Abstract
The genome-wide association study (GWAS) has become an established scientific method that provides an unbiased screen for genetic loci potentially associated with phenotypes of clinical interest such as chronic kidney disease (CKD). Thus, GWAS provides opportunities to gain new perspectives regarding the genetic architecture of CKD progression by identifying new candidate genes and targets for intervention. Thus it has become an important arm of translational science providing a complementary line of investigation to identify novel therapeutics to treat CKD. In this review, we describe the method and the challenges of performing GWAS in the pediatric CKD population. We also provide an overview of successful GWAS for kidney disease, and we discuss the established pediatric CKD cohorts in North America and Europe that are poised to identify genetic risk variants associated with CKD progression.
Keywords: GWAS, chronic kidney disease, translational
Introduction
Major multi-institutional investments have been made to study pediatric chronic kidney disease (CKD), better define its complications, and importantly to discover new treatment strategies that slow or halt the progressive decline in kidney function universal to CKD. Current efforts in pediatric nephrology are the Chronic Kidney Disease in Children (CKiD) study in North America [1]; and, in the European Union (EU), the Effect of Strict Blood Pressure Control and Angiotensin-Converting Enzyme (ACE) Inhibition on the Progression of CRF in Pediatric Patients (ESCAPE) study [2] as well as the Cardiovascular Comorbidity in Children with Chronic Kidney Disease (4C) study [3].
Major findings from these studies include: improved estimating equations for glomerular filtration rate (GFR) to help monitor the rate of kidney function decline [4]; an estimate of the annual percent decline in GFR among children with CKD as −4.2% (corresponding to a median decline of −1.8 ml/min/1.73m2) [5]; identification of risk factors for a more rapid onset of GFR loss [6]; and evidence that intensified blood-pressure control slows the progressive loss of kidney function [2]. These collective efforts are advancing our understanding of pediatric CKD in the domains of cardiovascular health, mineral bone disease, growth, and neurocognitive function. Complementing these efforts is a joint international collaboration of the CKiD, ESCAPE, and 4C studies, the Pediatric Investigation for Genetic Risk Factors Linked with Renal Progression (PediGFR), to identify novel genetic risk factors for CKD progression. The group is pursuing a genome-wide association study (GWAS) to identify genetic risk variants for CKD traits and progression.
GWAS offer novel opportunities to advance our understanding of the genetic epidemiology and molecular underpinnings of CKD progression. In light of the PediGFR initiative and anticipated results, this general review will describe how to conduct a GWAS, summarize results from prior kidney-related studies and provide a discussion of opportunities and challenges of GWAS in pediatric CKD.
The GWAS Method
GWAS is an approach to identify genetic variation contributing to human disease. Whereas monogenic disorders are caused by rare variant(s) in a single gene typically identified in studies of affected families, common genetic susceptibility variants can be examined in association studies using unrelated individuals. Prior to the advent of high throughput genotyping, candidate gene studies were used to investigate genes of known function for association in human disease. However, the gene-by-gene candidate approach was inefficient, suffered from lack of replication, and was conditioned on the current state of evidence-based science [7]. The GWAS method is applied to understand polygenic disorders where multiple gene variants, each with a potentially small effect, act in concert to cause common disorders, such as CKD. As GWAS use an unbiased genome-wide method, it can deliver insights impossible to gain using the traditional candidate gene approach.
An important consideration in the planning of a GWAS is identifying the phenotype of interest. A phenotype that is non-specific may require the study of a much larger cohort as compared to a more precise phenotype where a small to moderate cohort may be sufficient to provide adequate statistical power. In particular, a very strict phenotype allows enough reduction in heterogeneity of potential sub-phenotypes to permit a GWAS in small cohorts; which can make the GWAS approach feasible to study certain kidney conditions [8]. For example, the study by Gbadegesin et al. utilized a genome-wide approach using an exome array to study steroid sensitive nephrotic syndrome (SSNS). With a discovery set consisting of 214 children with SSNS and 149 controls of South Asian origin, they identified an association of missense variants in HLA-DQA1 with SSNS. Their replication cohort utilized 100 children with SSNS and 205 children of European ancestry as controls [9]. As another example, the study of membranous nephropathy (MN) by Stanescu et al. had a sample size of 556 subjects with MN and 2338 ethnically matched controls for their GWAS. Their study identified SNPs in HLA-DQA1 and PLA2R1 gene regions that associated with membranous nephropathy (MN), an unambiguous clinical phenotype based on histological findings [10]. Therefore a focused GWAS on a very specific phenotype may permit using a cohort of modest sample size by minimizing phenotypic heterogeneity.
In contrast, the condition of CKD encompasses several clinical entities. Because of the heterogeneity of causes for CKD, a GWAS of this particular phenotype would typically require a much larger sample size compared to a GWAS of a more specific phenotype (i.e. a kidney condition where all the cases are verified histologically). Although investigating this aim is worthwhile, it is also challenging given the relatively low frequency of CKD in children compared to adults; and the current cohorts to study CKD progression represent a conglomeration of children with many different causes of CKD. Given the heterogeneity of phenotypes that cause CKD, identifying variants for CKD progression rely on the assumption that the underlying process of CKD progression is common to all causes of pediatric CKD. At this time there are few large pediatric cohorts that have longitudinal assessments of GFR and ascertainment of end-stage renal disease.
The cohorts of the PediGFR are one of the largest collection of children with detailed CKD progression data with serial assessments of GFR and determination of events for dialysis and kidney transplant. CKiD is an epidemiologic cohort study of children with mild to moderate CKD among 43 nephrology centers across North America. There are 586 children in cohort 1 and approximately 305 in cohort 2. CKiD has collected detailed CKD progression data in order to identify novel and traditional risk factors for renal disease progression as well as correlate the evolution of co-morbid changes in: neurocognition and behavior; cardiovascular risk factors; and growth. In cohort 1, 444 subjects are genotyped. With 468 pediatric patients with mild to severe CKD across 33 European pediatric nephrology centers, the ESCAPE trial is a randomized controlled-trial on the effect of strict blood pressure control and angiotensin converting enzyme (ACE) inhibition on CKD progression in pediatric patients. Its goals were also to elucidate the genetic and molecular mechanisms that initiate and determine CKD progression [2]. It enrolled 468 children in two treatment arms (intensified vs. conventional blood pressure control) of which 315 were genotyped. The 4C study is an observational study to correlate measures of cardiovascular health with CKD progression in children [3]. It has recruited 705 pediatric patients across 40 pediatric nephrology units in 14 European countries; 691 subjects have been genotyped. We use our ongoing experience with the PediGFR to illustrate some of the key aspects of GWAS.
The steps in conducting a GWAS to study children with CKD are outlined in Box 1. After administrative agreements are in place and a genotyping laboratory is identified, one must choose a genotyping array. Depending on one's budget, available genotyping arrays (chips) contain between 500,000 to 5 million single nucleotide polymorphisms (SNPs), with newer arrays having more rare variant content and higher cost. The gene variants, some of them being tag-SNPs, represent a larger block of SNPs, called a haplotype [11]. SNPs within a haplotype block can be correlated with one another and are then said to be in linkage disequilibrium (LD). This allows cost-efficient screening of the genome with no need to genotype all variants for a genome wide scan. It is essential to perform genotyping at an established facility where quality is high and risks of genotyping errors are low [12]. The genotyping for the PediGFR study occurred over two years at two different genotyping centers; one in the US and one in the EU. Both laboratories followed a common genotyping protocol. The spread over 2 years permitted the coordination of the study given the time lag of observational follow-up time and collection of DNA samples between CKiD and 4C; the genetic samples for ESCAPE had been already collected. By the end of this time period, we had genotyping data on 1450 children with CKD ready for further analysis. With the available subjects with measured GFR, the PediGFR has >85% power to detect a change in average GFR of ≥16% or an odds ratio for CKD of ≥1.7 per risk allele at genome-wide significance [13].
Being necessary team members, genetic epidemiologists or genetic statisticians have expertise to perform the association analyses between the phenotype and the genotyped data. A typical GWAS involves a large volume of data in the magnitude of multiple gigabytes, that requires specific analytic techniques to insure rigorous data cleaning and quality control prior to performing association analyses. Given the importance of having high standards for data quality, the PediGFR has taken an additional quality control step by which we use known associations for data checks as another way to detect additional sources of computational error prior to performing a GWAS [14]. It is difficult to identify errors in computer coding in a GWAS from a global evaluation of summary statistics. We used data from the Chronic Renal Insufficiency Cohort (CRIC) where we could check our code for association analyses using their data on bilirubin levels to associate with rs6742078, a variant in the UGT1A gene that is strongly associated with bilirubin levels. The initial check of the association with the coding error led to an attenuated p-value for association between rs6742078 and bilirubin levels. When the coding error was identified and corrected, we found that the allele frequencies, effect direction and effect size were consistent with previously published results with an association P= 6 × 10−32; details for the technique are provided by Wuttke et al. [14].
A number of epidemiologic study designs can invoke the GWAS approach—case-control studies, cohort studies, and clinical trials--as long as the inherent strengths and weaknesses of the respective study paradigms are considered in the GWAS [12]. In a hypothetical case-control study, it is ideal that cases are as homogeneous as possible. Also, controls should be comparable to the source population from which cases are selected and any exclusion or restriction criteria used to identify cases should also apply to controls [15]. The differences in allele frequency between cases and controls at each marker locus are tested for all markers across the genome. Genome-wide statistical significance depends on the number of markers being tested. Given the issues of an extremely large number of comparisons, GWAS significance thresholds must be more stringent than the standard threshold for typical epidemiologic studies (p<0.05), and can be set according to the number of evaluated SNPs. For early commercially available arrays of 500,000 to 600,000 SNPs, a p-value of approximately 10−7 was considered significant; for current genotyping arrays of 2.5 million SNPs as well as imputed datasets, the suggested p-value threshold is 5 × 10−8 or less [16].
As with any epidemiologic association study, biased sampling or systematic errors might lead to false associations with the condition of interest. Thus, these issues must be addressed at both the GWAS planning stage and during statistical modeling. With consideration for the PediGFR, bias can arise as a consequence of differences among the three study groups. Although all studies followed the decline in GFR over time, each differed slightly in design, as indicated above.
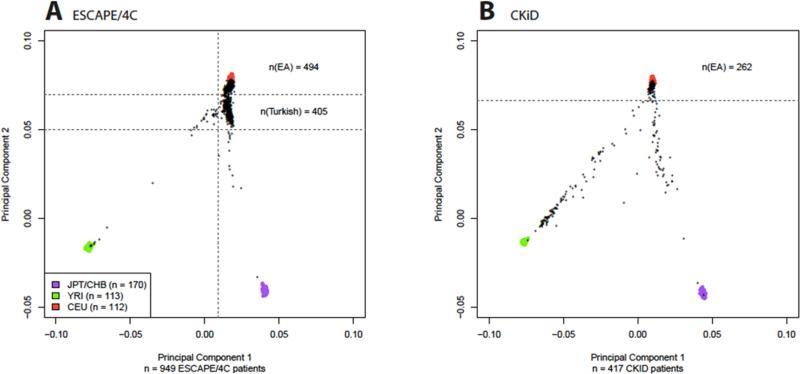
Another potential source of bias in PediGFR is population stratification, which may lead to a spurious association with disease when both disease risk as well as allele frequencies differ across subpopulations [17]. Figure 1 displays the principal component analyses for the CKiD cohort and the ESCAPE/4C cohorts. The plot separates study patients by their genetic ancestry [13]. This figure demonstrates presence of different sub-populations by ethnic origin, where in Panel A subjects from Turkey are represented in a cluster distinct from the central European subject cluster among the ESCAPE/4C cohorts. For CKiD in Panel B, European American and African American subjects can be distinguished.
Figure 1.
Red—CEU (Caucasian ancestry), Purple—YRI (African ancestry), Green—CHB/YPT (Asian ancestry). The genotyped individuals are shown black dots. Principal component analyses are shown in Panel A: the ESCAPE/4C cohorts (N= 934). We note the cluster near the CEU region for most participants recruited from Western and Central European countries. There is a sub-cluster just below, representing Turkish participants and in Panel B In Panel B, the CKiD cohort (n=444). Near the CEU region there are a cluster of subjects of European ancestry and there is a cluster near YRI representing African Americans. [13]
Given potential differences in ascertaining subjects, ancestry, and phenotyping, meta-analysis of study- and ethnicity-specific GWAS results should be preferred over pooling all subjects prior to GWAS [18]. Meta-analysis is an established statistical method of combining evidence across independent studies and can increase statistical power to examine a common research hypothesis. At the same time, it acknowledges potential differences across studies and allows testing for possible heterogeneity [19]. For PediGFR, meta-analysis was used to combine five distinct groups defined by European or Turkish ancestry and study origin. African American individuals were analyzed separately [13].
The discovery findings from an initial GWAS cannot distinguish between spurious and true associations. To lower the likelihood of a false positive finding, there is a standard expectation that discovery GWAS findings will be replicated in an independent cohort [7]. In the case of PediGFR, other characterized cohorts of children with CKD are lacking, although replication could be considered in adult CKD cohorts [20, 21]. Yet we must acknowledge that the genetic predisposition to pediatric CKD may differ from that of adult CKD, with congenital causes of CKD being the most common etiology in children; whereas diabetes and hypertension are the top causes of CKD in adults [22]. Despite differences in CKD initiation, the prevailing model of CKD progression suggests that propagation of tubulointerstitial injury and fibrosis may be similar in children and adults with CKD [23, 24].
As a complementary approach or in addition to replication, follow-up work for GWAS candidate loci of interest may include deep sequencing of the region as well as functional studies. Deep sequencing is used to refine the signal from the GWAS and may help to identify the causal variant giving rise to the observed association [25]. Follow-up projects are required to validate functional consequences of the variant such as modified gene expression. Functional studies performed usually in animal models can demonstrate that the molecular change introduced by the genetic variant is localized to the organ of interest or perturbs the model system in a way relevant to the disease process [25].
GWAS: Generating Novel Insights into the Mechanisms of Disease
Findings from GWAS of CKD phenotypes have opened new fields of inquiry for the translation of findings from epidemiological/clinical research to basic science investigations regarding the function of identified genes and their pathways. As mentioned above, SSNS associating with HLA-DQA1 and MN with HLA-DQA1 and PLA2R1 have provided insights into the possible mechanisms of disease that were not previously known. These and other examples of GWAS provide us with new directions for understanding disease mechanisms and motivate our current efforts of the PediGFR.
APOL1/MYH9 and CKD in African Americans
In 2008, two genome-wide investigations using mapping by admixture linkage disequilibrium identified an association between CKD (non-diabetic end stage renal disease [ESRD] and focal segmental glomerulosclerosis) and the myosin heavy chain 9, non-muscle (MYH9) gene located on chromosome 22q12 in African Americans [26, 27]. These initial studies and others had identified an association with MYH9; however, subsequent studies identified an even stronger association between CKD in African Americans and genetic variants in the nearby APOL1 gene. The Genovese et al. study provided evidence that gene variants of APOL1 are associated with increased CKD susceptibility among African Americans, rather than MYH9. The authors performed an initial case-control study between 205 subjects with focal segmental glomerulosclerosis (FSGS) without family history of the disease and 180 healthy controls. Despite the modest sample size, the investigators found highly significant associations between FSGS and APOL1 variants [28]. They were then able to replicate this finding in a bigger cohort of 1,030 patients with hypertensive ESRD and 1,025 suitably matched control subjects. Interestingly, the authors argued that APOL1 gene variants offer a survival advantage by conferring resistance to Trypanosoma brucei rhodensiense infections, which is endemic in sub-Saharan Africa, similar to the positive selection pressure for sickle cell trait offering resistance to malarial infections.
In African Americans, APOL1 variants are strongly associated with CKD outcomes. Parsa et al. demonstrated that the genetic risk variants in APOL1 are associated with CKD progression. In 693 hypertensive participants of the African American Study of Kidney Disease and Hypertension and 2,955 Caucasian and African American participants of the Chronic Renal Insufficiency Cohort study [29], APOL1 variants were found to confer higher risks of ESRD and CKD progression in African American patients when compared to their Caucasian counterparts, irrespective of their diabetic status. This is the first report associating APOL1 with CKD progression in longitudinal cohorts of sufficient duration.
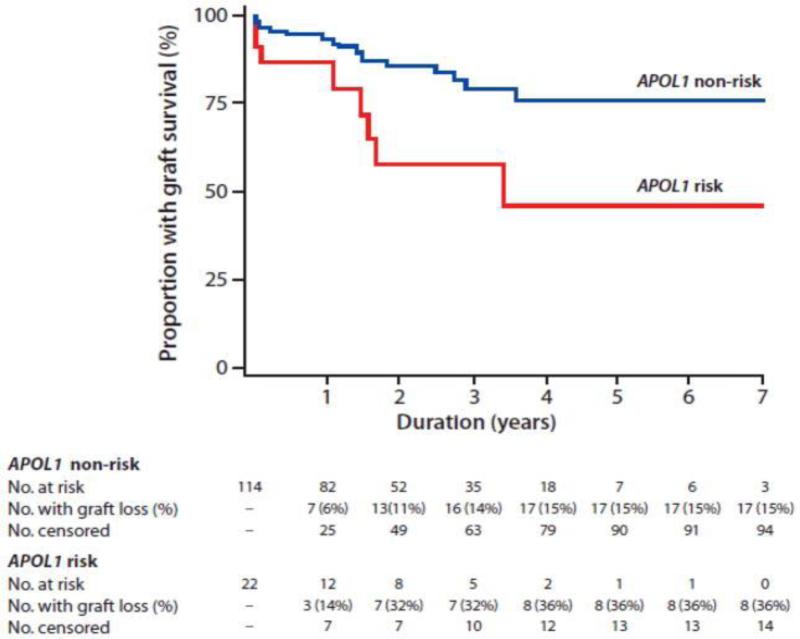
Not only is APOL1 associated with risk of CKD progression in patients with CKD, but it may also be predictive of kidney transplant outcome. There was a decrease in allograft survival associated with kidneys from deceased donors who were African-American and had two risk alleles for APOL1 when compared to transplanted kidneys obtained from donors with no or a single risk allele (Figure 2) [30]; these results were confirmed with a much larger sample size by expanding the initial cohort with additional centers [31]. It is important to note that these results may not be generalized to graft survival of living donor kidneys since these studies focused on deceased donor kidneys. These aforementioned studies did not examine the effect of recipient's genotype on decreased allograft survival. Looking at this question in particular Lee et al. found no association between kidney recipient's APOL1 genotype and allograft survival. [32]. It is clear that the discovery of APOL1 being associated with kidney outcomes has led to a significant paradigm shift in our understanding of kidney disease among those of African American ancestry and motivates further study of APOL1 and it's function in renal health.
Figure 2.
Kaplan-Meier kidney allograft survival curves in recipient of donor kidney with (red line) and without (blue line) two APOL1 risk variant alleles [30].
UMOD and CKD
The UMOD gene has gained significant attention being consistently associated with CKD and estimated GFR (eGFR) [33, 34]. The UMOD gene product is also known as Tamm-Horsfall protein. In 2009, Köttgen et al. identified an association between CKD—defined as eGFR <60 ml/min/1.73m2—and a gene locus containing SNP rs12917707 in a consortium of population-based studies (Figure 3). The SNP is located in a highly evolutionary conserved region upstream of UMOD on chromosome 16 (p=5×10−16 across discovery and replication samples). Furthermore, other independent cohorts have successfully replicated the association of UMOD and CKD in subjects of European ancestry [35, 36] as well as those of African [37] and of Asian ancestry [38]. Trudu et al. demonstrated that UMOD risk variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression [39].
Figure 3.
Meta-analysis of GWAS results –log10 (P value) versus genomic position plots for CKD (estimated GFR creatinine <60ml/min/1.73m2) in European-ancestry participants of four population-based cohorts (ARIC, CHS, FHS, and RS). Abbreviations: ARIC, Atherosclerosis Risk in Communities Study; CHS, Cardiovascular Health Study; CKD, chronic kidney disease; FHS, Framingham Heart Study; GFR, glomerular filtration rate; RS, Rotterdam Study. [34].
IgA Nephropathy
GWAS of IgA Nephropathy (IgAN) has certainly led to new hypotheses regarding the underlying pathophysiology of this important kidney disease. In the first GWAS of IgAN, Feehally et al. demonstrated association of SNPs in the major histocompatibility complex (MHC) locus on chromosome 6p with IgAN (p=1× 10−9) using a family-based association analysis (533 individuals, 187 affected children) as well as a case control analysis (244 cases and 4,980 controls) [40]. Subsequently, Gharavi and collaborators identified and replicated five gene regions associated with IgAN. Three signals mapped to the MHC locus on chromosome 6p21: the first and strongest signal coming from a region that harbors HLA-DRB1, -DQA1, and -DQB1 genes; a second signal was localized to a LD segment that contains TAP2, TAP1, PSMB8, and the PSMB9 genes; and the third signal was in a region that harbors the genes HLA-DPA1-DPB1-DPB2. A fourth signal, independent of the three MHC loci, was observed on 1q31-q32, a region that includes the complement factor H (CFH), CFHR3, CFHR1, CFHR4, CFHR2, and CFHR5 genes. The fifth signal, also independent of the three MHC loci, was on chromosome 22q12, which contains the HORMAD2 gene [41]. It is interesting to note that these IgA nephropathy associated loci also confer increased risk of other autoimmune disorders (e.g., Type 1 diabetes [T1D], inflammatory bowel disease, and multiple sclerosis) as well as infectious diseases (e.g., meningococcal infections).
An IgAN meta-analysis on a combined sample of 10,775 subjects confirmed associations at all five loci. The investigators further performed a geospatial analysis of an IgAN genetic risk score constructed from associated variants (Figure 4) [42]. Utilizing 6,319 healthy subjects across 85 world populations, investigators identified an East–West gradient with East Asians and Native Americans having the highest, and African ancestry having the lowest, mean standardized risk scores. Specific to Europe, a North–South gradient was observed. These findings aligned with the well known geographical East–West (and North–South in case of Europe) gradient in IgAN prevalence. Investigators also analyzed US Renal Data System data for IgAN-ESRD prevalence and found that Asian Americans and Caucasians were respectively 15- and 5-fold more prone to develop IgAN-ESRD as compared to African-Americans [42].
Figure 4.
Depiction of the worldwide geospatial distribution for an IgAN genetic risk score. Lowest risk scores (green) in Africa and highest (red) in Asia and the Americas (inset). [42].
Among the Han Chinese, Yu et al. performed an IgAN GWAS utilizing 1,434 cases and 4,270 controls followed by replication of top SNPs in 2,703 cases and 3,464 controls [43]. In addition to confirming previous associations above, the investigators identified two new loci: one on chromosome 17p13 that includes the gene tumor necrosis factor (TNFSF13) involved in the inflammatory response, and the other on chromosome 8p23 that includes genes coding for α-defensins (DEFA) involved in innate and adaptive immunity against microbes.
Demonstrating the importance of multinational cooperations to collect very large study samples for GWAS, Kiryluk et al. subsequently performed one of the largest IgAN GWAS with more than 20,000 subjects of Asian and European descent. They confirmed the nine previously reported signals and identified six new ones, including four in regions not identified previously, containing ITGAM-ITGAX, VAV3, and CARD9 [44]. The majority of IgAN loci identified were either related to autoimmune disorders or coded for proteins that were responsible for maintaining the integrity of the intestinal mucosal barrier or were involved in immune response to intestinal pathogens.
As with the previous study, a genetic risk score strongly correlated with distance from the African continent as well as with IgAN prevalence. Further analysis revealed that the genetic risk score was strongly associated with local pathogen load, specifically helminthic diversity and also with geographical location. This very important finding may explain the observed difference in worldwide IgAN prevalence. The authors suggested that, because helminthic infection is a known reason for selective pressure, the higher IgAN prevalence in some far eastern countries could be the unwanted result of intestinal adaptation to infection [44]. The evolving GWAS story of IgA nephropathy illustrates how such data provide insight into disease mechanisms. Furthermore, it stimulates new hypotheses for selection pressures that might trigger human diseases.
Diabetic Nephropathy
The study of DN emphasizes the importance of identifying very strict phenotype when conducting a GWAS. In 2003, the first DN GWAS was performed in a Japanese cohort of 94 Type 2 Diabetes (T2D) patients with retinopathy and nephropathy (cases) and 94 T2D patients with retinopathy but no nephropathy (controls) using a set of more than 56,000 SNPs covering the whole genome. The authors reported an association between DN and genetic variation in solute carrier family 12 member 3 gene (SLC12A3) with a p<10−5, and replicated the finding in a larger cohort of 553 case and 317 control subjects [45]. Subsequently, using the same cohort but a larger panel of 80,000 SNPs, the investigators identified an association between DN and the Engulfment and Cell Motility 1 gene (ELMO1) [46, 47]. This association was further validated in a large Caucasian cohort of 1,705 subjects with T1D (820 with DN [284 with proteinuria and 536 with ESRD], and 885 without DN) from the Genetics of Kidneys in Diabetes (GoKinD) study [48]; however the strongest associations this study uncovered were independent of those reported in Japanese cohorts. Although DN findings may have been nominally significant, no SNPs described above reached genome-wide statistical significance.
Unlike IgAN, where larger samples provide more statistical power to identify relevant disease-associated loci, no clear themes emerged with the meta-analytic efforts of GWAS for DN so far. Investigators from the GEnetics of Nephropathy—an International Effort (GENIE) consortium—performed a GWAS meta-analysis involving 11,847 subjects with T1D and close to 2.4 million SNPs (UK-ROI, FinnDiane, and GoKinD US). They identified two genome-wide significant loci associated with ESRD due to diabetes—one in the AFF3 gene and the other in the intergenic region between the genes RGMA and MCTP2. There were no genome-wide significant loci associated specifically with DN, although an intronic SNP in the gene ERBB4 was suggestive (p=2.1 × 10−7) [49]. As commented by Boger and Sedor, the lack of statistical genetic evidence for associated loci may be due to the use of cross-sectional designs and lack of individual follow-up for renal progression [50]. Further, they identify that misclassification with a clinical DN definition has the potential to reduce statistical power in a GWAS. Compared to IgAN where the diagnosis is specific and confirmed by kidney biopsy, DN is not as specific. Furthermore, type 1 DN and type 2 DN might be different phenotypically, which underscore that significant heterogeneity in the phenotype of interest may undermine the performance of a GWAS despite a significantly large sample size. For a GWAS of pediatric CKD, the DN experience illustrates potential limitations of cross-sectional data analysis and valid concerns of having heterogeneous causes of CKD in our GWAS of pediatric CKD progression.
Pathway-based Analyses of GWAS Data
Genotypes and summary statistics from GWAS can be used for complementary purposes, such as for pathway-based analyses. Pathways represent a network of genes working together to affect a biological function or a physiological pathway. Using GWAS data, pathway-based analyses test the joint contribution of multiple variants in multiple genes in the same pathway affecting the phenotype [51]. Pathway-based analyses of GWAS data might have some advantages with regard to statistical power as compared to testing millions of individual SNPs. Utilizing this approach, Chasman et al. identified and replicated six novel genes (FBXL20, INHBC, LRP2, PLEKHA1, SLC7A6, and SLC3A2) associated with eGFR [52]. The authors highlight the translational impact of GWAS by suggesting that the six associated loci could be a starting point for functional studies to elucidate which genes in an associated region may influence kidney function. Pathway-based analysis is another analytic approach to further utilize existing genotyping data, yet initial or discovery findings may still be susceptible to false-positive findings for association between the gene pathway and the phenotype of interest. Thus one must still rely on confirmation of initial results with replication of findings and functional studies.
Other Challenges for Pediatric CKD
Inherent in GWAS are challenges to balance the interests of primary investigators, regulatory and funding authorities, and the research community at large. As indicated by the large number of authors on GWAS publications, there has been an unprecedented need for international collaboration [53]. There is a need to coordinate regulatory paperwork, oversee effort for project management, and maintain trust among collaborators with the promise of future benefits by co-authorship on published works. At the same time, principal investigators must be responsive and serve stakeholder needs. For example, the National Institutes of Health (NIH) believes that the full value of GWAS to the public can be realized by making GWAS datasets available as rapidly as possible to a wide range of scientific investigators. Hence, NIH-funded GWAS are subject to mandatory data sharing submitted to the database for Genotypes and Phenotypes (dbGaP) with a limited period of publication exclusivity.
Although necessary for NIH grant recipients, additional responsibilities for such stewardship compound the challenges. As discussed by McGuire et al., institutional differences may exist, not only in policies for data sharing, but also in interpretation of data use limitations that are responsive to participant concerns [54]. They identified a need to develop significant resources for researchers and institutions to facilitate, educate, and harmonize data sharing requirements. At the same time, these policies must also assure protection of subject confidentiality. Despite challenges, the authors would like to emphasize that, with the recent abundance of such complex studies, the GWAS data-sharing requirement is becoming the norm.
Concluding Remarks
As highlighted by the examples above, GWAS may offer novel insights into kidney disease; and in particular pediatric CKD progression. At the same time, it requires a considerable investment of time and resources. In order to undertake a successful GWAS, great attention must be given in consideration of both the study design and the phenotype of interest. Unlike the study of a very well-defined phenotype, the broad investigation of CKD progression requires a large number of subjects to provide sufficient statistical power, motivating the need for a current collaborative effort by CKiD, ESCAPE, and 4C. Despite the potential weaknesses of using the general CKD phenotype in the PediGFR, there are benefits in having genotyped data available across all the cohorts to permit more focused investigations of other phenotypes; which are available in dbGaP with the published baseline data [13].
The advent of genome-wide screening technologies, related bioinformatics, and cooperative agreements among pediatric CKD cohorts has created new opportunities for gene discoveries through GWAS. We have reviewed examples of GWAS in nephrology that highlighted the motivation for a GWAS in the pediatric CKD population and identified the associated challenges—including a need for other pediatric CKD studies with genome-wide genotyping and well-characterized GFR measures to characterize renal disease progression. To meet this challenge, we hope that the steps detailing the GWAS technique will help others; and foster new collaborations to advance our understanding of pediatric CKD progression.
Box 1. Steps for Conducting GWAS.
Step 1. Identify an adequately phenotyped population of interest with consent for GWAS participation and genetic studies.
Step 2. Verify that the sample size and statistical power of the proposed population is sufficient to detect hypothesized effect sizes even after correcting for multiple testing.
Step 3. Have the DNA samples genotyped with a reliable genotyping laboratory (samples should be genotyped all at once to minimize batch effects).
Step 4. Verify the quality of the genotyping by performing careful data cleaning, quality control, and SNP imputation.
Step 5. Conduct statistical association testing with appropriate adjustment for potential confounders, including population stratification. (If using multiple cohorts with different study designs, consider meta-analyzing the association statistics across cohorts).
Step 6. Inference: an associated SNP itself or a correlated unknown variant influences disease risk.
Step 7. If possible, replicate the discovery findings externally in other independent cohorts that have the same phenotype available.
Step 8. Conduct follow-up projects to provide further evidence of causality of the association between the genetic locus (or loci) of interest and the disease or condition. These may be in the form of clinical or epidemiologic characterization, deep sequencing, or exome sequencing the region(s) of interest, functional studies in lower eukaryotes or cell-based systems.
Modified from [11]
Acknowledgements
Jayanta Gupta, Peter A. Kanetsky, Franz Schaefer, and Craig S. Wong were supported by NIH grant number DK082394. Anna Köttgen was supported by the Emmy Noether Program of the German Research Foundation (KO 3598/2-1). Craig S. Wong received additional support from DK066143. Franz Schaefer received additional support from the European Community 7th Framework Programme (grant 2012-305608, EURenOmics), The KfH Foundation for Preventive Medicine (http://www.kfh-stiftung-praeventivmedizin.de/) and the ERA-EDTA.
Glossary
- Allele
Alternative DNA sequences at the same physical position on homologous chromosomes.
- Ancestry informative markers
Single nucleotide polymorphisms that have large frequency differences across different continental populations; can be used to infer ancestry and adjust for population stratification in genetic association studies.
- Common variant
A genetic variant that is common in the population; frequently defined as one with a population frequency of 1% or more.
- Deep sequencing
The whole genome or a genomic region is sequenced multiple times using high throughput sequencing (next-generation sequencing) technology to reduce the number of sequencing errors and detect rare genetic variants.
- Genomic
Pertaining to the collection of all DNA in an organism.
- Genotype
The genetic makeup of an individual, which may refer to the whole genome or to specific genes or region of genes.
- Haplotype
A set of genetic variants (usually clustered together on the same physical position of the genome) that are inherited together.
- Imputation
In the context of genetic studies, a statistical procedure that uses linkage disequilibrium (i.e. correlation between individual genetic variants) to infer missing or untyped genotypes in an individual based on a reference panel of several individuals who have been fully genotyped.
- Linkage disequilibrium
A non-random association of two or more alleles
- Loci
Specific positions of genes or a genetic markers on a chromosome; the term locus is singular for loci.
- Phenotype
Measureable or observable physical characteristics or trait of an organism.
- Population stratification
presence of a systematic difference in allele frequencies between subpopulations in a population, can be introduced by different ancestries.
- Principal Component Analysis
A statistical procedure for reducing high dimensional data into fewer dimensions. Used in population genetics to determine how genetic diversity across the genome varies according to geographical location and race/ethnicity. Widely used to adjust for population stratification in GWAS.
- Rare variant
A genetic variant with a population frequency of less than 1%.
- Replication
Validation of a gene-disease association from a discovery genetic association study by a follow-up study in an independent sample of subjects.
- Single nucleotide polymorphisms (SNPs)
DNA sequence variation resulting from a change of a single base pair (nucleotide), commonly defined as having a frequency of at least 1% in a given population.
- Tag-SNP
a SNP that is highly correlated with other SNPs and used to serve as a marker for a haplotype block. Tag-SNPs can be utilized for SNP-based genotyping.
- Variant
a variation in the DNA sequence; e.g a SNP. Other forms of genetic variation include structural variations (e.g. deletions and insertions, copy number variations etc.)
Contributor Information
Jayanta Gupta, Department of Biomedical Sciences, Texas Tech University Health Sciences Center, El Paso, TX.
Peter A. Kanetsky, Department of Cancer Epidemiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL
Matthias Wuttke, Renal Division, Medical Center - University of Freiburg, Freiburg, Germany.
Anna Köttgen, Renal Division, Medical Center - University of Freiburg, Freiburg, Germany.
Franz Schaefer, Pediatric Nephrology Division, University of Heidelberg, Heidelberg, Germany.
Craig S. Wong, Department of Pediatrics, University of New Mexico Children's Hospital, Albuquerque, NM
References
- 1.Furth SL, Cole SR, Moxey-Mims M, Kaskel F, Mak R, Schwartz G, Wong C, Munoz A, Warady BA. Design and methods of the Chronic Kidney Disease in Children (CKiD) prospective cohort study. Clin J Am Soc Nephrol. 2006;1:1006–1015. doi: 10.2215/CJN.01941205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wuhl E, Trivelli A, Picca S, Litwin M, Peco-Antic A, Zurowska A, Testa S, Jankauskiene A, Emre S, Caldas-Afonso A, Anarat A, Niaudet P, Mir S, Bakkaloglu A, Enke B, Montini G, Wingen AM, Sallay P, Jeck N, Berg U, Caliskan S, Wygoda S, Hohbach-Hohenfellner K, Dusek J, Urasinski T, Arbeiter K, Neuhaus T, Gellermann J, Drozdz D, Fischbach M, Moller K, Wigger M, Peruzzi L, Mehls O, Schaefer F. Strict blood-pressure control and progression of renal failure in children. N Engl J Med. 2009;361:1639–1650. doi: 10.1056/NEJMoa0902066. [DOI] [PubMed] [Google Scholar]
- 3.Querfeld U, Anarat A, Bayazit AK, Bakkaloglu AS, Bilginer Y, Caliskan S, Civilibal M, Doyon A, Duzova A, Kracht D, Litwin M, Melk A, Mir S, Sozeri B, Shroff R, Zeller R, Wuhl E, Schaefer F. The Cardiovascular Comorbidity in Children with Chronic Kidney Disease (4C) study: objectives, design, and methodology. Clin J Am Soc Nephrol. 2010;5:1642–1648. doi: 10.2215/CJN.08791209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz GJ, Schneider MF, Maier PS, Moxey-Mims M, Dharnidharka VR, Warady BA, Furth SL, Munoz A. Improved equations estimating GFR in children with chronic kidney disease using an immunonephelometric determination of cystatin C. Kidney Int. 2012;82:445–453. doi: 10.1038/ki.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furth SL, Abraham AG, Jerry-Fluker J, Schwartz GJ, Benfield M, Kaskel F, Wong C, Mak RH, Moxey-Mims M, Warady BA. Metabolic abnormalities, cardiovascular disease risk factors, and GFR decline in children with chronic kidney disease. Clin J Am Soc Nephrol. 2011;6:2132–2140. doi: 10.2215/CJN.07100810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warady BA, Abraham AG, Schwartz GJ, Wong CS, Munoz A, Betoko A, Mitsnefes M, Kaskel F, Greenbaum LA, Mak RH, Flynn J, Moxey-Mims MM, Furth S. Predictors of Rapid Progression of Glomerular and Nonglomerular Kidney Disease in Children and Adolescents: The Chronic Kidney Disease in Children (CKiD) Cohort. Am J Kidney Dis. 2015;65:878–888. doi: 10.1053/j.ajkd.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NCI-NHGRI Working Group on Replication in Association Studies. Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr., Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 8.Manchia M, Cullis J, Turecki G, Rouleau GA, Uher R, Alda M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One. 2013;8:e76295. doi: 10.1371/journal.pone.0076295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gbadegesin RA, Adeyemo A, Webb NJ, Greenbaum LA, Abeyagunawardena A, Thalgahagoda S, Kale A, Gipson D, Srivastava T, Lin JJ, Chand D, Hunley TE, Brophy PD, Bagga A, Sinha A, Rheault MN, Ghali J, Nicholls K, Abraham E, Janjua HS, Omoloja A, Barletta GM, Cai Y, Milford DD, O'Brien C, Awan A, Belostotsky V, Smoyer WE, Homstad A, Hall G, Wu G, Nagaraj S, Wigfall D, Foreman J, Winn MP, Mid-West Pediatric Nephrology C. HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol. 2015;26:1701–1710. doi: 10.1681/ASN.2014030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, Voinescu C, Patel N, Pearce K, Hubank M, Stephens HA, Laundy V, Padmanabhan S, Zawadzka A, Hofstra JM, Coenen MJ, den Heijer M, Kiemeney LA, Bacq-Daian D, Stengel B, Powis SH, Brenchley P, Feehally J, Rees AJ, Debiec H, Wetzels JF, Ronco P, Mathieson PW, Kleta R. Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med. 2011;364:616–626. doi: 10.1056/NEJMoa1009742. [DOI] [PubMed] [Google Scholar]
- 11.Kottgen A. Genome-wide association studies in nephrology research. American journal of kidney diseases : the official journal of the National Kidney Foundation. 2010;56:743–758. doi: 10.1053/j.ajkd.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nat Rev Genet. 2005;6:847–859. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- 13.Wuttke M, Wong CS, Wühl E, Epting D, Luo L, Hoppmann A, Doyon A, Li Y, Consortium C, Sözeri B, Thurn D, Helmstädter M, Huber TB, Blydt-Hansen TD, Kramer-Zucker A, Mehls O, Melk A, Querfeld U, Furth SL, Warady BA, Schaefer F, Köttgen A. Genetic loci associated with renal function measures and chronic kidney disease in children: the Pediatric Investigation for Genetic Factors Linked with Renal Progression Consortium. Nephrology Dialysis Transplantation. 2015 doi: 10.1093/ndt/gfv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuttke M, Schaefer F, Wong CS, Kottgen A. Genome-wide association studies in nephrology: using known associations for data checks. American journal of kidney diseases : the official journal of the National Kidney Foundation. 2015;65:217–222. doi: 10.1053/j.ajkd.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennekens, Buring . Epidemiology in Medicine. Williams, and Wilkins; Lippincot: 1987. [Google Scholar]
- 16.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 18.Panagiotou OA, Willer CJ, Hirschhorn JN, Ioannidis JP. The power of meta-analysis in genome-wide association studies. Annu Rev Genomics Hum Genet. 2013;14:441–465. doi: 10.1146/annurev-genom-091212-153520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorski M, Tin A, Garnaas M, McMahon GM, Chu AY, Tayo BO, Pattaro C, Teumer A, Chasman DI, Chalmers J, Hamet P, Tremblay J, Woodward M, Aspelund T, Eiriksdottir G, Gudnason V, Harris TB, Launer LJ, Smith AV, Mitchell BD, O'Connell JR, Shuldiner AR, Coresh J, Li M, Freudenberger P, Hofer E, Schmidt H, Schmidt R, Holliday EG, Mitchell P, Wang JJ, de Boer IH, Li G, Siscovick DS, Kutalik Z, Corre T, Vollenweider P, Waeber G, Gupta J, Kanetsky PA, Hwang SJ, Olden M, Yang Q, de Andrade M, Atkinson EJ, Kardia SL, Turner ST, Stafford JM, Ding J, Liu Y, Barlassina C, Cusi D, Salvi E, Staessen JA, Ridker PM, Grallert H, Meisinger C, Muller-Nurasyid M, Kramer BK, Kramer H, Rosas SE, Nolte IM, Penninx BW, Snieder H, Fabiola Del Greco M, Franke A, Nothlings U, Lieb W, Bakker SJ, Gansevoort RT, van der Harst P, Dehghan A, Franco OH, Hofman A, Rivadeneira F, Sedaghat S, Uitterlinden AG, Coassin S, Haun M, Kollerits B, Kronenberg F, Paulweber B, Aumann N, Endlich K, Pietzner M, Volker U, Rettig R, Chouraki V, Helmer C, Lambert JC, Metzger M, Stengel B, Lehtimaki T, Lyytikainen LP, Raitakari O, Johnson A, Parsa A, Bochud M, Heid IM, Goessling W, Kottgen A, Kao WH, Fox CS, Boger CA. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 2014 doi: 10.1038/ki.2014.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lash JP, Go AS, Appel LJ, He J, Ojo A, Rahman M, Townsend RR, Xie D, Cifelli D, Cohan J, Fink JC, Fischer MJ, Gadegbeku C, Hamm LL, Kusek JW, Landis JR, Narva A, Robinson N, Teal V, Feldman HI. Chronic Renal Insufficiency Cohort (CRIC) Study: baseline characteristics and associations with kidney function. Clin J Am Soc Nephrol. 2009;4:1302–1311. doi: 10.2215/CJN.00070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chadha V, Warady BA. Epidemiology of pediatric chronic kidney disease. Adv Chronic Kidney Dis. 2005;12:343–352. doi: 10.1053/j.ackd.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Eddy AA. Progression in chronic kidney disease. Adv Chronic Kidney Dis. 2005;12:353–365. doi: 10.1053/j.ackd.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Hodgkins KS, Schnaper HW. Tubulointerstitial injury and the progression of chronic kidney disease. Pediatr Nephrol. 2012;27:901–909. doi: 10.1007/s00467-011-1992-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signa ls. Nat Rev Genet. 2009;10:318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nature genetics. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, Berns JS, Briggs W, Cho ME, Dart RA, Kimmel PL, Korbet SM, Michel DM, Mokrzycki MH, Schelling JR, Simon E, Trachtman H, Vlahov D, Winkler CA. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT, Jr., Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369:2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, Adams PL, Langefeld CD, Bowden DW, Hicks PJ, Stratta RJ, Lin JJ, Kiger DF, Gautreaux MD, Divers J, Freedman BI. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011;11:1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freedman BI, Julian BA, Pastan SO, Israni AK, Schladt D, Gautreaux MD, Hauptfeld V, Bray RA, Gebel HM, Kirk AD, Gaston RS, Rogers J, Farney AC, Orlando G, Stratta RJ, Mohan S, Ma L, Langefeld CD, Hicks PJ, Palmer ND, Adams PL, Palanisamy A, Reeves-Daniel AM, Divers J. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant. 2015;15:1615–1622. doi: 10.1111/ajt.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee BT, Kumar V, Williams TA, Abdi R, Bernhardy A, Dyer C, Conte S, Genovese G, Ross MD, Friedman DJ, Gaston R, Milford E, Pollak MR, Chandraker A. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant. 2012;12:1924–1928. doi: 10.1111/j.1600-6143.2012.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O'Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer HK, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Kramer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. New loci associated with kidney function and chronic kidney disease. Nature genetics. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kottgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Pare G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS. Multiple loci associated with indices of renal function and chronic kidney disease. Nature genetics. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudbjartsson DF, Holm H, Indridason OS, Thorleifsson G, Edvardsson V, Sulem P, de Vegt F, d'Ancona FC, den Heijer M, Wetzels JF, Franzson L, Rafnar T, Kristjansson K, Bjornsdottir US, Eyjolfsson GI, Kiemeney LA, Kong A, Palsson R, Thorsteinsdottir U, Stefansson K. Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet. 2010;6:e1001039. doi: 10.1371/journal.pgen.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boger CA, Gorski M, Li M, Hoffmann MM, Huang C, Yang Q, Teumer A, Krane V, O'Seaghdha CM, Kutalik Z, Wichmann HE, Haak T, Boes E, Coassin S, Coresh J, Kollerits B, Haun M, Paulweber B, Kottgen A, Li G, Shlipak MG, Powe N, Hwang SJ, Dehghan A, Rivadeneira F, Uitterlinden A, Hofman A, Beckmann JS, Kramer BK, Witteman J, Bochud M, Siscovick D, Rettig R, Kronenberg F, Wanner C, Thadhani RI, Heid IM, Fox CS, Kao WH. Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD. PLoS Genet. 2011;7:e1002292. doi: 10.1371/journal.pgen.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu CT, Garnaas MK, Tin A, Kottgen A, Franceschini N, Peralta CA, de Boer IH, Lu X, Atkinson E, Ding J, Nalls M, Shriner D, Coresh J, Kutlar A, Bibbins-Domingo K, Siscovick D, Akylbekova E, Wyatt S, Astor B, Mychaleckjy J, Li M, Reilly MP, Townsend RR, Adeyemo A, Zonderman AB, de Andrade M, Turner ST, Mosley TH, Harris TB, Rotimi CN, Liu Y, Kardia SL, Evans MK, Shlipak MG, Kramer H, Flessner MF, Dreisbach AW, Goessling W, Cupples LA, Kao WL, Fox CS. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet. 2011;7:e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, Takahashi A, Maeda S, Tsunoda T, Chen P, Lim SC, Wong TY, Liu J, Young TL, Aung T, Seielstad M, Teo YY, Kim YJ, Lee JY, Han BG, Kang D, Chen CH, Tsai FJ, Chang LC, Fann SJ, Mei H, Rao DC, Hixson JE, Chen S, Katsuya T, Isono M, Ogihara T, Chambers JC, Zhang W, Kooner JS, Albrecht E, Yamamoto K, Kubo M, Nakamura Y, Kamatani N, Kato N, He J, Chen YT, Cho YS, Tai ES, Tanaka T. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nature genetics. 2012;44:904–909. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell'Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S, Kumar A, Peden JF, Maxwell PH, Morris DL, Padmanabhan S, Vyse TJ, Zawadzka A, Rees AJ, Lathrop M, Ratcliffe PJ. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21:1791–1797. doi: 10.1681/ASN.2010010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nature genetics. 2011;43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiryluk K, Li Y, Sanna-Cherchi S, Rohanizadegan M, Suzuki H, Eitner F, Snyder HJ, Choi M, Hou P, Scolari F, Izzi C, Gigante M, Gesualdo L, Savoldi S, Amoroso A, Cusi D, Zamboli P, Julian BA, Novak J, Wyatt RJ, Mucha K, Perola M, Kristiansson K, Viktorin A, Magnusson PK, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boland A, Metzger M, Thibaudin L, Wanner C, Jager KJ, Goto S, Maixnerova D, Karnib HH, Nagy J, Panzer U, Xie J, Chen N, Tesar V, Narita I, Berthoux F, Floege J, Stengel B, Zhang H, Lifton RP, Gharavi AG. Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet. 2012;8:e1002765. doi: 10.1371/journal.pgen.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, Sun LD, Sim KS, Li Y, Foo JN, Wang W, Li ZJ, Yin XY, Tang XQ, Fan L, Chen J, Li RS, Wan JX, Liu ZS, Lou TQ, Zhu L, Huang XJ, Zhang XJ, Liu ZH, Liu JJ. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nature genetics. 2012;44:178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- 44.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Battaglia GG, Garozzo M, Maixnerova D, Tesar V, Eitner F, Rauen T, Floege J, Kovacs T, Nagy J, Mucha K, Paczek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Barratt J, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, Gharavi AG. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nature genetics. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka N, Babazono T, Saito S, Sekine A, Tsunoda T, Haneda M, Tanaka Y, Fujioka T, Kaku K, Kawamori R, Kikkawa R, Iwamoto Y, Nakamura Y, Maeda S. Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphisms. Diabetes. 2003;52:2848–2853. doi: 10.2337/diabetes.52.11.2848. [DOI] [PubMed] [Google Scholar]
- 46.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, Kawai K, Iiizumi T, Imanishi M, Shinosaki T, Yanagimoto T, Ikeda M, Omachi S, Kashiwagi A, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakajima M, Nakamura Y, Maeda S. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54:1171–1178. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 47.Maeda S, Kobayashi MA, Araki S, Babazono T, Freedman BI, Bostrom MA, Cooke JN, Toyoda M, Umezono T, Tarnow L, Hansen T, Gaede P, Jorsal A, Ng DP, Ikeda M, Yanagimoto T, Tsunoda T, Unoki H, Kawai K, Imanishi M, Suzuki D, Shin HD, Park KS, Kashiwagi A, Iwamoto Y, Kaku K, Kawamori R, Parving HH, Bowden DW, Pedersen O, Nakamura Y. A single nucleotide polymorphism within the acetyl coenzyme A carboxylase beta gene is associated with proteinuria in patients with type 2 diabetes. PLoS Genet. 2010;6:e1000842. doi: 10.1371/journal.pgen.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pezzolesi MG, Katavetin P, Kure M, Poznik GD, Skupien J, Mychaleckyj JC, Rich SS, Warram JH, Krolewski AS. Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes. 2009;58:2698–2702. doi: 10.2337/db09-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, McKay GJ, Williams WW, Sadlier DM, Makinen VP, Swan EJ, Palmer C, Boright AP, Ahlqvist E, Deshmukh HA, Keller BJ, Huang H, Ahola AJ, Fagerholm E, Gordin D, Harjutsalo V, He B, Heikkila O, Hietala K, Kyto J, Lahermo P, Lehto M, Lithovius R, Osterholm AM, Parkkonen M, Pitkaniemi J, Rosengard-Barlund M, Saraheimo M, Sarti C, Soderlund J, Soro-Paavonen A, Syreeni A, Thorn LM, Tikkanen H, Tolonen N, Tryggvason K, Tuomilehto J, Waden J, Gill GV, Prior S, Guiducci C, Mirel DB, Taylor A, Hosseini SM, Parving HH, Rossing P, Tarnow L, Ladenvall C, Alhenc-Gelas F, Lefebvre P, Rigalleau V, Roussel R, Tregouet DA, Maestroni A, Maestroni S, Falhammar H, Gu T, Mollsten A, Cimponeriu D, Ioana M, Mota M, Mota E, Serafinceanu C, Stavarachi M, Hanson RL, Nelson RG, Kretzler M, Colhoun HM, Panduru NM, Gu HF, Brismar K, Zerbini G, Hadjadj S, Marre M, Groop L, Lajer M, Bull SB, Waggott D, Paterson AD, Savage DA, Bain SC, Martin F, Hirschhorn JN, Godson C, Florez JC, Groop PH, Maxwell AP. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8:e1002921. doi: 10.1371/journal.pgen.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boger CA, Sedor JR. GWAS of diabetic nephropathy: is the GENIE out of the bottle? PLoS Genet. 2012;8:e1002989. doi: 10.1371/journal.pgen.1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28:323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chasman DI, Fuchsberger C, Pattaro C, Teumer A, Boger CA, Endlich K, Olden M, Chen MH, Tin A, Taliun D, Li M, Gao X, Gorski M, Yang Q, Hundertmark C, Foster MC, O'Seaghdha CM, Glazer N, Isaacs A, Liu CT, Smith AV, O'Connell JR, Struchalin M, Tanaka T, Li G, Johnson AD, Gierman HJ, Feitosa MF, Hwang SJ, Atkinson EJ, Lohman K, Cornelis MC, Johansson A, Tonjes A, Dehghan A, Lambert JC, Holliday EG, Sorice R, Kutalik Z, Lehtimaki T, Esko T, Deshmukh H, Ulivi S, Chu AY, Murgia F, Trompet S, Imboden M, Coassin S, Pistis G, Harris TB, Launer LJ, Aspelund T, Eiriksdottir G, Mitchell BD, Boerwinkle E, Schmidt H, Cavalieri M, Rao M, Hu F, Demirkan A, Oostra BA, de Andrade M, Turner ST, Ding J, Andrews JS, Freedman BI, Giulianini F, Koenig W, Illig T, Meisinger C, Gieger C, Zgaga L, Zemunik T, Boban M, Minelli C, Wheeler HE, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Nothlings U, Jacobs G, Biffar R, Ernst F, Homuth G, Kroemer HK, Nauck M, Stracke S, Volker U, Volzke H, Kovacs P, Stumvoll M, Magi R, Hofman A, Uitterlinden AG, Rivadeneira F, Aulchenko YS, Polasek O, Hastie N, Vitart V, Helmer C, Wang JJ, Stengel B, Ruggiero D, Bergmann S, Kahonen M, Viikari J, Nikopensius T, Province M, Ketkar S, Colhoun H, Doney A, Robino A, Kramer BK, Portas L, Ford I, Buckley BM, Adam M, Thun GA, Paulweber B, Haun M, Sala C, Mitchell P, Ciullo M, Kim SK, Vollenweider P, Raitakari O, Metspalu A, Palmer C, Gasparini P, Pirastu M, Jukema JW, Probst-Hensch NM, Kronenberg F, Toniolo D, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Siscovick DS, van Duijn CM, Borecki IB, Kardia SL, Liu Y, Curhan GC, Rudan I, Gyllensten U, Wilson JF, Franke A, Pramstaller PP, Rettig R, Prokopenko I, Witteman J, Hayward C, Ridker PM, Parsa A, Bochud M, Heid IM, Kao WH, Fox CS, Kottgen A. Integration of genome-wide association studies with biological knowledge identifies six novel genes related to kidney function. Hum Mol Genet. 2012;21:5329–5343. doi: 10.1093/hmg/dds369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bulik-Sullivan BK, Sullivan PF. The authorship network of genome-wide association studies. Nature genetics. 2012;44:113. doi: 10.1038/ng.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGuire AL, Basford M, Dressler LG, Fullerton SM, Koenig BA, Li R, McCarty CA, Ramos E, Smith ME, Somkin CP, Waudby C, Wolf WA, Clayton EW. Ethical and practical challenges of sharing data from genome-wide association studies: the eMERGE Consortium experience. Genome Res. 2011;21:1001–1007. doi: 10.1101/gr.120329.111. [DOI] [PMC free article] [PubMed] [Google Scholar]