomes of 6 German strains and compared them to previously reported MYXV and RFV genomes. In our analyses, we focused on genes encoding proteins that play a role in immunomodulation, virulence, host range, and envelope composition. We showed that vaccine strain MAV and virulent field strain Munich-1 revealed the most mutations and deletions among the sequenced strains. These mutations and deletions were located mostly in the TIR and in genes encoding immunomodulatory proteins. Some envelope proteins were also affected. These numerous mutations and deletions probably resulted in the attenuation of vaccine strain MAV, whereas strain Munich-1 stayed virulent. Strain Munich-1 has not yet been phenotyped for virulence (grades 1 to 5). Despite these mutations, strain MAV fully protects rabbits against lethal MYXV challenges. Furthermore, we were able to show that vaccine strain MAV is a recombinant virus from European and Californian strains that reveals unequivocal signs of recombination throughout the whole genome. Understanding the mechanism of successful protection against MYXV infections is important for the evaluation of already existing MYXV vaccine strains and the construction of new vaccine strains.
MATERIALS AND METHODS
Viruses and cells.
The commercially available vaccine strain MAV was isolated in 1952 in California by McKercher and Saito. Attenuation of this virulent strain, referred to as MSD, was achieved by passaging the virus 44 times on rabbit kidney cells (47). The virulent challenge strain ZA was isolated from MYXV outbreaks in Poland in 1985 (48). The 4 German MYXV field strains were isolated from rabbits that died of myxomatosis. Strain Munich-1 was isolated from an unvaccinated wild rabbit in Munich (English Garden), Germany, in 1985. Strain FLI-H was isolated from a vaccinated rabbit kept as a pet in Greifswald, Germany, in 2004. Vaccination may have taken place within the incubation time of MYXV infection. Strains 2604 and 3207 were isolated in 2004 and 2007 from unvaccinated rabbits that tested positive for MYXV infection in accredited laboratories. Rabbit kidney cells (RK13; ATCC CCL-37) and African green monkey kidney cells (MA-104; ATCC CRL-2378.1) were grown in Dulbecco's modified Eagle medium (DMEM; Pan Biotech GmbH, Aidenbach, Germany) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen GmbH, Darmstadt, Germany), 100 U/ml penicillin, and 100 μg/ml streptomycin. Strains MAV, FLI-H, 2604, 3207, and ZA were propagated in RK13 cells cultured under the same conditions except for 2% FBS. Field isolate Munich-1 was propagated in MA-104 cells with 2% FBS. All MYXV strains were subjected to 5 rounds of plaque purification and passaged 5 times to prepare working stocks for sequencing and further analyses. The isolates of MYXV and RFV used in this study are described in Tables 1 and 2.
Preparation of genomic DNA.
Virus purification was performed as described previously (49–51), with modifications. MYXV-infected RK13 or MA-104 cells were harvested and collected by centrifugation at 13,000 × g for 2 h in a GS-3 rotor at 4°C. The cell pellet was homogenized in 10 mM Tris (pH 9.0) (Tris-EDTA [TE] buffer, pH 9.0 [TE9]) and subjected to 3 cycles of freeze-thawing in liquid nitrogen at −80°C and 37°C and sonication (20 kHz for 1 min at 4°C) (Sonoplus HD 2200; Bandelin Electronic, Berlin, Germany). Cell debris and nuclei were removed by centrifugation at 1,000 × g for 10 min at 4°C. The supernatant fluid was layered onto 10-ml sucrose cushions (36% [wt/vol] in TE9) and centrifuged at 110,000 × g for 90 min in an SW 28 rotor at 4°C. The pellet was homogenized in TE9 by sonication and purified by centrifugation at 40,000 × g for 90 min in an SW 40 rotor at 4°C on a 20%-40%-60% (wt/wt) (in TE9) sucrose step gradient. The virus was concentrated in the 40% sucrose layer, harvested, diluted in TE9, sonicated, and centrifuged again at 110,000 × g for 1 h in an SW 40 rotor at 4°C. The pellet was homogenized in TE9 by sonication and purified by centrifugation at 40,000 × g for 90 min in an SW 40 rotor at 4°C on a 20 to 60% (wt/wt) (in TE9) linear sucrose density gradient. The viral band appeared with ∼40% (wt/wt) sucrose and was harvested by lateral puncture and diluted in DNase I buffer (10 mM Tris-HCl, 2.5 mM MgCl2, 0.5 mM CaCl2 [pH 7.6]). Viruses were recovered by centrifugation at 110,000 × g for 1 h in an SW 40 rotor at 4°C. The pellet was homogenized in DNase I buffer. For digestion of contaminating mammalian DNA, dithiothreitol and DNase I were added to final concentrations of 5 mM and 100 U/ml, respectively, and the mixture was incubated at 37°C for 1 h. DNase I was then heat inactivated at 75°C for 10 min. Lysis of virions was performed for 6 h with gentle agitation, as described previously (52). DNA purification was performed by classical phenol-chloroform extraction and isopropanol precipitation, as described previously (53). Genomic DNA was resuspended in a total volume of 500 μl 10 mM Tris-HCl (pH 8.5).
DNA sequencing and assembly.
Whole-genome shotgun sequencing of the MYXV genomes was performed by using a 454 GS-FLX system (Roche 454 Life Science, Mannheim, Germany). The initial assembly was performed with Roche Newbler 2.3 FLX assembler software (454 Life Sciences, Roche Applied Science, Branford, CT), using MYXV Lausanne as the reference genome. In addition, viral DNA was prepared (ZR Viral DNA kit; Zymo Research, Irvine, CA, USA) and sent for Sanger sequencing to close the remaining gaps. Sequence editing was performed by using the GAP4 program in the Staden software package (54). Automatic gene prediction and annotation were carried out with the aid of the Genome Annotation Transfer Utility (GATU) (55) and the Artemis software package (56). The annotated ORFs were aligned with the corresponding ORFs in strain Lausanne (GenBank accession number AF170726) by using MegAlign from the Lasergene DNASTAR version 10.1.2.20 software package (DNASTAR Inc., Madison, WI, USA).
Analysis of envelope proteins.
The MYXV orthologs of VACV envelope proteins were analyzed by multiple-sequence alignments using MegAlign. First, potentially antigenic regions of the protein sequence, with a minimum length of 7 aa, were predicted by using the method of Kolaskar and Tongaonkar (57). This method is based on the occurrence of hydrophobic residues (Cys, Leu, and Val) on the surface of a protein. Sites composed of 1 of these amino acids are more likely to form part of the antigenic sites. In a second step, the propensities to form transmembrane regions were calculated by using an algorithm described previously by Persson and Argos (58). Prediction of potentially antigenic regions and calculation of propensities to form transmembrane regions were conducted with EMBOSS 1.5 (59).
Multiple-sequence alignment and similarity analysis.
Multiple-sequence alignments were performed with nucleotide sequences by using Clustal Omega (45, 46). To perform similarity analysis (pairwise comparison of viral sequences), we used a sliding-window approach as employed in Simplot (3). As opposed to Simplot, the R script that we used indicates the genome regions in which a sequence is located compared to a large fraction composed of gaps, and the sequence is therefore excluded from the analysis of the respective genome region. We used a window size of 1,000 to smooth the distances, employing the Jukes-Cantor (JC) distance and excluding gaps for the calculation of distance. The step size for plotting was set to 200. A sequence was excluded from analysis in a window when the number of positions at which it has a gap exceeds 20% in that window.
Phylogenetic analysis.
We used the MEGA 6.0 package to build phylogenetic trees of the putatively recombination-free regions of the genome. In preparation, we employed jModelTest 2.0 (60) to determine the site-wise substitution model, using Bayesian information criterion (BIC) as a selection criterion. The model selected as the best model was the transversional model plus gamma distribution (TVM+G), i.e., a substitution model with variable base frequencies, variable transversion rates, equal transition rates, and gamma-distributed site-wise variation rates. As MEGA 6 does not allow the choice of a TVM+G model as a site-wise substitution model, a general time-reversible model with gamma-distributed rates (GTR+G model) was used as the best approximation. To assess the influence of model uncertainty on the phylogenetic inference (5), we constructed two trees for each of the six recombination-free regions, one using the JC model as a substitution model and the other using the GTR+G model. As we observed a difference in the tree topology for only the shortest region (2,000 bp; nt 140601 to 142600) that we identified as being recombination free, and this difference was a minor one, we conclude that the choice of the substitution model has a negligible influence on the results of our analysis. We constructed maximum likelihood trees using a JC model with uniform rates and a GTR+G model with five categories for the gamma distribution. The number of bootstrap replications was set to 500, and partial deletion with a site coverage cutoff of 95% was used for the gap treatment.
Ethics statement.
The animal experiments for approval of the vaccine Cunivak Myxo were performed in accordance with the regulations of the German Animals Protection Act as amended on 8 August 1986 (64), which complies with European Union guidelines on the welfare of animals used in research (65). The experiments were authorized by the government of the federal state of Sachsen-Anhalt (Ministry of Food, Agriculture, and Forestry) under reference numbers 25-Dr.Ju/Gr A12/91, 18 June 1991 (assessment of protection dosage; Cunivak Myxo; rabbit 01/91), and 25-Dr.Ju/Gr A13/91, 18 June 1991 (potency testing; Cunivak Myxo; rabbit 02/91).
Vaccine efficacy.
Vaccine strain MAV is an approved MYXV vaccine and widely used in Germany (Cunivak Myxo). For drug approval, the recommended dose of vaccine strain MAV as well as the onset and duration of immunity were established experimentally (data not shown). Therefore, the efficacy of strain MAV in New Zealand White rabbits (Charles River GmbH, Germany) was tested in vaccinated animals (group I) compared to nonvaccinated rabbits (group II), and the test was repeated several times. The animals of group I (n = 2 to 4) received 1 vaccine dose (103.1 to 104.68 TCID50) of Cunivak Myxo dissolved in 1 ml of diluent for viral vaccines, which was inoculated subcutaneously into the chest wall of each rabbit. Rabbits of group II (n = 2) received no vaccination. The TCID50 was determined as described previously by Spearman and Kärber (61, 62). Daily physical examinations were conducted on each rabbit for clinical signs of myxomatosis, e.g., conjunctival inflammation, swollen head, closed eyelids, and impaired respiration. Both groups received 1 ml (106.0 TCID50) of challenge strain ZA inoculated into the chest wall of each rabbit subcutaneously at 17 to 24 days postvaccination. Additionally, 0.1 ml of challenge strain ZA was applied to each conjunctiva of the lower eyelid. Daily physical examinations were conducted on each rabbit until 42 days postvaccination.
Accession number(s).
The six new MYXV genome assemblies were submitted to GenBank and assigned accession numbers KP723386 to KP723391.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Lynne Riddles for completing the critical review of the manuscript.
Research reported in this publication was supported by Deutsche Forschungsgemeinschaft fellowship BU 2685/4-1. This work was funded by IDT Biologika GmbH. The funders had no role in study design, data collection and analysis (except for vaccine efficacy testing), decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01570-16.
REFERENCES
- 1.Fenner F, Ratcliffe FN. 1965. Myxomatosis. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 2.Kerr PJ, Rogers MB, Fitch A, Depasse JV, Cattadori IM, Twaddle AC, Hudson PJ, Tscharke DC, Read AF, Holmes EC, Ghedin E. 2013. Genome scale evolution of myxoma virus reveals host-pathogen adaptation and rapid geographic spread. J Virol 87:12900–12915. doi: 10.1128/JVI.02060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr PJ, Liu J, Cattadori I, Ghedin E, Read AF, Holmes EC. 2015. Myxoma virus and the leporipoxviruses: an evolutionary paradigm. Viruses 7:1020–1061. doi: 10.3390/v7031020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton KP, Nicieza I, de Llano D, Gullon J, Inza M, Petralanda M, Arroita Z, Parra F. 2015. Vaccine breaks: outbreaks of myxomatosis on Spanish commercial rabbit farms. Vet Microbiol 178:208–216. doi: 10.1016/j.vetmic.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Skinner MA, Buller RM, Damon IK, Lefkowitz EJ, McFadden G, McInnes CJ, Mercer AA, Moyer RW, Upton C. 2012. Family: Poxviridae, p 291–309. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 6.Kerr PJ, Ghedin E, DePasse JV, Fitch A, Cattadori IM, Hudson PJ, Tscharke DC, Read AF, Holmes EC. 2012. Evolutionary history and attenuation of myxoma virus on two continents. PLoS Pathog 8:e1002950. doi: 10.1371/journal.ppat.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenner F. 1983. Biological control, as exemplified by smallpox eradication and myxomatosis. Proc R Soc Lond B Biol Sci 218:259–285. doi: 10.1098/rspb.1983.0039. [DOI] [PubMed] [Google Scholar]
- 8.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao JX, Macaulay C, Willer D, Evans D, McFadden G. 1999. The complete DNA sequence of myxoma virus. Virology 264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 9.Purcell DA, Clarke JK. 1972. Some aspects of the morphogenesis of myxoma virus in vivo. Arch Gesamte Virusforsch 39:369–375. doi: 10.1007/BF01241016. [DOI] [PubMed] [Google Scholar]
- 10.Stanford MM, Werden SJ, McFadden G. 2007. Myxoma virus in the European rabbit: interactions between the virus and its susceptible host. Vet Res 38:299–318. doi: 10.1051/vetres:2006054. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Honeychurch KM, Yang G, Byrd CM, Harver C, Hruby DE, Jordan R. 2009. Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis. Virol J 6:44. doi: 10.1186/1743-422X-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moss B. 2011. Smallpox vaccines: targets of protective immunity. Immunol Rev 239:8–26. doi: 10.1111/j.1600-065X.2010.00975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irwin CR, Evans DH. 2012. Modulation of the myxoma virus plaque phenotype by vaccinia virus protein F11. J Virol 86:7167–7179. doi: 10.1128/JVI.06936-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firth C, Kitchen A, Shapiro B, Suchard MA, Holmes EC, Rambaut A. 2010. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol 27:2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller A, Silva E, Abrantes J, Esteves PJ, Ferreira PG, Carvalheira JC, Nowotny N, Thompson G. 2010. Partial sequencing of recent Portuguese myxoma virus field isolates exhibits a high degree of genetic stability. Vet Microbiol 140:161–166. doi: 10.1016/j.vetmic.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 16.Marlier D. 2010. Vaccination strategies against myxomavirus infections: are we really doing the best? Tijdschr Diergeneeskd 135:194–198. [PubMed] [Google Scholar]
- 17.Morales M, Ramirez MA, Cano MJ, Parraga M, Castilla J, Perez-Ordoyo LI, Torres JM, Barcena J. 2009. Genome comparison of a nonpathogenic myxoma virus field strain with its ancestor, the virulent Lausanne strain. J Virol 83:2397–2403. doi: 10.1128/JVI.02189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camus-Bouclainville C, Gretillat M, Py R, Gelfi J, Guerin JL, Bertagnoli S. 2011. Genome sequence of SG33 strain and recombination between wild-type and vaccine myxoma viruses. Emerg Infect Dis 17:633–638. doi: 10.3201/eid1704.101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr PJ, Rogers MB, Fitch A, Depasse JV, Cattadori IM, Hudson PJ, Tscharke DC, Holmes EC, Ghedin E. 2013. Comparative analysis of the complete genome sequence of the California MSW strain of myxoma virus reveals potential host adaptations. J Virol 87:12080–12089. doi: 10.1128/JVI.01923-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boomker JM, Luttikhuizen DT, Veninga H, de Leij LF, The TH, de Haan A, van Luyn MJ, Harmsen MC. 2005. The modulation of angiogenesis in the foreign body response by the poxviral protein M-T7. Biomaterials 26:4874–4881. doi: 10.1016/j.biomaterials.2004.11.059. [DOI] [PubMed] [Google Scholar]
- 21.Macen JL, Upton C, Nation N, McFadden G. 1993. SERP1, a serine proteinase inhibitor encoded by myxoma virus, is a secreted glycoprotein that interferes with inflammation. Virology 195:348–363. doi: 10.1006/viro.1993.1385. [DOI] [PubMed] [Google Scholar]
- 22.Blanie S, Mortier J, Delverdier M, Bertagnoli S, Camus-Bouclainville C. 2009. M148R and M149R are two virulence factors for myxoma virus pathogenesis in the European rabbit. Vet Res 40:11. doi: 10.1051/vetres:2008049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camus-Bouclainville C, Fiette L, Bouchiha S, Pignolet B, Counor D, Filipe C, Gelfi J, Messud-Petit F. 2004. A virulence factor of myxoma virus colocalizes with NF-kappaB in the nucleus and interferes with inflammation. J Virol 78:2510–2516. doi: 10.1128/JVI.78.5.2510-2516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacNeill AL, Turner PC, Moyer RW. 2006. Mutation of the myxoma virus SERP2 P1-site to prevent proteinase inhibition causes apoptosis in cultured RK-13 cells and attenuates disease in rabbits, but mutation to alter specificity causes apoptosis without reducing virulence. Virology 356:12–22. doi: 10.1016/j.virol.2006.07.049. [DOI] [PubMed] [Google Scholar]
- 25.Guerin JL, Gelfi J, Camus C, Delverdier M, Whisstock JC, Amardeihl MF, Py R, Bertagnoli S, Messud-Petit F. 2001. Characterization and functional analysis of Serp3: a novel myxoma virus-encoded serpin involved in virulence. J Gen Virol 82:1407–1417. doi: 10.1099/0022-1317-82-6-1407. [DOI] [PubMed] [Google Scholar]
- 26.Guerin JL, Gelfi J, Boullier S, Delverdier M, Bellanger FA, Bertagnoli S, Drexler I, Sutter G, Messud-Petit F. 2002. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J Virol 76:2912–2923. doi: 10.1128/JVI.76.6.2912-2923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gedey R, Jin XL, Hinthong O, Shisler JL. 2006. Poxviral regulation of the host NF-kappaB response: the vaccinia virus M2L protein inhibits induction of NF-kappaB activation via an ERK2 pathway in virus-infected human embryonic kidney cells. J Virol 80:8676–8685. doi: 10.1128/JVI.00935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramelot TA, Cort JR, Yee AA, Liu F, Goshe MB, Edwards AM, Smith RD, Arrowsmith CH, Dever TE, Kennedy MA. 2002. Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha. J Mol Biol 322:943–954. doi: 10.1016/S0022-2836(02)00858-6. [DOI] [PubMed] [Google Scholar]
- 29.Smith GL, Benfield CT, Maluquer de Motes C, Mazzon M, Ember SW, Ferguson BJ, Sumner RP. 2013. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol 94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 30.Schreiber M, Rajarathnam K, McFadden G. 1996. Myxoma virus T2 protein, a tumor necrosis factor (TNF) receptor homolog, is secreted as a monomer and dimer that each bind rabbit TNFalpha, but the dimer is a more potent TNF inhibitor. J Biol Chem 271:13333–13341. doi: 10.1074/jbc.271.23.13333. [DOI] [PubMed] [Google Scholar]
- 31.Everett H, Barry M, Sun X, Lee SF, Frantz C, Berthiaume LG, McFadden G, Bleackley RC. 2002. The myxoma poxvirus protein, M11L, prevents apoptosis by direct interaction with the mitochondrial permeability transition pore. J Exp Med 196:1127–1139. doi: 10.1084/jem.20011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barry M, Hnatiuk S, Mossman K, Lee SF, Boshkov L, McFadden G. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239:360–377. doi: 10.1006/viro.1997.8894. [DOI] [PubMed] [Google Scholar]
- 33.Hnatiuk S, Barry M, Zeng W, Liu L, Lucas A, Percy D, McFadden G. 1999. Role of the C-terminal RDEL motif of the myxoma virus M-T4 protein in terms of apoptosis regulation and viral pathogenesis. Virology 263:290–306. doi: 10.1006/viro.1999.9946. [DOI] [PubMed] [Google Scholar]
- 34.Zuñiga MC. 2002. A pox on thee! Manipulation of the host immune system by myxoma virus and implications for viral-host co-adaptation. Virus Res 88:17–33. doi: 10.1016/S0168-1702(02)00118-1. [DOI] [PubMed] [Google Scholar]
- 35.Mansouri M, Bartee E, Gouveia K, Hovey Nerenberg BT, Barrett J, Thomas L, Thomas G, McFadden G, Fruh K. 2003. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J Virol 77:1427–1440. doi: 10.1128/JVI.77.2.1427-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng C, Haller SL, Rahman MM, McFadden G, Rothenburg S. 2016. Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc Natl Acad Sci U S A 113:3855–3860. doi: 10.1073/pnas.1515613113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalani AS, Graham K, Mossman K, Rajarathnam K, Clark-Lewis I, Kelvin D, McFadden G. 1997. The purified myxoma virus gamma interferon receptor homolog M-T7 interacts with the heparin-binding domains of chemokines. J Virol 71:4356–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston JB, Barrett JW, Nazarian SH, Goodwin M, Ricciuto D, Wang G, McFadden G. 2005. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity 23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Rahman MM, Mohamed MR, Kim M, Smallwood S, McFadden G. 2009. Co-regulation of NF-kappaB and inflammasome-mediated inflammatory responses by myxoma virus pyrin domain-containing protein M013. PLoS Pathog 5:e1000635. doi: 10.1371/journal.ppat.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinthong O, Jin XL, Shisler JL. 2008. Characterization of wild-type and mutant vaccinia virus M2L proteins' abilities to localize to the endoplasmic reticulum and to inhibit NF-kappaB activation during infection. Virology 373:248–262. doi: 10.1016/j.virol.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufman RJ. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 42.Zocchi MR, Rubartelli A, Morgavi P, Poggi A. 1998. HIV-1 Tat inhibits human natural killer cell function by blocking L-type calcium channels. J Immunol 161:2938–2943. [PubMed] [Google Scholar]
- 43.Barrett JW, Cao JX, Hota-Mitchell S, McFadden G. 2001. Immunomodulatory proteins of myxoma virus. Semin Immunol 13:73–84. doi: 10.1006/smim.2000.0298. [DOI] [PubMed] [Google Scholar]
- 44.Spiesschaert B, McFadden G, Hermans K, Nauwynck H, Van de Walle GR. 2011. The current status and future directions of myxoma virus, a master in immune evasion. Vet Res 42:76. doi: 10.1186/1297-9716-42-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKercher DG, Saito JK. 1964. An attenuated live virus vaccine for myxomatosis. Nature 202:933–934. doi: 10.1038/202933a0. [DOI] [PubMed] [Google Scholar]
- 48.Gorski J, Mizak B, Chrobocinska M. 1994. Control of rabbit myxomatosis in Poland. Rev Sci Tech 13:869–879. [DOI] [PubMed] [Google Scholar]
- 49.Czerny CP, Mahnel H. 1990. Structural and functional analysis of orthopoxvirus epitopes with neutralizing monoclonal antibodies. J Gen Virol 71(Part 10):2341–2352. [DOI] [PubMed] [Google Scholar]
- 50.Dalton KP, Ringleb F, Martin Alonso JM, Parra F. 2009. Rapid purification of myxoma virus DNA. J Virol Methods 162:284–287. doi: 10.1016/j.jviromet.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Smallwood SE, Rahman MM, Smith DW, McFadden G. 2010. Myxoma virus: propagation, purification, quantification, and storage. Curr Protoc Microbiol Chapter 14:Unit 14A.1. doi: 10.1002/9780471729259.mc14a01s17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petit F, Boucraut-Baralon C, Py R, Bertagnoli S. 1996. Analysis of myxoma virus genome using pulsed-field gel electrophoresis. Vet Microbiol 50:27–32. doi: 10.1016/0378-1135(96)00014-4. [DOI] [PubMed] [Google Scholar]
- 53.Moore D, Dowhan D. 2002. Purification and concentration of DNA from aqueous solutions. Curr Protoc Mol Biol Chapter 2:Unit 2.1A. doi: 10.1002/0471142727.mb0201as59. [DOI] [PubMed] [Google Scholar]
- 54.Staden R, Beal KF, Bonfield JK. 2000. The Staden package, 1998. Methods Mol Biol 132:115–130. [DOI] [PubMed] [Google Scholar]
- 55.Tcherepanov V, Ehlers A, Upton C. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 57.Kolaskar AS, Tongaonkar PC. 1990. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 276:172–174. doi: 10.1016/0014-5793(90)80535-Q. [DOI] [PubMed] [Google Scholar]
- 58.Persson B, Argos P. 1994. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol 237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 59.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 60.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spearman C. 1908. The method of ‘right and wrong cases’ (‘constant stimuli’) without Gauss's formula. Br J Psychol 2:227–242. doi: 10.1111/j.2044-8295.1908.tb00176.x. [DOI] [Google Scholar]
- 62.Kärber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch Exp Pathol Pharmakol 162:480–483. [Google Scholar]
- 63.Willer DO, McFadden G, Evans DH. 1999. The complete genome sequence of Shope (rabbit) fibroma virus. Virology 264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 64.Bundesgesetzblatt. 18 August 1986. Bundesgesetzblatt, Jahrgang 1986, Teil I. Bekanntmachung der Neufassung des Tierschutzgesetzes, p 1319–1329. [Google Scholar]
- 65.European Union. 24 November 1986. Council Directive 86/609/EEC on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes, p 1–28. http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:31986L0609. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.