Abstract
Precision oncology relies on targeted drugs, such as kinase inhibitors, that are presently administered based on molecular profiles obtained from surgical or bioptic tissue samples. The inherent ability of human tumors to molecularly evolve in response to drug pressures represents a daunting diagnostic challenge. Circulating free DNA (cfDNA) released from primary and metastatic lesions can be used to draw molecular maps that can be continuously updated to match each tumor’s evolution. We will present evidence that liquid biopsies can effectively interrogate how targeted therapies drive lesion-specific drug-resistance mechanisms. The impact of drug-induced molecular heterogeneity on subsequent lines of treatment will also be discussed.
Circulating free DNA released from primary and metastatic tumors can capture the molecular complexity of a patient’s cancer and can be used to monitor the effectiveness of treatment over time.
The availability of detailed maps of cancer genomes and the development of oncogene- specific inhibitors have transformed clinical oncology treatment into what is commonly known as personalized therapy or, perhaps more appropriately, precision medicine. According to this innovative paradigm, treatment should be administered based on patient-specific tumor profiles that are presently derived from surgical or bioptic tumor samples.
Unfortunately, the effectiveness of targeted therapies is often transient and, almost invariably, drug resistance develops (Kobayashi et al. 2005; Pao et al. 2005; Montagut et al. 2008; Diaz et al. 2012; Misale et al. 2012). When solid tumors recur, further treatment is administered based on analyses of single needle biopsies or surgical excisions that are unlikely to accurately capture the comprehensive genomic landscape of a patient’s cancer. This is because the molecular profiles of drug-resistant tumors are often extremely complex (Greaves and Maley 2012; Burrell et al. 2013). Several evidences suggest that resistance can be driven by outgrowth of low-frequency—often multiple and concomitant—subclones rather than by evolution of an increasingly aggressive dominant clone (Meacham and Morrison 2013). This phenomenon is particularly relevant when evolution is triggered by resistance to kinase inhibitors (Diaz et al. 2012; Ding et al. 2012; Shi et al. 2014; Landau et al. 2015; Murtaza et al. 2015; Siravegna et al. 2015; Russo et al. 2016b).
In this review, we will argue that, although precision oncology incorporates diagnostic strategies designed to interrogate interpatient heterogeneity, our capability to probe and monitor intrapatient molecular heterogeneity remains to be established. We will provide evidence that this is relevant during emergence of drug resistance, which determines lesion-specific molecular profiles. We will describe how molecular characterization of cell-free circulating tumor DNA (ctDNA) allows monitoring of clonal dynamics throughout the course of a patient’s disease, and can identify lesion-specific mechanisms of therapeutic resistance.
CLINICAL IMPACT OF TUMOR HETEROGENEITY
Molecular Heterogeneity and Clonal Evolution during Tumor Development
Nowell first proposed a clonal evolutionary model of cancer development in 1976 (Nowell 1976). Elaborating on Darwinian models of natural selection, Nowell proposed that initiation and progression of cancers occur through several clonal expansions, triggered by the stepwise acquisition of genetic and epigenetic alterations affecting oncogenes and tumor-suppressor genes. The application of Nowell’s theory to precision oncology predicts that drug treatments shape the growth and survival of the mutant clones with a biological fitness advantage (Sprouffske et al. 2012). There is now increasing evidence that molecular heterogeneity and clonal evolution are of pivotal relevance when metastatic tumors are challenged with targeted drugs such as inhibitors of oncogenic-signaling molecules (Shi et al. 2014; Landau et al. 2015; Murtaza et al. 2015; Russo et al. 2015b; Siravegna et al. 2015).
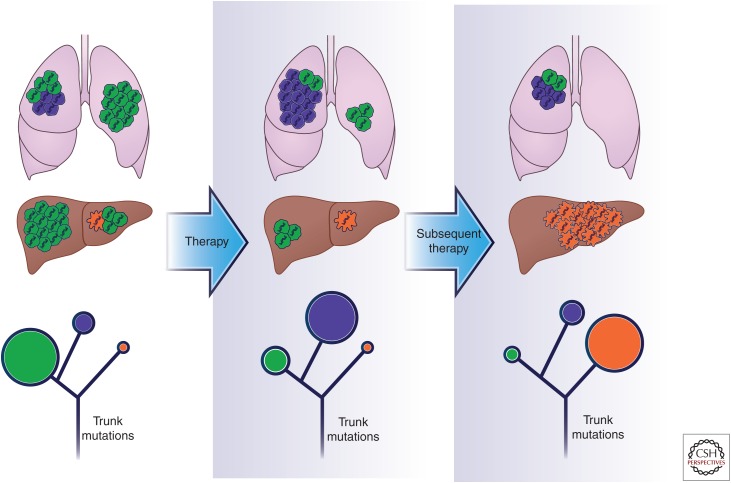
Morphological variation between distinct neoplastic regions has long been familiar to pathologists, and multiple sections of the same tumor are often routinely examined in clinical practice. It has also long been appreciated that cancers that originate from different tissues show distinct degrees of genetic and epigenetic diversity, and this affects prognosis and response to therapies. Moreover, each tumor contains an individual assortment of genomic aberrations, few of which are shared between patients with the same histopathological subtype (interpatient heterogeneity) (Weigelt and Reis-Filho 2009; Vogelstein et al. 2013). Accordingly, treatment decisions are currently based on the site of origin of the primary tumor (Burrell et al. 2013). Intrapatient heterogeneity also exists between the primary tumor and metastatic lesions, as well as between separated regions of the same tumor lesion, which show coexistence of genetically distinct clonal subpopulations (Fig. 1, upper panel) (Gerlinger et al. 2012; Shah et al. 2012; Burrell et al. 2013; Landau et al. 2013; Sottoriva et al. 2013; Brastianos et al. 2015; Yates et al. 2015).
Figure 1.
Targeted therapies drive clonal evolution. The figure portrays clonal evolution of a human cancer during drug treatment. At diagnosis the tumor is composed of a dominant (green) clone and two low-frequency subclonal populations, indicated in purple and orange, present in two separate tumor lesions. Blue DNA strands in each cell are ubiquitous trunk alterations present in each tumor cell. Frontline therapy, chosen based on the predominant population identified at the diagnosis (green, in this case), induces tumor shrinkage and drives the outgrowth of a preexistent clone intrinsically resistant to treatment (purple). When secondary resistance arises, a subsequent line of treatment is chosen based on the predominant resistant population. In this case, a subclone (orange clone) gains fitness and rapidly grows, leading to treatment failure. In the lower part of the figure, phylogenetic trees of the patient’s tumor over the course of therapy are depicted. In each stage, circles indicate the tumor subclonal composition. Dimensions of the circles are proportional to the population present in each time point.
The extent of intratumor heterogeneity has been revealed by analyses of multiple spatially separated regions. Gerlinger and colleagues (2012) used whole-exome sequencing (WES) of renal cell carcinomas to reveal that only about a third of nonsynonymous somatic mutations are detectable in all regions of the same tumor; the majority of the alterations are instead “private” and occur in distinct tumor regions. Such a high level of intratumor heterogeneity indicates that the molecular landscape diverged early during tumor evolution and continued to evolve following a branched rather than a linear evolution (Fig. 1, lower panel).
Indeed, as described by Yap and colleagues, heterogeneity can be graphically described with evolutionary trees, in which the trunk represents driver events, found in all cancer cells, essential for the initiation and the progression of the disease in the first phases, whereas branches represent parallel evolution of different tumor regions or subclones with “private” mutations that are not shared by all the tumor areas. Moreover, the role of somatic mutations as driver or passenger is dynamic. They are subject to environmental and treatment-selective pressures whereby passenger mutations can become driver events in the branches, and vice versa, as a result of changes in the environment (Fig. 1, lower panel) (Yap et al. 2012).
Darwinian dynamics regulate this process. Environmental forces that can vary in space and time provide strong selective pressures. In this way, the altered landscape selects cells that are able to survive in the new microenvironment, as mutations that were previously phenotypically silent (passengers) confer a selective advantage in the new conditions allowing the outgrowth of minor subclones (Bedard et al. 2013). Indeed, the occurrence of new lesions may be driven by selection of preexisting rare clones that become dominant (Sottoriva et al. 2013).
Molecular Heterogeneity and Clonal Evolution during Drug Treatment
Although we are beginning to unveil the levels of molecular heterogeneity during tumor development, much less is known regarding the impact of tumor heterogeneity during drug treatment. Studies on leukemias provided initial evidence that the genomic landscape of tumors evolve rapidly during drug treatment.
Ding and colleagues, for instance, showed that relapse on chemotherapy in patients with acute myeloid leukemia (AML) is correlated with the emergence of new mutations and clonal evolution, determined in part by the chemotherapy that the patients received. Deep mutational analysis of one patient’s tumor samples led to the identification of four clonal subpopulations in the primary tumor with different patterns of mutations. Of the four clones identified in tumor samples before clinical treatment started, only one was detected also in the chemotherapy-relapsed sample. Notably, this clone (clone #4) was present at a lower frequency before initiation of the chemotherapy treatment. An additional new clone was detected at relapse (clone #5), which contained all the mutations detected in clone #4, but gained 78 new somatic alterations, which probably provided a strong selective advantage allowing tumor cells to survive during chemotherapy and evolve into the dominant clone at relapse (Ding et al. 2012). The same pattern of clonal dynamics—under drug pressure—was confirmed in seven additional patients.
Landau and colleagues used WES and copy-number analysis to examine 18 chronic lymphocytic leukemia (CLL) patients (12 treated and six untreated) with two temporally separated samples (pre- and posttreatment). Ten out of the 12 treated cases underwent clonal evolution, in contrast to one of the six untreated cases. Five of the cases undergoing clonal evolution displayed branched evolution, with outgrowth of low-frequency subclones containing driver mutations (e.g., SF3B1 and TP53) that expanded over time (Landau et al. 2013). The results suggest that chemotherapy-treated CLLs often undergo clonal evolution, resulting in the expansion of previously minor subclones.
Similarly, exome sequences of 40 tumor regions from eight patients with operable esophageal adenocarcinomas revealed dynamic mutational processes associated with neoadjuvant platinum-based chemotherapy (Murugaesu et al. 2015). WES analysis revealed a high level of heterogeneity with nearly 47% of putative driver events occurring in subclonal populations, suggesting these occur later in tumor evolution (Murugaesu et al. 2015).
A nice example of alternating clonal dominance during clinical treatment comes from a deep analysis of a patient with multiple myeloma (MM). The patient received multiple therapeutic regimens, and then progressed after an initial partial response to first line therapy with lenalidomide and dexamethasone. Serial genomic analysis of seven time points over the entire disease course identified subclonal populations that alternate for dominance under treatment pressure until one clone took over. Although, at diagnosis, the tumor population was primarily composed of clone 1.1, after 1 year of initial treatment, when the patient was in a partial remission, the subclonal population 1.2 increased from 11% to 31%. Seven months later, when the patient progressed on lenalidomide, another subclone (2.1) became the dominant clone representing <60% of the population. At the fourth clinical relapse, the tumor clonal population definitively shifted with an additional clone that emerged later in tumor evolution (Keats et al. 2012). These findings suggest that often a clinical partial response, rather than reflecting substantial suppression of the entire tumor population, most likely represent the suppression of the sensitive population, whereas a refractory preexistent subclone remains stable and emerges leading to treatment failure.
Resistance to Targeted Therapies Drives Tumor Clonal Evolution and Lesion-Specific Heterogeneity
Cancer heterogeneity, including the relationships between subpopulations within and among tumor lesions, has profound implications for therapy. Targeted drugs have clearly shown efficacy on multiple tumor subtypes. However, in the majority of cases, even when clinical benefit is observed, it is often short-lived (Kobayashi et al. 2005; Montagut et al. 2008; Diaz et al. 2012).
If all the cancer cells were equally sensitive to a given therapy, any treatment that kills tumor cells quicker than they divide would eventually eradicate the tumor (Marusyk et al. 2012). Unfortunately, molecular heterogeneity prevents this outcome in most, if not all the cases. When targeted therapies are applied, resistant subclones are selected for and eventually lead to disease progression (Keats et al. 2012; Misale et al. 2012; Shi et al. 2014; Russo et al. 2015b).
Indeed, most cancers that initially respond to treatment, relapse with the outgrowth of cancer cells that are genetically and epigenetically different from those that were present at the diagnosis. Acquisition of resistance to targeted therapies has been shown in several tumor types, including lung (Kosaka et al. 2006; Turke et al. 2010), colorectal (Diaz et al. 2012), gastrointestinal stromal tumor (GIST) (Liegl et al. 2008), as well as chronic myeloid leukemia (Shah et al. 2002).
The observation that drug-resistant disease is composed of multiple subclones in AML, CLL, MM, GIST, and colorectal cancer (CRC) suggests that, in response to targeted therapy, polyclonal resistance occurs frequently. The mechanisms of resistance often involve multiple signal transduction pathways, resulting in distinct drug-resistance events (Burrell et al. 2013), and this poses considerable challenges for the design and selection of effective drug combinations.
Clonal evolution has been analyzed by WES of BRAF mutant melanoma patients at progression on BRAF inhibitors for whom multiple geographically and/or temporally separated biopsies were available (Shi et al. 2014). An analysis of one patient unveiled temporal and spatial tumor heterogeneity with all nine progressive sites of disease that had followed branched rather than linear evolutionary pattern. Each lesion harbored unique private mutations, and different lesions also harbored distinct drug-resistance mechanisms. In detail, at least five drivers of acquired BRAF inhibitor resistance accounted for a single patient’s clinical relapse. Thus, a single disease-progressive biopsy would have revealed only one of five (20%) mechanisms of acquired resistance. Branched evolution at relapse after treatment was also shown in three additional patients, again revealing distinct drug-resistance mechanisms in different lesions (Shi et al. 2014).
In 12 of 32 chronic myelogenous leukemia (CML) patients who developed resistance to imatinib, more than one resistant clone was identified, with two to four clones identified per patient (Shah et al. 2002). In ALK rearranged lung cancer, resistance to crizotinib treatment appears to be driven by multiple mechanisms within the same patient, including secondary ALK kinase domain mutations, as well as amplification of the mutant fusion gene or KIT (Choi et al. 2010; Katayama et al. 2012).
Importantly, there is evidence for phenotypic convergence within and across tumor types, suggesting that genetic events driving resistance and disease progression may funnel down a limited number of signaling pathways that may be therapeutically druggable (Gerlinger et al. 2012; Fisher et al. 2013; Johnson et al. 2014; Misale et al. 2014; Swanton 2014).
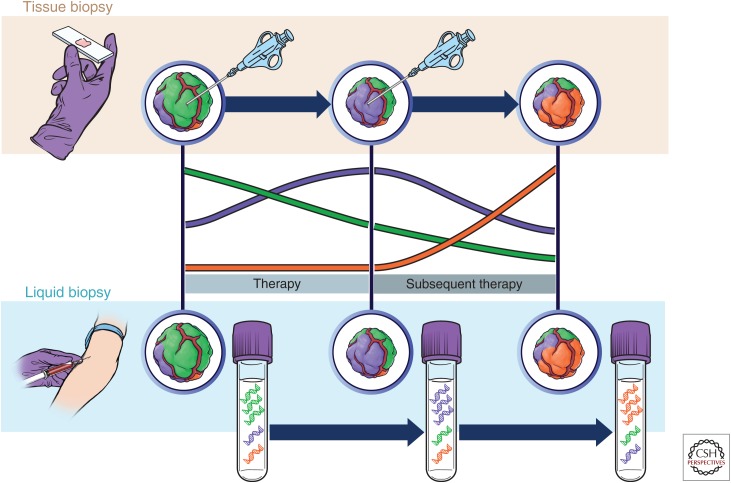
Serial tumor sampling (repeated tissue biopsies) may unveil the emergence of previously undetectable clones responsible for therapeutic failure. Tissue biopsies are not devoid of risks when related to solid tumors. Furthermore, biopsies may not be clinically practicable or unfeasible because of anatomic locations. Moreover, several lesions are often involved, precluding comprehensive sampling (Fig. 2, upper part).
Figure 2.
Applications of liquid biopsies to monitor tumor evolution. Treatment with targeted agents triggers clonal evolution that is missed by a tissue biopsy but can be captured in circulating free DNA (cfDNA). At diagnosis and on resistance to frontline therapy, a needle biopsy is taken, which usually detects only the most representative clonal population (green in first instance and purple on resistance) and misses the lower frequency subpopulations. This can result in suboptimal choice of further lines of treatment. Liquid biopsy, instead, better captures tumor heterogeneity at each stage, facilitating the identification of multiple resistance alterations present simultaneously. This information may be critical to define subsequent lines of therapy.
MONITORING TUMOR EVOLUTION
Liquid Biopsies to Measure Tumor Heterogeneity
As discussed above, evidence that tumor heterogeneity affects targeted therapies underscores that standard prognostic procedures, such as tissue biopsies, are inadequate to capture the molecular landscape of solid cancers.
Sampling different portions of the primary tumor, distinct metastatic lesions or even subsets of the same metastases often reveal remarkably different genomic profiles. This, in turn, may be highly relevant to determine the choice of treatment options. Furthermore, the inherent evolving nature of cancer requires longitudinal updates of the molecular landscapes during treatment and this cannot be provided by tissue biopsies (Fig. 2).
Recent technological advances have allowed the genotyping of DNA that circulates in the blood of patients (ctDNA). The ability to detect and quantify somatic mutations has proven effective in tracking tumor dynamics in real time, as well as serving as a “liquid biopsy” that can be used for a variety of clinical and investigational applications not previously possible. Indeed, many studies have shown the potential of liquid biopsies to assess tumor burden, determine the genomic profile of cancer patients, monitor response to treatment, assess the emergence of resistance, and quantify minimal residual disease (Beaver et al. 2014; Siravegna and Bardelli 2014; Siravegna et al. 2015; Tie et al. 2015; Alix-Panabières and Pantel 2016; Chabon et al. 2016; Imamura et al. 2016).
The blood of cancer patients contains tumor-derived proteins, circulating tumor cells (CTCs), cell-free DNA (cfDNA), and several types of vesicles, including exosomes. Initially detected in an 1869 autopsy within the blood of a patient with widespread breast cancer (Ashworth et al. 1869), CTCs are released from primary or metastatic tumor lesions and are thought to be enriched for metastatic precursors (Yu et al. 2011; Alix-Panabières and Pantel 2013). The process underlying the extravasation of tumor cells is not well understood and may involve both active invasion of cells, as well as passive shedding of individual cells or tumor cell clusters as a result of compromised tumor vasculature (Alix-Panabières and Pantel 2013; Alix-Panabières et al. 2016). Once in the circulation, CTCs seem to persist for a short time; in patients with localized cancer who have detectable CTCs, there is evidence of such cells at 24 h following surgical resection (Stott et al. 2010). Moreover, there is evidence that some CTCs travel in clusters, ranging from two cells to large microemboli with >50 cells detectable in the peripheral vasculature (Stott et al. 2010). Single-cell analyses have revealed heterogeneity in signaling pathways among CTCs from individual patients (Powell et al. 2012; Heitzer et al. 2013).
Cell-free DNA in the circulation was first described in 1948 (Mandel and Metais 1948) as non-cell-bound nucleic acids observed in the bloodstream of healthy subjects. It took many years to show that this DNA bore identical mutations to those observed in the tumors (Sorenson et al. 1994; Vasioukhin et al. 1994). As for CTCs, the exact mechanisms by which cfDNA is released in the blood remain to be clarified. In addition to apoptosis, cancer cells can release DNA through necrosis (Jahr et al. 2001); macrophages usually destroy all debris from normal cells, but tumors are often too large and their cells multiply so quickly that effective clearance is incomplete. Indeed, in patients with cancer a fraction of cfDNA is tumor derived and is termed circulating tumor DNA (ctDNA) (Jen et al. 2000). In individuals with very advanced cancers, tumors might be the source of most of the circulating DNA in the blood, but more commonly, ctDNA makes up barely 1% of the total and possibly as little as 0.01% (Diaz and Bardelli 2014). Conceptually, ctDNA may be derived from primary tumors, metastatic lesions, or CTCs.
In principle and in practice, the analysis of somatic (tumor-specific) mutations in ctDNA outperforms commonly used protein biomarkers. For example, prostate-specific antigen (PSA), a biomarker for prostate cancer, can give false positives because there are reasons—other than prostate cancer—that can cause an increase of PSA in the blood (Welch and Albertsen 2009; Brawley et al. 2016). False positives should be rarer with ctDNA because it is defined by mutations and other genomic changes that are hallmarks of cancer cells. Indeed, several studies have shown that mutations in ctDNA exactly recapitulate mutations identified in the tumor, including both point mutations and structural alterations such as copy-number changes and rearrangements (Leary et al. 2012; Bardelli et al. 2013; Murtaza et al. 2013; Bettegowda et al. 2014; Siravegna et al. 2015; Russo et al. 2016a). Rapid increases in ctDNA levels are also known to correlate with disease progression, and declines in ctDNA levels are associated with successful pharmacological or surgical treatment (Diehl et al. 2008; Misale et al. 2012; Beaver et al. 2014).
Detection of somatic genetic alterations in the circulation remains challenging. New approaches, such as next-generation sequencing (NGS) and droplet digital polymerase chain reaction (ddPCR), have facilitated sensitive and specific detection of alleles present at low frequency in cfDNA (Alix-Panabières and Pantel 2016).
Using CRC as a model system, Tie and colleagues explored the potential of ctDNA in evaluating tumor burden and monitoring response to chemotherapy at an early stage of treatment. Their study shows a close correlation between ctDNA quantification and initial tumor burden assessed by computed tomography (CT) scan. Importantly, this study emphasized that modulation of cancer-mutated alleles in ctDNA appears to anticipate response to therapy assessed by RECIST criteria (Tie et al. 2015).
Recent studies suggest that the allelic variance of trunk (stem) genetic alterations in the blood of a cancer patient throughout the course of disease reflects variations of tumor burden. Indeed, an increase or decrease of APC- or TP53-mutated alleles in the blood of CRC patients correlates with progression (increase in tumor burden) and response (tumor shrinkage) to clinical treatment, respectively (Russo et al. 2015b; Siravegna et al. 2015). Moreover, genetic translocations peculiar to a patient’s tumor have been shown to be reliable markers of disease burdens during treatment with targeted agents, and to predict/anticipate the tumor’s relapse (Russo et al. 2015a).
Liquid Biopsy to Monitor the Evolution of Drug Resistance
Precision oncology is based on the availability of accurate molecular maps of individual tumors. Accordingly, a seminal application of liquid biopsies is to detect and monitor the emergence of secondary resistance to targeted therapies.
As an example, in CRC, acquired resistance to the anti-epidermal growth factor receptor (EGFR) antibodies cetuximab and panitumumab is associated with the emergence of RAS mutations, and these mutations have been detected in ctDNA before disease progression can be documented by standard imaging (Diaz et al. 2012; Misale et al. 2012). Indeed, patients with CRC who acquired resistance to EGFR antibodies displayed a heterogeneous pattern of mutations in KRAS, NRAS, BRAF, and EGFR (Misale et al. 2012; Bettegowda et al. 2014; Arena et al. 2015; Siravegna et al. 2015).
Along the same line, longitudinal genetic profiling of the ctDNA of a CRC patient treated with the TRK inhibitor entrectinib allowed the identification of drug-resistance mechanisms to the inhibitor in parallel with clinical treatment, before resistance was clinically assessed by CT scan (Russo et al. 2015a).
Mohan and colleagues (2014) performed whole-genome sequencing of ctDNA in patients with CRC treated with anti-EGFR therapy and found several copy-number changes in all samples, including loss of the chromosomal 5q22 region harboring the APC gene, loss of chromosome arms 17p and 18q, as well as amplifications in known genes involved in the resistance to EGFR blockade such as MET, ERBB2, and KRAS. Similarly, serial assessment of plasma samples from lung patients allowed the detection of increasing levels of the EGFR secondary mutation T790M, responsible for secondary resistance to the EGFR tyrosine kinase inhibitor (TKI) erlotinib, weeks or even months before clinical development of resistance (Oxnard et al. 2014).
Overall, these studies indicate that clonal evolution during targeted therapies can be monitored by longitudinal assessment of ctDNA (Fig. 2).
Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy
As discussed above, analysis of individual tissue biopsies in patients with multiple sites of disease may not fully reflect the complex subclonal landscape and the diversity of resistance mechanisms that ensue at the time of progression on therapy (Fig. 2). This is particularly relevant in situations in which a tumor contains multiple subclonal events that become dominant on acquisition of drug resistance. For example, Russo and colleagues (2016b) investigated a patient with metastatic CRC and acquired resistance to cetuximab. The investigators reported that distinct metastatic biopsies from the same CRC patient displayed genetically distinct mechanisms of resistance to EGFR blockade. An initial liver biopsy identified a MEK1 K57T mutation following prolonged response to cetuximab. Based on preclinical modeling and functional characterization of the resistance mechanism, combinatorial treatment with the anti-EGFR antibody panitumumab and the MEK inhibitor trametinib was chosen as a subsequent line of therapy. Imaging revealed that the lesion harboring the MEK1 mutation responded to the treatment; however, a neighboring metastasis progressed and was found to harbor a completely distinct resistance mechanism (KRAS p.Q61H). By assessing tissue biopsies in parallel with ctDNA analysis, the investigators confirmed that separate metastases could independently evolve distinctive resistance mechanisms, resulting in striking differences in lesion-specific response to targeted therapy.
Most notably, this study highlights how the analysis of a single-lesion biopsy can inadequately capture the molecular heterogeneity of the patient’s cancer, and fails to detect the simultaneous presence of multiple resistance mechanism (KRAS mutation) that ultimately led to treatment failure. This highlights the potential pitfalls of selecting a targeted therapy strategy based on the molecular profile of a single resistant lesion. ctDNA profiles, instead, not only allowed the detection of concomitant resistance mechanisms residing in separate metastases, but it did so while the patient was receiving therapy, thereby predicting both the timing and cause of impending treatment failure.
A further seminal example of how liquid biopsies allow effective monitoring of tumor evolution was provided by Murtaza and colleagues (2015) who performed extensive comparative analysis of eight tumor biopsies and nine plasma samples collected from a metastatic breast cancer patient, treated with tamoxifen and trastuzumab initially and lapatinib afterward, over a period of 3 years. Using WES, it was found that common trunk mutations have the highest circulating levels in plasma; in contrast, plasma levels of private somatic mutations dynamically change according to disease progression. Among these, a mutation in the tyrosine kinase domain of ERBB4, most likely a key determinant of resistance to lapatinib, increased during treatment and reached the highest levels at the time of disease progression. In contrast, an actionable mutation in PIK3CA (p.E542K) was identified in plasma at the time of progression on trastuzumab and tamoxifen but declined during lapatinib treatment and then became undetectable. This mutation was only marginally detectable in two tumor biopsies (Murtaza et al. 2015).
Overall, these analyses highlight how ctDNA allow real-time monitoring of multifocal clonal evolution during therapy. These examples provide a glimpse of future applications of precision medicine, whereby we envision adaptive therapy in response to evolving polyclonal mechanisms of drug resistance.
Exploiting Clonal Evolution: Adaptive and Rechallenge Therapies
Understanding how tumor clonal heterogeneity impacts therapeutic outcomes is still an unmet clinical and scientific need. Innovative therapeutic strategies may have the potential to overcome the challenges posed by clonal heterogeneity. Gatenby and colleagues proposed the concept of adaptive therapy, whereby cancer treatment should continuously adapt to the tumor clonal evolution in space and time to maintain a fixed population of drug-sensitive cells, which can in turn suppress or limit the growth of drug-resistant cells (Gatenby et al. 2009).
In general, retreatment of patients with a therapy on which they previously progressed is avoided because of the assumption of persistent drug resistance. However, studies investigating the role of intermittent chemotherapy in CRC patients provide evidence that rechallenge with chemotherapy can be effective not only among patients who previously responded, but also in some patients who previously progressed to the same clinical regimen (Maindrault-Goebel et al. 2004; Tournigand et al. 2006).
May intermittent therapy schedules lead to reemergence of sensitive subclones, thus, providing the rationale for rechallenge therapy? Building on this concept, Siravegna and colleagues recently showed that the percentage of mutated KRAS clones, responsible for acquisition of resistance to anti-EGFR therapy in CRC patients, declines in the blood of the patients when EGFR-specific antibodies are withdrawn. Importantly, the drop of RAS mutant levels in the blood is associated with further clinical benefit with the same therapeutic regimen (Siravegna et al. 2015). These results indicate that the CRC genome adapts dynamically to intermittent drug schedules and provides a molecular explanation for the efficacy of rechallenge therapies based on EGFR blockade. These findings also provide the rationale for adaptive therapy strategies, allowing patients to benefit from multiple rounds of EGFR blockade. Similar results were observed in lung cancer patients who regained sensitivity to gefitinib after a drug holiday period (Kurata et al. 2004; Yoshimoto et al. 2007) and in metastatic renal cell carcinoma patients treated with sunitinib (Zama et al. 2010).
Why would resistant mutated subclones decline on drug withdrawl? Thakur and colleagues showed that BRAF-mutated melanoma cells resistant to vemurafenib remain oncogene dependent despite the onset of resistance. Importantly, the fitness benefit provided by the resistance alleles (e.g., expression of splice variant of BRAF V600E) becomes a fitness deficit when the drug is removed. This phenomenon may be responsible for vemufarenib-resistant melanomas regression in vivo on drug withdrawal (Das Thakur et al. 2013). Essentially, without the selective pressure of the drug, the resistant clones lose fitness and are outgrown by faster growing drug-sensitive clones.
Noteworthy, the same type of clonal fitness and dynamic on drug withdrawl has been observed in human immunodeficiency virus (HIV), in which drug-resistant viruses frequently have poor “replicative fitness” compared with wild-type HIV (Balduin et al. 2005; Martinez-Picado and Martínez 2008).
Knowledge of the existence of low-frequency-resistant subclones at diagnosis supports the use of combined therapeutic regimens targeting mechanisms of resistance from the start, therefore, delaying or preventing the emergence of resistance (Misale et al. 2015).
CONCLUDING REMARKS
Presently, therapy decisions are guided by molecular analysis of primary tumors obtained by single biopsy specimens that can fail to detect clinically relevant resistance alterations present in separate metastases or even in different regions of the same metastasis. In other words, tissue biopsies are unable to properly uncover intratumor heterogeneity, which is driven by therapeutic pressures. On the contrary, liquid biopsies may better capture the molecular heterogeneity of a patient’s tumor, and facilitate identification of multiple resistance alterations that, in turn, can profoundly affect clinical response to therapies.
Furthermore, there is clear evidence that therapy reshapes the tumor mutational landscape by promoting outgrowth of distinct resistant subclones. To tackle this, clinical trials and biomarker studies should include longitudinal analyses of tumor evolution. We propose that longitudinal ctDNA profiles should be integrated with serial tumor biopsies and lesion-specific radiographic responses to delineate mechanisms of drug resistance and guide selection of subsequent lines of treatment.
ACKNOWLEDGMENTS
This study is supported by the European Community’s Seventh Framework Programme under Grant Nos. 602901 MErCuRIC (A.B.) and 635342-2 MoTriColor (A.B.), IMI Contract No. 115749 CANCER-ID (A.B.), AIRC 2010 Special Program Molecular Clinical Oncology 5 per mille, Project No. 9970 (A.B.), Associazione Italiana Ricerca Cancro (AIRC) IG No. 16788 (A.B.), and Fondazione Piemontese per la Ricerca sul Cancro-ONLUS 5 per mille 2011 Ministero della Salute (A.B.).
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
- Alix-Panabières C, Pantel K. 2013. Circulating tumor cells: Liquid biopsy of cancer. Clin Chem 59: 110–118. [DOI] [PubMed] [Google Scholar]
- Alix-Panabières C, Pantel K. 2016. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 6: 479–491. [DOI] [PubMed] [Google Scholar]
- Alix-Panabières C, Bartkowiak K, Pantel K. 2016. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol Oncol 10: 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arena S, Bellosillo B, Siravegna G, Martinez A, Canadas I, Lazzari L, Ferruz N, Russo M, Misale S, Gonzalez I, et al. 2015. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res 21: 2157–2166. [DOI] [PubMed] [Google Scholar]
- Ashworth TR. 1869. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J 14: 146–149. [Google Scholar]
- Balduin M, Sierra S, Däumer MP, Rockstroh JK, Oette M, Fätkenheuer G, Kupfer B, Beerenwinkel N, Hoffmann D, Selbig J, et al. 2005. Evolution of HIV resistance during treatment interruption in experienced patients and after restarting a new therapy. J Clin Virol 34: 277–287. [DOI] [PubMed] [Google Scholar]
- Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D, et al. 2013. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 3: 658–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver JA, Jelovac D, Balukrishna S, Cochran RL, Croessmann S, Zabransky DJ, Wong HY, Valda Toro P, Cidado J, Blair BG, et al. 2014. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 20: 2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard PL, Hansen AR, Ratain MJ, Siu LL. 2013. Tumour heterogeneity in the clinic. Nature 501: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, et al. 2015. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov 5: 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley OW, Thompson IM, Grönberg H. 2016. Evolving recommendations on prostate cancer screening. Am Soc Clin Oncol Educ Book 35: e80–e87. [DOI] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C. 2013. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501: 338–345. [DOI] [PubMed] [Google Scholar]
- Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, Kurtz DM, Stehr H, Scherer F, Karlovich CA, et al. 2016. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 7: 11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, et al. 2010. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 363: 1734–1739. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, Stuart DD. 2013. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA, Bardelli A. 2014. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol 32: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. 2012. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486: 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. 2008. Circulating mutant DNA to assess tumor dynamics. Nat Med 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R, Pusztai L, Swanton C. 2013. Cancer heterogeneity: Implications for targeted therapeutics. Br J Cancer 108: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Silva AS, Gillies RJ, Frieden BR. 2009. Adaptive therapy. Cancer Res 69: 4894–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. 2012. Clonal evolution in cancer. Nature 481: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, et al. 2013. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res 73: 2965–2975. [DOI] [PubMed] [Google Scholar]
- Imamura F, Uchida J, Kukita Y, Kumagai T, Nishino K, Inoue T, Kimura M, Oba S, Kato K. 2016. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 94: 68–73. [DOI] [PubMed] [Google Scholar]
- Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. 2001. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 61: 1659–1665. [PubMed] [Google Scholar]
- Jen J, Wu L, Sidransky D. 2000. An overview on the isolation and analysis of circulating tumor DNA in plasma and serum. Ann NY Acad Sci 906: 8–12. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. 2014. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343: 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, Jessop NA, Wain JC, Yeo AT, Benes C, et al. 2012. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 4: 120ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, Van Wier S, Blackburn PR, Baker AS, Dispenzieri A, et al. 2012. Clonal competition with alternating dominance in multiple myeloma. Blood 120: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. 2005. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352: 786–792. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Tada H, Kuwano H, Mitsudomi T. 2006. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res 12: 5764–5769. [DOI] [PubMed] [Google Scholar]
- Kurata T, Tamura K, Kaneda H, Nogami T, Uejima H, Asai Go G, Nakagawa K, Fukuoka M. 2004. Effect of re-treatment with gefitinib (“Iressa,” ZD1839) after acquisition of resistance. Ann Oncol 15: 173–174. [DOI] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, et al. 2015. Mutations driving CLL and their evolution in progression and relapse. Nature 526: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O’Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, et al. 2012. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 4: 162ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegl B, Kepten I, Le C, Zhu M, Demetri GD, Heinrich MC, Fletcher CD, Corless CL, Fletcher JA. 2008. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol 216: 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maindrault-Goebel F, Tournigand C, André T, Carola E, Mabro M, Artru P, Louvet C, de Gramont A. 2004. Oxaliplatin reintroduction in patients previously treated with leucovorin, fluorouracil and oxaliplatin for metastatic colorectal cancer. Ann Oncol 15: 1210–1214. [DOI] [PubMed] [Google Scholar]
- Mandel P, Metais P. 1948. Les acides nucléiques du plasma sanguin chez l’homme [Nucleic acids in the blood of humans]. C R Seances Soc Biol Fil 142: 241–243. [PubMed] [Google Scholar]
- Martinez-Picado J, Martínez MA. 2008. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: A view from the clinic and ex vivo. Virus Res 134: 104–123. [DOI] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, Polyak K. 2012. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer 12: 323–334. [DOI] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. 2013. Tumour heterogeneity and cancer cell plasticity. Nature 501: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. 2012. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486: 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, Russo M, Buscarino M, Lazzari L, Sartore-Bianchi A, et al. 2014. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med 6: 224ra226. [DOI] [PubMed] [Google Scholar]
- Misale S, Bozic I, Tong J, Peraza-Penton A, Lallo A, Baldi F, Lin KH, Truini M, Trusolino L, Bertotti A, et al. 2015. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nat Commun 6: 8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S, Heitzer E, Ulz P, Lafer I, Lax S, Auer M, Pichler M, Gerger A, Eisner F, Hoefler G, et al. 2014. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet 10: e1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, Dias-Santagata D, Stubbs H, Lee DY, Singh A, et al. 2008. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res 68: 4853–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. 2013. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497: 108–112. [DOI] [PubMed] [Google Scholar]
- Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, Chin SF, Tsui DW, Marass F, Gale D, et al. 2015. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 6: 8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaesu N, Wilson GA, Birkbak NJ, Watkins TB, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, et al. 2015. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 5: 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. 1976. The clonal evolution of tumor cell populations. Science 194: 23–28. [DOI] [PubMed] [Google Scholar]
- Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O’Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P, Jackman DM, et al. 2014. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 20: 1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. 2005. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et al. 2012. Single cell profiling of circulating tumor cells: Transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 7: e33788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo M, Misale S, Wei G, Siravegna G, Crisafulli G, Lazzari L, Corti G, Rospo G, Novara L, Mussolin B, et al. 2016a. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov 6: 36–44. [DOI] [PubMed] [Google Scholar]
- Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, Mussolin B, Kwak EL, Buscarino M, Lazzari L, et al. 2016b. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov 6: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. 2002. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2: 117–125. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. 2014. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siravegna G, Bardelli A. 2014. Minimal residual disease in breast cancer: In blood veritas. Clin Cancer Res 20: 2505–2507. [DOI] [PubMed] [Google Scholar]
- Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C, et al. 2015. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21: 827. [DOI] [PubMed] [Google Scholar]
- Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. 1994. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 3: 67–71. [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci 110: 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske K, Merlo LM, Gerrish PJ, Maley CC, Sniegowski PD. 2012. Cancer in light of experimental evolution. Curr Biol 22: R762–R771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, et al. 2010. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med 2: 25ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C. 2014. Cancer evolution: The final frontier of precision medicine? Ann Oncol 25: 549–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, Wong R, Singh M, Karapetis CS, et al. 2015. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 26: 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournigand C, Cervantes A, Figer A, Lledo G, Flesch M, Buyse M, Mineur L, Carola E, Etienne PL, Rivera F, et al. 2006. OPTIMOX1: A randomized study of FOLFOX4 or FOLFOX7 with oxaliplatin in a stop-and-go fashion in advanced colorectal cancer—A GERCOR study. J Clin Oncol 24: 394–400. [DOI] [PubMed] [Google Scholar]
- Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, et al. 2010. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. 1994. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol 86: 774–779. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Reis-Filho JS. 2009. Histological and molecular types of breast cancer: Is there a unifying taxonomy? Nat Rev Clin Oncol 6: 718–730. [DOI] [PubMed] [Google Scholar]
- Welch HG, Albertsen PC. 2009. Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986–2005. J Natl Cancer Inst 101: 1325–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. 2012. Intratumor heterogeneity: Seeing the wood for the trees. Sci Transl Med 4: 127ps110. [DOI] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto A, Inuzuka K, Kita T, Kawashima A, Kasahara K. 2007. Remarkable effect of gefitinib retreatment in a patient with nonsmall cell lung cancer who had a complete response to initial gefitinib. Am J Med Sci 333: 221–225. [DOI] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M, Maheswaran S, Haber DA. 2011. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol 192: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zama IN, Hutson TE, Elson P, Cleary JM, Choueiri TK, Heng DY, Ramaiya N, Michaelson MD, Garcia JA, Knox JJ, et al. 2010. Sunitinib rechallenge in metastatic renal cell carcinoma patients. Cancer 116: 5400–5406. [DOI] [PubMed] [Google Scholar]