Abstract
Fosfomycin, a natural product antibiotic, has been in use for >20 years in Spain, Germany, France, Japan, Brazil, and South Africa for urinary tract infections (UTIs) and other indications and was registered in the United States for the oral treatment of uncomplicated UTIs because of Enterococcus faecalis and Escherichia coli in 1996. It has a broad spectrum, is bactericidal, has very low toxicity, and acts as a time-dependent inhibitor of the MurA enzyme, which catalyzes the first committed step of peptidoglycan synthesis. Whereas resistance to fosfomycin arises rapidly in vitro through loss of active transport mechanisms, resistance is rarely seen during therapy of UTIs, seemingly because of the low fitness of the resistant organisms. Recently, interest has grown in the use of fosfomycin against multidrug-resistant (MDR) pathogens in other indications, prompting the advent of development in the United States of a parenteral formulation for use, initially, in complicated UTIs. Whereas resistance has not been problematic in the uncomplicated UTI setting, it remains to be seen whether resistance remains at bay with expansion to other indications.
Fosfomycin is a broad-spectrum antibiotic that is mainly used to treat urinary tract infections. Interest in its use for other, more complex indications is growing; it is unknown whether resistance will be a concern.
Fosfomycin, originally called phosphonomycin, is a broad spectrum antibiotic first found in fermentation broths of Streptomyces fradiae (ATCC 21096) in Spain through a collaborative effort of Merck and the Compañía Española de Penicilina y Antibióticos (CEPA) (Hendlin et al. 1969). Fosfomycin was initially developed in Europe by CEPA and has been in use since the early 1970s, initially as an IV preparation of the disodium salt and later as an oral formulation of fosfomycin trometamol. Its primary use in Spain, Germany, France, Japan, Brazil, and South Africa has been as an oral treatment for urinary tract infections (UTIs) but it has also been used more broadly in other indications (Falagas et al. 2008, 2009, 2010a). Fosfomycin was approved for use in the United States (as Monurol, fosfomycin tromethamine [same as trometamol]) in 1996 for treatment by single-dose oral therapy of uncomplicated UTIs (acute cystitis) in women caused by Escherichia coli and Enterococcus faecalis. With the problem of increasing resistance to other antibiotics, parenteral use of fosfomycin has been studied in therapy of a variety of infections because it is active against many multidrug-resistant (MDR) pathogens (Falagas et al. 2009) and is now under development in the United States for parenteral treatment of complicated UTIs (Zavante 2016).
DISCOVERY AND SPECTRUM
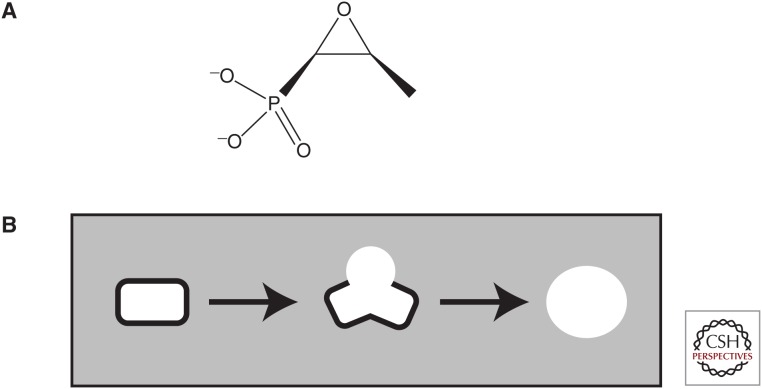
Fosfomycin (Fig. 1A) is a phosphonic acid antibiotic discovered in Spain in a fermentation broth of S. fradiae by means of a Merck screen for inhibition of peptidoglycan synthesis, the SPHERO assay (Gadebusch et al. 1992). In this morphological assay, schematized in Figure 1B, Gram-negative bacilli are grown in osmotically protective medium and treated for several doublings with test samples. Inhibitors of steps in the synthesis of peptidoglycan will lead to the production of microscopically recognizable refractile spheroplasts. Fosfomycin is also produced by several other Streptomyces, including Streptomyces viridochromogenes (ATCC21240) and Streptomyces wedmorensis (ATCC 21239), as well as Pseudomonas syringae (Shoji et al. 1986), Pseudomonas viridiflava, and Pseudomonas fluorescens (Katayama et al. 1990).
Figure 1.
Fosfomycin and the screen in which it was discovered. (A) Structure of fosfomycin. (B) Schematic of SPHERO assay. Gram-negative rods grown in osmotically protective medium, when treated with inhibitors of peptidoglycan synthesis, will show morphological changes leading to the production of round cells called spheroplasts, which are highly differentiable microscopically from normal rods and debris. The assay was run in plastic trays with wells for growth of cells and the wells viewed, after incubation, with a stereomicroscope.
Early tests indicated that fosfomycin was efficacious via IV dosing against intraperitoneal murine infections by specific isolates of E. coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus vulgaris, Salmonella schottmuelleri, Staphylococcus aureus, and Streptococcus pyogenes—although ED50s (median effective dose) against K. pneumoniae, P. aeruginosa, and S. pyogenes were high (although attainable and safe) (Hendlin et al. 1969). More recent reports note its broad spectrum, including activity against many important pathogens such as S. aureus (including methicillin-resistant S. aureus [MRSA]), Staphylococcus epidermidis, Streptococcus pneumoniae, E. faecalis, E. coli, Proteus species, K. pneumoniae, Enterobacter species, Serratia marcescens, and Salmonella typhi. Whereas P. aeruginosa shows variable susceptibility, fosfomycin has shown anti-pseudomonal efficacy especially in combinations with cefepime, aztreonam, and meropenem (Falagas et al. 2008). Acinetobacter, Vibrio fischeri, Chlamydia trachomatis, and Bacteroides species are resistant to fosfomycin (Falagas et al. 2008; Karageorgopoulos et al. 2012). Fosfomycin is not cross-resistant with other antibiotics because of its unique structure and mechanism of action.
MECHANISM OF ACTION
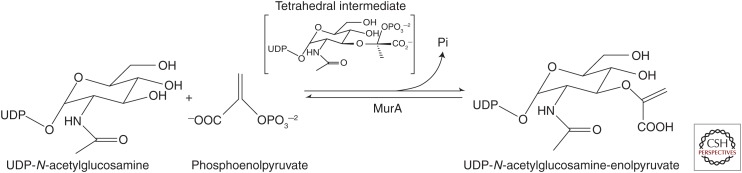
Fosfomycin is an inhibitor of the MurA enzyme, UDP-N-acetylglucosamine-enolpyruvyltransferase, that catalyzes the first committed step in peptidoglycan synthesis, the reaction of UDP-N-acetylglucosamine (UDP-GlcNAc) with phosphoenolpyruvate (PEP) to form UDP-GlcNAc-enoylpyruvate plus inorganic phosphate (shown in Fig. 2). The finding that competition of fosfomycin inhibition by PEP implied that fosfomycin was acting as a PEP analog (Kahan et al. 1974). Fosfomycin covalently binds, in a time- dependent reaction involving nucleophilic attack by the cysteine 115 residue of E. coli MurA on the epoxide of fosfomycin (Marquardt et al. 1994), and C115 is considered responsible for the catalytic action on PEP in the synthetic reaction (Wanke and Amrhein 1993; Brown et al. 1994).
Figure 2.
Overall reaction performed by the MurA enzyme.
In vitro, the MurA reaction proceeds with UDP-GlcNAc binding to the so-called open form of MurA, leading to a structural change to a closed form to which PEP binds (Skarzynski et al. 1996) leading to a tetrahedral intermediate and product release (Eschenburg et al. 2005). Even though it acts as an analog of PEP, fosfomycin’s interaction with MurA is highly selective (Kahan et al. 1974; Marquardt et al. 1994) and shows extremely low toxicity. Its LD50 (median lethal dose) in mice by intraperitoneal dosing of Na2-fosfomycin was 4 g/kg, lethality likely being because of sodium content (Hendlin et al. 1969).
The existence of the MurA analog of Mycobacterium tuberculosis that is naturally resistant to fosfomycin and has an asparagine in the position comparable to the C115 led to studies revealing further details of MurA reactions. Replacement of the C115 of the E. coli enzyme by asparagine (to yield a C115D enzyme) leads to fosfomycin resistance (Kim et al. 1996) and, as expected, replacement of asparagine in the M. tuberculosis enzyme with cysteine endows it with fosfomycin sensitivity (De Smet et al. 1999). This indicates that C115 is not necessarily required for the catalytic activity of the enzyme, but this activity can be supplied by the asparagine acting as a general acid (Kim et al. 1996). Substitution of C115 of Enterobacter cloacae MurA by serine (C115S) yielded an enzyme with the reaction products, UDP-GlcNAc-enoylpyruvate and Pi, bound in the active site (Eschenburg et al. 2005), leading the authors to conclude that C115 is necessary for turnover and release of the products. They reasoned that, in C115S, other residues could carry out the catalytic step and that, in the C115D enzyme, the flexible asparagine should be able to catalyze the release of the reaction products.
Recent work (Zhu et al. 2012) indicates that, in the cell, the activity of MurA is subject to regulation by the binding of UDP-NAc-muramic acid (UDP-MurNAc), the product of the MurB enzyme, to MurA. In the presence of bound UDP-MurNAc, PEP covalently attaches to C115, leading to the formation of a “locked” dormant tertiary complex (MurA:PEP-UDP-MurNAc), which is the predominant form of cellular MurA. This had apparently been missed previously because much crystallographic work with MurA had been done by diluting samples into phosphate buffer, which releases the ligands. Extensive crystallographic findings by these authors led them to a model for the regulatory and reaction schemes: when the UDP-MurNAc to UDP-GlcNAc ratio in the cell decreases, UDP-GlcNAc replaces UDP-MurNAc in the dormant complex in a rapidly reversible manner, leading to formation of the tetrahedral reaction intermediate that then yields the products, still bound in the active site. PEP enters, reacts with C115, and displaces Pi, stimulating the reversible exchange of UDP-GlcNAc with UDP-GlcNAc-enoylpyruvate, leading to its release to restart the cycle. The authors speculate that the existence of the locked dormant complex may explain why discovery of reversible, noncovalent inhibitors of MurA has been difficult. In this model, then, the covalent interaction of PEP with C115 is necessary for formation of the dormant complex and also for release of the reaction products (as had been proposed by Eschenburg et al. 2005) but is not necessary for catalysis by the enzyme that may be supplied by other residues (as had been proposed by Kim et al. 1996).
In low GC Gram-positives, there are two “murA” genes, murA and murZ, the products of which are structurally very similar (Du et al. 2000; Blake et al. 2009). In the case of the S. pneumoniae and S. aureus enzymes, each gene product is capable of sustaining peptidoglycan synthesis and both isozymes are sensitive to fosfomycin in vitro (Blake et al. 2009). Neither gene is essential but a double deletion is not viable. In Bacillus subtilis (Kobayashi et al. 2003; Kock et al. 2004) and Bacillus anthracis (Kedar et al. 2008), however, only the murA gene is essential and, evidently, the second enzyme cannot substitute.
FOSFOMYCIN UPTAKE
In E. coli, two carrier-dependent systems can actively transport fosfomycin. Initial work showed that fosfomycin could be transported by the system for uptake of α-glycerophosphate and that mutants in the glpT gene, encoding the α-glycerophosphate permease, were 30-fold more resistant than the isogenic parent strain. Whereas α-glycerophosphate will induce expression of GlpT, the basal levels of GlpT are sufficient for fosfomycin uptake (Kahan et al. 1974). The GlpT system is widespread, found at least in P. aeruginosa, E. coli, Salmonella, Shigella flexneri, Klebsiella, Haemophilus influenzae, S. aureus, B. subtilis, E. faecalis, and Rickettsia prowazekii (Kahan et al. 1974; Lemieux et al. 2004). It was also shown that the blood component, glucose-6-phosphate (G-6-P), could induce the hexose-phosphate uptake system, UhpT, to levels sufficient for fosfomycin transport (Zimmerman et al. 1969; Kahan et al. 1974). The UhpT system is limited to Enterobacteriaceae (with the exception of Proteus species) and S. aureus (Winkler 1973). Importantly, Pseudomonas does not have a UhpT system and so solely uses glpT for uptake (Winkler 1973; Castañeda-García et al. 2009).
FOSFOMYCIN RESISTANCE
The mechanisms of resistance to fosfomycin have been recently reviewed (Karageorgopoulos et al. 2012; Castañeda-García et al. 2013; Nikolaidis et al. 2014). It should be noted that the breakpoints for fosfomycin susceptibility have been formalized for few species and are different for the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST break points for Enterobacteriaceae (oral or IV) and S. aureus (IV only) are S ≤ 32 mg/L and R > 32 mg/L (in the presence of 25 mg/L G-6-P). Wild-type isolates of Pseudomonas species minimum inhibitory concentrations (MICs) ≤128 mg/L have been treated with combinations (European Committee on Antimicrobial Susceptibility Testing 2016). CLSI breakpoints for E. coli and E. faecalis are S ≤ 64 mg/L, I 128 mg/L, R ≥ 256 mg/L (also in the presence of G-6-P) (Performance Standards for Antimicrobial Susceptibility Testing 2012).
Mutational Resistance
Resistance to fosfomycin arises in E. coli at high frequencies in vitro, because of loss of transport systems required for uptake. As noted above, in E. coli, uptake of fosfomycin can be mediated by the GlpT permease system, whose main substrate is α-glycerophosphate, and also, when induced by G-6-P, by the UhpT system (Kahan et al. 1974). Both of these systems are positively regulated by cAMP, and cAMP levels can be lowered by mutations in the ptsI or cyaA genes (which will also affect catabolism of a variety of carbohydrates) (Alper and Ames 1978; Tsuruoka et al. 1978; Castañeda-García et al. 2009). For E. coli grown in the absence of G-6-P, in which only the GlpT permease is active, the frequency of resistance (ascertained by fluctuation test) is 10−7, while in the presence of G-6-P the frequency is 10−8 (Nilsson et al. 2003). Most of the mutations seen in the Nilsson study were located in genes leading to a decrease in cAMP. Nilsson noted that despite this finding of high frequencies of resistance in vitro the rates of clinical resistance to fosfomycin in E. coli throughout Europe from 1999 to 2000 (Kahlmeter 2000) were uniformly low, in the range of 0.7% to 1.5%, with no differences between countries with a long history of fosfomycin use and those not using it. As opposed to their in vitro findings, 13 resistant clinical isolates tested by Nilsson were found to have mutations almost exclusively in glpT and/or uhpA or uhpT, with none found in the cAMP regulatory loci, perhaps indicating a selective disadvantage in vivo. Whereas mutants grew well in the absence of fosfomycin, it was found that growth rates of the three tested in vitro mutants and 12 of the 13 clinical isolates were severely reduced in growth rate in LB medium or urine in the presence of ≥8 mg/L of fosfomycin, which is significantly lower than the fosfomycin concentration in urine during treatment, normally greater than 128 mg/L. Mathematical modeling indicated that the growth rate retardation seen should be enough to prevent resistant strains from establishment in the bladder (Nilsson et al. 2003). Slow growth and the absence of the cAMP-related mutations in vivo could explain the discrepancy between high-frequency mutational resistance to fosfomycin in vitro and rates of resistance seen in the clinic.
Interestingly, very few instances have been reported of mutations in the fosfomycin target gene, murA. A mutation in murA of E. coli was isolated after mutagenesis (Wu and Venkateswaran 1974) and counterselection against transport mutants and two murA mutants of E. coli were reported among clinical isolates in a Japanese study (Takahata et al. 2010) It is likely that, in vitro, the frequency of transport mutants is so great relative to target mutants that they are not generally seen. The finding of murA mutants with clinical isolates of E. coli might reflect the low fitness of in vitro transport mutants.
Recent in vitro experiments (Couce et al. 2012) showed that overexpression of the murA gene by induction of a regulated promoter can lead to greatly increased MICs, to levels that would afford clinical resistance, while having relatively low effects on fitness (relative to mutations to fosfomycin resistance found in clinical isolates). However, this has not been noted yet in clinical isolates.
A study of a set of 441 Italian Gram-negative urinary isolates (Marchese et al. 2003) showed very high susceptibility to fosfomycin of E. coli isolates (99%), 87.5% susceptibility for Proteus species, whereas other species showed variable susceptibilities. Fosfomycin-resistant mutants (≥2000 mg/L) of sensitive E. coli, K. pneumoniae, and P. aeruginosa were obtained through stepwise selection on agar containing fosfomycin, and the growth rates of these resistant mutants were compared to their parental strains. In contrast to the mutants tested by Nilsson et al., these all showed significantly reduced growth rates in a variety of conditions (in the absence of fosfomycin) as well as reduced adhesion to uroepithelial cells and to urinary catheters.
A study of fosfomycin resistance in Pseudomonas (Rodríguez-Rojas et al. 2010b) showed that a glpT null mutant was equal in virulence to its glp+ parent in a mouse lung infection survival model, had equivalent biofilm-forming ability, and caused similar inflammation in histological studies. This and other data were interpreted by the authors to show that mutational resistance in P. aeruginosa may have no obvious fitness cost in vivo. It may well be that the site of infection contributes to the difference in apparent fitness seen between P. aeruginosa in the lung and E. coli in the bladder.
As there is interest in use of parenteral fosfomycin for treatment of indications in addition to uncomplicated UTI, recent hollow fiber pharmacokinetic/pharmacodynamic (PK/PD) studies were undertaken by the Ambrose group (VanScoy et al. 2015) to determine the PK/PD index for IV fosfomycin and to determine requirements for stasis, and 1- and 2-log reductions of colony-forming units. The results showed that, at least for the E. coli isolates studied, there was a large resistant subpopulation from the inception of infection. Thus, a new PK/PD index was instituted, %T > RIC—the percentage of the dosing interval in which the concentration was above the “resistance inhibitory concentration.” For stasis, 1- and 2-log reductions, %T > RIC were 11.9%, 20.9%, and 32.8%, respectively. The authors note some caveats with this type of in vitro work: in vivo fitness of the mutants is unknown and longer treatment will be required to model clinical treatment duration.
In vitro pharmacodynamics of Na2-fosfomycin against clinical 64 P. aeruginosa isolates was studied by an Australian group (Walsh et al. 2015). MICs from 1 to >512 were seen with 61% of isolates susceptible (MIC ≤ 64 mg/L), but all isolates tested had a resistant subpopulation as revealed by population analysis profiling. At low inocula (∼106 cfu/mL), there was moderate killing with regrowth by 24 h at most dosages. No killing was seen with high inocula (∼108 cfu/mL). This indicates that monotherapy of P. aeruginosa with fosfomycin may be problematic if this finding translates to in vivo conditions; furthermore, study of combination therapy is warranted.
Fosfomycin-Modifying Enzymes
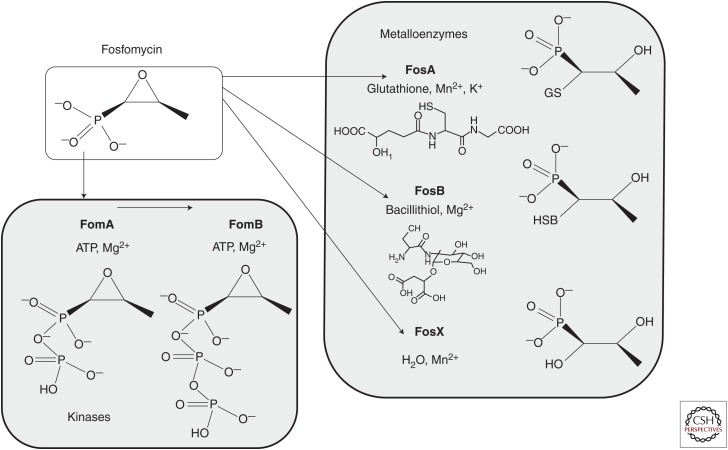
Several fosfomycin-modifying enzymes have been found that inactivate the drug. The main enzymes described are three types of metalloenzymes, FosA, FosB, and FosX, and two kinases, FomA and FomB, as shown in Figure 3.
Figure 3.
Fosfomycin-modifying enzymes leading to inactivation of fosfomycin and responsible for fosfomycin resistance. Enzyme names are in bold and are shown with their substrates, metal cofactors, and the end products of their modifying activity. (This figure is adapted from Castañeda-García et al. 2013 and is used under license from Creative Commons, creativecommons.org/licenses/by/3.0.)
The metalloenzymes open the epoxide (oxirane ring) by the addition of various substrates as recently reviewed (Castañeda-García et al. 2013). FosA is a glutathione-S-transferase present on transposon TN2921 originally found on a plasmid in S. marcescens (Suárez and Mendoza 1991), using Mn2+ and K+ as metal cofactors; other related glutathione transferase (FosA type) enzymes found to be plasmid-borne are FosA3, FosA4, FosA5, and FosC2. A related FosA enzyme is found encoded in the chromosome of P. aeruginosa.
Recent studies in China (Li et al. 2015) found that 7.8% of nonduplicate E. coli clinical isolates collected from 20 geographically dispersed hospitals from July 2009 to June 2010 were nonsusceptible to fosfomycin (MIC > 64 mg per liter). Of these, 80% carried the fosA3 gene; the fosA3 gene of 42% of those isolates was transferable, presumably via a conjugative plasmid. With growing interest in the possible use of fosfomycin to combat resistant Gram-negative infections, a study was done to ascertain the prevalence of fosfomycin resistance in 278 K. pneumoniae isolates carrying the K. pneumoniae carbapenemase (KPC) and in 80 extended-spectrum β-lactams (ESBLs) (non-KPC) K. pneumoniae from 12 hospitals in China (Jiang et al. 2015). Fosfomycin resistance was 60.8% in KPC producers, compared to 12.5% among ESBL producers. Ninety-four of the KPC isolates were found to carry fosA3 genes and 92 of those appeared clonally related, with 71% belonging to a single clonal group. The distribution of fosfomycin resistance levels varied greatly among the different hospitals, but there was general correlation between levels of resistance and presence of fos3 genes. Rather alarming was the finding that, in a representative isolate of the predominant clone, the fosA3 and blaKPC-2 genes were colocalized on a plasmid designated pFOS18. The blaKPC-2 gene, located on a Tn3–Tn4401 structure, and fosA3, bracketed by IS26 sequences, are normally transmitted on plasmids. This data indicates that they can be linked on a single plasmid. The data from this recent study in China contrasts with earlier data from a U.S. study (Endimiani et al. 2010) in which fosfomycin susceptibility of 68 K. pneumoniae KPC-possessing strains was measured and 93% were found susceptible. It is likely that the Chinese study represents a clonal outbreak that has not (yet) spread to the United States. It will be necessary to monitor the spread of such clones.
FosA3 was also found on CTX-M plasmids of E. coli in Japan, flanked there as well by IS26 elements (Sato et al. 2013). An unexpected increase in fosfomycin resistance of E. coli CTX-M-15-carrying strains in Madrid was studied and found to be caused by two main clonal types (Oteo et al. 2009). In the larger clone, none of the normal mutational resistance alleles were found, whereas in the second clone of five strains, there was a small deletion in the uhpA gene and a single IS26 linked to blaCTX-M-15. There was no mention of genes associated with the IS26. Is it possible that these clones could contain fosfomycin-modifying enzymes, perhaps Fos3 associated with CTX-M as in the Japanese isolates (Sato et al. 2013)?
The regulation of expression of the fosA gene that is resident in the genome of P. aeruginosa is not well studied, if at all. It would seem critically important to study the conditions under which fosA is expressed in P. aeruginosa, to ascertain what its effect is on basal MICs and whether sufficient expression to yield a resistant phenotype can be achieved by mutation or induction. One reference (De Groote et al. 2011) notes that fosfomycin resistance in P. aeruginosa can be caused by (engineered) overexpression of the resident FosA because of an inserted promoter. Whether turn-on of FosA in P. aeruginosa is a possible source of mutational resistance is not known. However, the main mutational resistance in P. aeruginosa is clearly mutations in the GlpT system (Castañeda-García et al. 2009), because no spontaneous fosfomycin-resistant colonies grew on plates containing α-glycerophosphate as a carbon source, and all 10 sequenced mutations in this study were missense mutations in glpT.
FosB type enzymes, first discovered in S. epidermidis (Zilhao and Courvalin 1990), are found in low GC Gram-positive bacteria such as B. subtilis, B. anthracis, S. epidermidis, and S. aureus, species that do not make glutathione but use bacillithiol (shown in Fig. 3) as a substitute thiol donor. These bacillithiol-S-transferases use Mg2+ as a cofactor. The FosB of S. aureus is chromosomally located and is responsible for the innate level of fosfomycin activity—as deletion of either the fosB gene or the bacillithiol synthetic machinery greatly increases the sensitivity to fosfomycin (Thompson et al. 2014).
FosX is related to the FosA and FosB enzymes, but it is an Mn2+-dependent epoxide hydrolase, using water to break the ring. FosX enzymes are found in Listeria monocytogenes, Clostridium botulinum, and Brucella melitensis (Castañeda-García et al. 2013).
FomA and FomB are kinases from S. wedmorensis, a species that produces fosfomycin (Kuzuyama et al. 1996), that sequentially add phosphates to the phosphonate moiety of fosfomycin from ATP with Mg2+ as a cofactor. Presumably, these enzymes act to provide autoresistance to the producers. Another such kinase, originally called FosC and found in the fosfomycin producer P. syringae, is actually an ortholog of FomA (Kim et al. 2012).
CLINICAL CONSIDERATIONS ON RESISTANCE
As fosfomycin has low toxicity and allergenicity, a broad spectrum of activity, including MDR organisms, good pharmacokinetics, and is available in parenteral as well as oral formulations, its use in non-UTI indications for hard-to-treat infections has become attractive. But there is not a large body of data on results of controlled trials in other indications. The Falagas group has published a large number of meta-analyses of small studies of fosfomycin use in various settings. Initial data supported use of fosfomycin for treatment of UTIs caused by MDR Enterobacteriaceae, including those carrying ESBLs (Falagas et al. 2010a). Resistance data from studies after 2010 (Falagas et al. 2016) was mostly on pathogens from urine samples and showed that E. coli susceptibility ranged from 82% to 100% and K. pneumoniae from 15% to 100%. There was less data on other Enterobacteriaceae, but fosfomycin was active against 72% to 97.5%. Fosfomycin was 90.5% to 100% active against MDR Enterobacteriaceae. One study showed 80.6% susceptibility to fosfomycin of carbapenem-resistant P. aeruginosa strains. Carbapenem-resistant Acinetobacter baumannii was resistant. No major differences in resistance were seen between data published before 2010 and that published after for Gram-negatives and S. aureus.
The main question is whether resistance to fosfomycin will compromise its use in non-UTI indications. There is no adequate answer as yet. It is clear that the single-dose oral treatment of cystitis with fosfomycin trometamol/tromethamine is highly effective and equivalent to other therapies with low evolution of resistance seen (Falagas et al. 2010b). Most of these infections are caused by E. coli, and it may be that the high urinary levels of fosfomycin attainable and the lowered fitness of mutants with impaired fosfomycin uptake (as discussed above) contribute to the efficacy of fosfomycin in this setting.
Combinations
Checkerboard assays, kill curves, and in vitro models have been used to predict the efficacy and potential synergy of combinations of fosfomycin. Whereas monotherapy against E. coli and E. faecalis cystitis has been successful over the years, there is concern that treatment of other pathogens at other sites where local drug concentrations will likely be lower than in urine may lead to resistance selection. Thus, combination therapy is being evaluated.
Combinations with underused cell-wall inhibitors were tested against E. coli and K. pneumoniae (Hickman et al. 2013), including MDR strains, leading to the finding that a fosfomycin/aztreonam and a fosfomycin/aztreonam/mecillinam combination were effective in reducing the population in a variety of models, including a new in vitro kinetic model.
An examination of resistance selection by fosfomycin in combination with a number of drugs in both wild-type and a mutator strain (Rodríguez-Rojas et al. 2010a) gave the interesting result that, in wild-type, combinations with tobramycin, amikacin, meropenem, ceftazidime, ciprofloxacin, and colistin gave frequencies below the limit of detection (<1 × 10−10), but the combination of fosfomycin and imipenem had a frequency of 1.1 × 10−9, higher than the product of the individual frequencies (3.5 × 10−13). For the mutator strain, in which frequencies for the single drugs were ∼100 fold higher than in wild-type, all combinations yielded frequencies below the limit of detection except for fosfomycin plus imipenem or ceftazidime, which were 1.1 × 10−7 and 1 × 10−8, respectively. There was, apparently, some sort of antagonistic effect occurring.
A recent publication (Walsh et al. 2016) reported testing combinations of fosfomycin with several drugs in killing P. aeruginosa. Against fosfomycin-susceptible isolates, fosfomycin monotherapy led to efficient killing but rapid regrowth. Combination of fosfomycin with polymyxin B or tobramycin against these susceptible isolates and combination of ciprofloxacin plus fosfomycin against fosfomycin-resistant isolates led to increased killing (versus monotherapy), but did not prevent regrowth of resistant mutants. Certainly, repeat dosing should be tried to ascertain whether rebound could be prevented with further treatment, but this study is sobering.
CONCLUSIONS
Fosfomycin is an old antibiotic, but it has proven useful. In this age of increasing antibiotic resistance, fosfomycin is being reconsidered for use against MDR pathogens within its spectrum of action. But, if it is true that fosfomycin has proven effective for uncomplicated UTIs because of fitness cost of mutants and high urinary drug concentrations, then these are variables that must be addressed to move on to indications involving other body sites. The use of combinations may help to keep resistance at bay, but that will likely rely on matched pharmacokinetics. Furthermore, data from various tests of combinations against P. aeruginosa show the need to choose combinations carefully. Dosing of drug at “resistance-inhibitory” or “mutant-prevention” concentrations may be a more realistic approach. Whereas the frequencies of fosfomycin resistance seen over time have not changed very much, that is likely because of the fact that most of the present resistance seen is a result of mutations in the pathogen. The reports on plasmid-borne fosfomycin resistance forewarn of increased levels of resistance, likely through spread of problematic clones in which resistance to fosfomycin is linked to resistance to other drugs, especially β-lactams. The Chinese report of FosA3 and KPC-2 on a single plasmid and its likely clonal spread (Jiang et al. 2015) are very worrying. Whereas drugs to treat the growing threat of MDR bacteria are sorely needed, and fosfomycin has some attractive properties that could favor its use for treatment of certain life-threatening infections, to retain its productive use as a single-dose oral therapy for cystitis, stewardship and monitoring the spread of plasmid-borne resistance will be needed.
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Alper MD, Ames BN. 1978. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: Positive selection of Salmonella typhimurium cya and crp mutants. J Bacteriol 133: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake KL, O’Neill AJ, Mengin-Lecreulx D, Henderson PJ, Bostock JM, Dunsmore CJ, Simmons KJ, Fishwick CW, Leeds JA, Chopra I. 2009. The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis inhibitors. Mol Microbiol 72: 335–343. [DOI] [PubMed] [Google Scholar]
- Brown ED, Marquardt JL, Lee JP, Walsh CT, Anderson KS. 1994. Detection and characterization of a phospholactoyl-enzyme adduct in the reaction catalyzed by UDP-N-acetylglucosamine enolpyruvoyl transferase, MurZ. Biochemistry 33: 10638–10645. [DOI] [PubMed] [Google Scholar]
- Castañeda-García A, Rodríguez-Rojas A, Guelfo JR, Blázquez J. 2009. The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J Bacteriol 191: 6968–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda-García A, Blázquez J, Rodríguez-Rojas A. 2013. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2: 217–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couce A, Briales A, Rodríguez-Rojas A, Costas C, Pascual A, Blázquez J. 2012. Genomewide overexpression screen for fosfomycin resistance in Escherichia coli: MurA confers clinical resistance at low fitness cost. Antimicrob Agents Chemother 56: 2767–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote VN, Fauvart M, Kint CI, Verstraeten N, Jans A, Cornelis P, Michiels J. 2011. Pseudomonas aeruginosa fosfomycin resistance mechanisms affect non-inherited fluoroquinolone tolerance. J Med Microbiol 60: 329–336. [DOI] [PubMed] [Google Scholar]
- De Smet KAL, Kempsell KE, Gallagher A, Duncan K, Young DB. 1999. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology 145: 3177–3184. [DOI] [PubMed] [Google Scholar]
- Du W, Brown JR, Sylvester DR, Huang J, Chalker AF, So CY, Holmes DJ, Payne DJ, Wallis NG. 2000. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in Gram-positive bacteria. J Bacteriol 182: 4146–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endimiani A, Patel G, Hujer KM, Swaminathan M, Perez F, Rice LB, Jacobs MR, Bonomo RA. 2010. In vitro activity of fosfomycin against blaKPC-containing Klebsiella pneumoniae isolates, including those nonsusceptible to tigecycline and/or colistin. Antimicrob Agents Chemother 54: 526–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenburg S, Priestman M, Schonbrunn E. 2005. Evidence that the fosfomycin target Cys115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J Biol Chem 280: 3757–3763. [DOI] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoint tables for interpretation of MICs and zone diameters. Version 6. www.eucast.org/clinical_breakpoints. [Google Scholar]
- Falagas ME, Giannopoulou KP, Kokolakis GN, Rafailidis PI. 2008. Fosfomycin: Use beyond urinary tract and gastrointestinal infections. Clin Inf Dis 46: 1069–1077. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. 2009. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: A systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents 34: 111–120. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. 2010a. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum β-lactamase producing, Enterobacteriaceae infections: A systematic review. Lancet Inf Dis 10: 43–50. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Vouloumanou EK, Togias AG, Karadima M, Kapaskelis AM, Rafailidis PI, Athanasiou S. 2010b. Fosfomycin versus other antibiotics for the treatment of cystitis: A meta-analysis of randomized controlled trials. J Antimicrob Chemother 65: 1862–1877. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. 2016. Fosfomycin. Clin Microbiol Rev 29: 321–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadebusch HH, Stapley EO, Zimmerman SB. 1992. The discovery of cell wall active antibacterial antibiotics. Crit Rev Biotechnol 12: 225–243. [DOI] [PubMed] [Google Scholar]
- Hendlin D, Stapley EO, Jackson M, Wallick H, Miller AK, Wolf FJ, Miller TW, Chaiet L, Kahan FM, Foltz EL, et al. 1969. Phosphonomycin, a new antibiotic produced by strains of Streptomyces. Science 166: 122–123. [DOI] [PubMed] [Google Scholar]
- Hickman RA, Hughes D, Cars T, Malmberg C, Cars O. 2013. Cell-wall-inhibiting antibiotic combinations with activity against multidrug-resistant Klebsiella pneumoniae and Escherichia coli. Clin Microbiol Inf 20: O267–O273. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Shen P, Wei Z, Liu L, He F, Shi K, Wang Y, Wang H, Yu Y. 2015. Dissemination of a clone carrying a fosA3-harbouring plasmid mediates high fosfomycin resistance rate of KPC-producing Klebsiella pneumoniae in China. Int J Antimicrob Agents 45: 66–70. [DOI] [PubMed] [Google Scholar]
- Kahan FM, Kahan JS, Cassidy PJ, Kropp H. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann NY Acad Sci 235: 364–386. [DOI] [PubMed] [Google Scholar]
- Kahlmeter G. 2000. The ECO•SENS Project: A prospective, multinational, multicentre epidemiological survey of the prevalence and antimicrobial susceptibility of urinary tract pathogens—Interim report. J Antimicrob Chemother 46: 15–22. [PubMed] [Google Scholar]
- Karageorgopoulos DE, Wang R, Yu XH, Falagas ME. 2012. Fosfomycin: Evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J Antimicrob Chemother 67: 255–268. [DOI] [PubMed] [Google Scholar]
- Katayama N, Tsubotani S, Nozaki Y, Harada S, Ono H. 1990. Fosfadecin and fosfocytocin, new nucleotide antibiotics produced by bacteria. J Antibiot (Tokyo) 43: 238–246. [DOI] [PubMed] [Google Scholar]
- Kedar GC, Brown-Driver V, Reyes DR, Hilgers MT, Stidham MA, Shaw KJ, Finn J, Haselbeck RJ. 2008. Comparison of the essential cellular functions of the two murA genes of Bacillus anthracis. Antimicrob Agents Chemother 52: 2009–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Lees WJ, Kempsell KE, Lane WS, Duncan K, Walsh CT. 1996. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35: 4923–4928. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ju K-S, Metcalf WW, Evans BS, Kuzuyama T, van der Donk WA. 2012. Different biosynthetic pathways to fosfomycin in Pseudomonas syringae and Streptomyces species. Antimicrob Agents Chemother 56: 4175–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P. 2003. Essential Bacillus subtilis genes. Proc Natl Acad Sci 100: 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock H, Gerth U, Hecker M. 2004. MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol Microbiol 51: 1087–1102. [DOI] [PubMed] [Google Scholar]
- Kuzuyama T, Kobayashi S, O’Hara K, Hidaka T, Seto H. 1996. Fosfomycin monophosphate and fosfomycin diphosphate, two inactivated fosfomycin derivatives formed by gene products of fomA and fomB from a fosfomycin producing organism Streptomyces wedmorensis. J Antibiot (Tokyo) 49: 502–504. [DOI] [PubMed] [Google Scholar]
- Lemieux MJ, Huang Y, Wang DN. 2004. Glycerol-3-phosphate transporter of Escherichia coli: Structure, function and regulation. Res Microbiol 155: 623–629. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng B, Li Y, Zhu S, Xue F, Liu J. 2015. Antimicrobial susceptibility and molecular mechanisms of fosfomycin resistance in clinical Escherichia coli isolates in mainland China. PLoS ONE 10: e0135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, Gualco L, Debbia EA, Schito GC, Schito AM. 2003. In vitro activity of fosfomycin against Gram-negative urinary pathogens and the biological cost of fosfomycin resistance. Int J Antimicrob Agents 22: 53–59. [DOI] [PubMed] [Google Scholar]
- Marquardt JL, Brown ED, Lane WS, Haley TM, Ichikawa Y, Wong CH, Walsh CT. 1994. Kinetics, stoichiometry, and Identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry 33: 10646–10651. [DOI] [PubMed] [Google Scholar]
- Nikolaidis I, Favini-Stabile S, Dessen A. 2014. Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci 23: 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AI, Berg OG, Aspevall O, Kahlmeter G, Andersson DI. 2003. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob Agents Chemother 47: 2850–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteo J, Orden B, Bautista V, Cuevas O, Arroyo M, Martinez-Ruiz R, Perez-Vazquez M, Alcaraz M, Garcia-Cobos S, Campos J. 2009. CTX-M-15-producing urinary Escherichia coli O25b-ST131-phylogroup B2 has acquired resistance to fosfomycin. J Antimicrob Chemother 64: 712–717. [DOI] [PubMed] [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. 2012. 22nd informational supplement M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- Rodríguez-Rojas A, Couce A, Blázquez J. 2010a. Frequency of spontaneous resistance to fosfomycin combined with different antibiotics in Pseudomonas aeruginosa. Antimicrob Agents Chemother 54: 4948–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rojas A, Maciá MD, Couce A, Gómez C, Castañeda-García A, Oliver A, Blázquez J. 2010b. Assessing the emergence of resistance: The absence of biological cost in vivo may compromise fosfomycin treatments for P. aeruginosa infections. PLoS ONE 5: e10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Kawamura K, Nakane K, Wachino JI, Arakawa Y. 2013. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb Drug Res 19: 477–482. [DOI] [PubMed] [Google Scholar]
- Shoji J, Kato T, Hinoo H, Hattori T, Hirooka K, Matsumoto K, Tanimoto T, Kondo E. 1986. Production of fosfomycin (phosphonomycin) by Pseudomonas syringae. J Antibiot (Tokyo) 39: 1011–1012. [DOI] [PubMed] [Google Scholar]
- Skarzynski T, Mistry A, Wonacott A, Hutchinson SE, Kelly VA, Duncan K. 1996. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure 4: 1465–1474. [DOI] [PubMed] [Google Scholar]
- Suárez JE, Mendoza MC. 1991. Plasmid-encoded fosfomycin resistance. Antimicrob Agents Chemother 35: 791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Ida T, Hiraishi T, Sakakibara S, Maebashi K, Terada S, Muratani T, Matsumoto T, Nakahama C, Tomono K. 2010. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int J Antimicrob Agents 35: 333–337. [DOI] [PubMed] [Google Scholar]
- Thompson MK, Keithly ME, Goodman MC, Hammer ND, Cook PD, Jagessar KL, Harp J, Skaar EP, Armstrong RN. 2014. Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry 53: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruoka T, Miyata A, Yamada Y. 1978. Two kinds of mutants defective in multiple carbohydrate utilization isolated from in vitro fosfomycin-resistant strains of Escherichia coli K-12. J Antibiot (Tokyo) 31: 192–201. [DOI] [PubMed] [Google Scholar]
- VanScoy BD, McCauley J, Ellis-Grosse EJ, Okusanya OO, Bhavnani SM, Forrest A, Ambrose PG. 2015. Exploration of the pharmacokinetic–pharmacodynamic relationships for fosfomycin efficacy using an in vitro infection model. Antimicrob Agents Chemother 59: 7170–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CC, McIntosh MP, Peleg AY, Kirkpatrick CM, Bergen PJ. 2015. In vitro pharmacodynamics of fosfomycin against clinical isolates of Pseudomonas aeruginosa. J Antimicrob Chemother 70: 3042–3050. [DOI] [PubMed] [Google Scholar]
- Walsh CC, Landersdorfer CB, McIntosh MP, Peleg AY, Hirsch EB, Kirkpatrick CM, Bergen PJ. 2016. Clinically relevant concentrations of fosfomycin combined with polymyxin B, tobramycin or ciprofloxacin enhance bacterial killing of Pseudomonas aeruginosa, but do not suppress the emergence of fosfomycin resistance. J Antimicrob Chemother 10.1093/jac/dkw115. [DOI] [PubMed] [Google Scholar]
- Wanke C, Amrhein N. 1993. Evidence that the reaction of the UDP-N-acetylglucosamine 1-carboxyvinyltransferase proceeds through the O-phosphothioketal of pyruvic acid bound to Cys115 of the enzyme. Eur J Biochem 218: 861–870. [DOI] [PubMed] [Google Scholar]
- Winkler HH. 1973. Distribution of an inducible hexose-phosphate transport system among various bacteria. J Bacteriol 116: 1079–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Venkateswaran PS. 1974. Fosfomycin-resistant mutant of Escherichia coli. Ann NY Acad Sci 235: 587–592. [DOI] [PubMed] [Google Scholar]
- Zavante Therapeutics. 2016. Zavante initiates the ZEUS study for ZTI-01 for the treatment of complicated urinary tract infections, Zavante Therapeutics, Inc., www.zavante.com/news/zavante-initiates-the-zeus-study-for-zti-01-for-the-treatment-of-complicated-urinary-tract-infections. [Google Scholar]
- Zhu JY, Yang Y, Han H, Betzi S, Olesen SH, Marsilio F, Schönbrunn E. 2012. Functional consequence of covalent reaction of phosphoenolpyruvate with UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA). J Biol Chem 287: 12657–12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilhao R, Courvalin P. 1990. Nucleotide sequence of the fosB gene conferring fosfomycin resistance in Staphylococcus epidermidis. FEMS Microbiol Lett 68: 267–272. [DOI] [PubMed] [Google Scholar]
- Zimmerman S, Stapley E, Wallick H, Baldwin R. 1969. Phosphonomycin. IV: Susceptibility testing method and survey. Antimicrob Agents Chemother (Bethesda) 9: 303–309. [PubMed] [Google Scholar]