ABSTRACT
An impediment to the understanding of cancer is the heterogeneous nature of cell populations within a tumor microenvironment. We reported a method to query protein signaling in single epithelial cells from formalin-fixed paraffin-embedded (FFPE) colorectal cancer tissues. Here, we discuss the feasibility and limitations of this approach for investigating signaling state heterogeneity.
KEYWORDS: Colorectal cancer, disaggregation, epithelial cells, FFPE, mass cytometry, solid tissues, single cell, systems biology, signal transduction, tumor heterogeneity
Intratumoral heterogeneity, as defined by the presence of cancer cells in different functional states, is an emerging hallmark of cancer that plays significant roles in therapeutic resistance, cancer recurrence, and metastasis.1 Single-cell analysis holds the promise to deconstruct this heterogeneity. Current technologies enable the profiling of individual cells in a quantitative manner, followed by mathematical grouping of high-dimensional data points into cellular subsets. The functional state of a cell is dictated by its signaling network interpretation of external cues to initiate a transcriptional program. While this program can be indirectly inferred by mapping transcribed gene sets to their corresponding transcription factors and upstream signaling pathways, a more attractive strategy is to directly measure the activation states of signaling proteins central to these pathways. Although bulk assays can achieve this goal in a multiplex fashion,2,3 a major obstacle in adapting signaling assays to single-cell studies, especially for solid tissues, is the substantial perturbation to cellular signaling pathways when single cells are disaggregated from an intact tissue. We recently developed an approach called DISSECT (Disaggregation for Intracellular Signaling in Single Epithelial Cells from Tissue) that enables evaluation of the native activation states of signaling proteins in single epithelial cells4 using multi-parameter flow or mass cytometry (Fig. 1A – Bottom).5 Using intestinal epithelium as a stereotypical example, we demonstrated the validity of DISSECT-generated single-cell results against those from conventional bulk assays. Mass cytometry represents a relatively high-throughput technique to query millions of cells that, when coupled to lower throughput but spatially resolved imaging approaches,6,7 is a powerful tool to address intratumoral heterogeneity.
Figure 1.
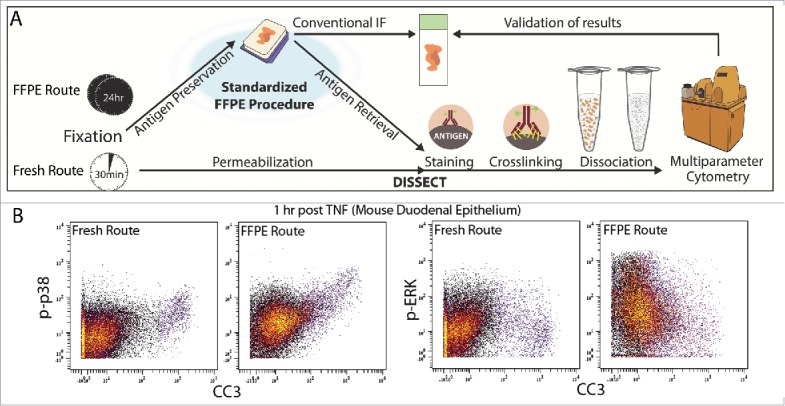
Mass cytometry-based single-cell analysis of epithelial signaling. (A) Comparative outline of the DISSECT (Disaggregation for Intracellular Signaling in Single Epithelial Cells from Tissue) approach applied to formalin-fixed paraffin-embedded (FFPE) (top) or freshly isolated (bottom) tissues. (B) Representative comparisons of signaling data acquired from fresh or FFPE tissues using DISSECT coupled to mass cytometry. CC3, cleaved caspase-3; p-ERK, phosphorylated extracellular signal–regulated kinases; p-p38, phosphorylated p38 mitogen-activated protein kinase; TNF, tumor necrosis factor.
While freshly isolated tissues are relatively easy to obtain from animal models, acquisition of fresh clinical specimens from human patients is more difficult. In this regard, human studies may involve the prospective collection of large numbers of specimens over time, storage of such specimens in biobanks, and then waiting, possibly for years, for outcome results. Using biobanks with flash-frozen specimens may be a viable option, but in these studies, paired specimens designed to test clinically meaningful hypotheses may exist only in low numbers (for instance, responders vs. refractory cases). An attractive alternative is to leverage the vast number of clinically annotated specimens stored in formalin-fixed paraffin-embedded (FFPE) archives from decades of collection.8 With electronic medical record database mining to preselect annotated FFPE specimens, one can rapidly plan and conduct appropriately powered and balanced studies. Significant progress can potentially be made toward the understanding of human tumor heterogeneity if FFPE repositories can be leveraged for single-cell analysis.
In a recent report in Science Signaling, we made additional advances with DISSECT for its application to FFPE human colon and colorectal cancer (CRC) specimens (Fig. 1A - Top).9 With data generated by FFPE-DISSECT coupled to mass cytometry, we used pairwise correlation between signaling analytes across all single cells in each sample to derive a metric of coordination between signaling pathways, and then used hierarchical clustering over this metric to define signaling modules. Using quantitative tree comparisons, we concluded that signaling pathways in normal human colon were organized into well-defined modules whereas this organization was lost in CRC. Furthermore, the single-cell landscape can classify CRCs by their microsatellite and BRAF mutation status, while differentiated cell populations were suppressed in the majority of, but not all, CRCs. Our study highlighted the feasibility of FFPE-DISSECT for single-cell signaling studies in a small number of well-controlled FFPE specimens that can potentially be extended to larger clinical cohorts.
Although FFPE-DISSECT can potentially address human tumor heterogeneity at the functional level, there are several limitations that must be considered. First, proper tissue handling and pre-analytical processing are crucial to maintain the integrity of protein antigens. Specimens to be analyzed must come from well-documented sources with standardized collection, fixation, and storage procedures, with additional screening for artifacts of improper handling. Considerations of surface area and volume should be made with regard to penetration of fixatives. Second, antibodies that are validated for another application may not perform in FFPE. In our experience of using “well-known” antibodies for different applications (flow cytometry or immunofluorescence), we have found that about 20% of antibodies are still able to recognize their epitopes effectively in FFPE-DISSECT. Furthermore, for reagents that give positive results, the dynamic ranges and signal-to-noise ratios achieved by FFPE-DISSECT tend to be lower than those of DISSECT on freshly isolated tissues, although the relative trends remain the same (Fig. 1B). This is not unexpected given the substantial masking of antigens during FFPE processing. Third, we have not considered whether FFPE-DISSECT retrieves all cell types equally. Because we focus on the epithelial compartment, other cell populations that are more imbedded, such as collagen-secreting fibroblasts, may not be completely recovered as single cells. A rigorous, standardized study to quantify the cell types retrieved by FFPE-DISSECT compared with the intact tissue has not been conducted. As such, we find that coupled immunofluorescence (IF) imaging is a vital component of the validation process. For example, IF of tissue sections for cell types can be used to confirm the yield of various cell types by FFPE-DISSECT. Furthermore, IF can be performed on pre-disaggregated tissue to visually confirm proper localization of a signal, which provides confidence that a positive signal is not an artifact from nonspecific staining. Imaging of single-cell suspensions should be performed for every study to confirm successful disaggregation.
Adaptation of DISSECT to a variety of tissues, including pancreatic, liver, gastric, brain, and lymphoma tissues, is ongoing. We hope that these efforts will enable the quantification of intact signaling states in single cells in order to unravel the complex heterogeneity that exists in cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Vanderbilt Epithelial Biology Center for helpful discussions, experimental guidance, and reagents.
Funding
The original study was supported in part by NIH grants R01DK103831, 2UL1TR000445, P30CA068485, P30DK058404, P50CA095103, U24CA159988, as well as an Innovator Award from the AACR-Landon Foundation (15–20–27-LAUK) and a CCFA CDA (308221).
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2.Lau KS, Cortez-Retamozo V, Philips SR, Pittet MJ, Lauffenburger DA, Haigis KM. Multi-scale in vivo systems analysis reveals the influence of immune cells on TNF-α-induced apoptosis in the intestinal epithelium. PLoS Biol 2012; 10:e1001393; PMID:23055830; http://dx.doi.org/ 10.1371/journal.pbio.1001393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau KS, Schrier SB, Gierut J, Lyons J, Lauffenburger DA, Haigis KM. Network analysis of differential Ras isoform mutation effects on intestinal epithelial responses to TNF-α. Integr Biol (Camb) 2013; 5:1355-65; PMID:24084984; http://dx.doi.org/ 10.1039/c3ib40062j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons AJ, Banerjee A, McKinley ET, Scurrah CR, Herring CA, Gewin LS, Masuzaki R, Karp SJ, Franklin JL, Gerdes MJ, et al.. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo. Mol Syst Biol 2015; 11:835; PMID:26519361; http://dx.doi.org/ 10.15252/msb.20156282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al.. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011; 332:687-96; PMID:21551058; http://dx.doi.org/ 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenmiller B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst 2016; 2:225-38; PMID:27135535; http://dx.doi.org/ 10.1016/j.cels.2016.03.008 [DOI] [PubMed] [Google Scholar]
- 7.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, et al.. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A 2013; 110:11982-7; PMID:23818604; http://dx.doi.org/ 10.1073/pnas.1300136110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank 2013; 11:101-6; PMID:24845430; http://dx.doi.org/ 10.1089/bio.2012.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmons AJ, Scurrah CR, Mckinley ET, Herring CA, Irish JM, Washington MK, Coffey RJ, Lau KS. Impaired coordination between signaling pathways is revealed in human colorectal cancer using single-cell mass cytometry of archival tissue blocks. 2016; 11:1-13; PMID:27729552; http://dx.doi.org/ 10.1126/scisignal.aah4413 [DOI] [PMC free article] [PubMed] [Google Scholar]


